Abstract
Glucocorticoids inhibit proliferation of many cell types, but the relationship between the glucocorticoid receptor (GR) and the proteins regulating cell cycle progression is not fully understood. We previously found that during fibrosarcoma (FS) progression, GR displays only modest transcriptional activity in the preneoplastic stages, whereas it is highly active in FS cells. Now, we report that glucocorticoids reduce proliferation throughout FS development. The cyclin-dependent kinase inhibitor p16INK4a is frequently absent in many cancers, including FSs. We observed that p16INK4a protein expression is lost at the tumor stage of FS progression. Treatment with the demethylating agent 5-aza-2′-deoxycytidine restores p16INK4a expression and reverts the phenotype of FS cells to low GR transcriptional activity, similar to that of the p16INK4a-expressing preneoplastic stages. Importantly, exogenous p16INK4a introduced by cotransfection is sufficient to reduce GR activity in FS cells, without affecting GR activity in p16-positive aggressive fibromatosis cells. Furthermore, GR transcriptional activity is elevated in mouse embryo fibroblasts derived from INK4a−/− mice compared with those derived from WT mice, implying that the difference in p16INK4a expression is sufficient to modulate GR activity. These results suggest a relationship between steroid hormone receptor activity and cell cycle inhibition, whereby absence of p16INK4a protein leads to higher GR transactivation activity and reduced cell sensitivity to dexamethasone. This observation might have important implications for current cancer therapies.
Glucocorticoids influence many fundamental biological processes, from development and homeostasis, to proliferation, differentiation, and apoptosis (1, 2). In many cell types, they promote arrest in the G1/S transition of the cell cycle, resulting in a decrease in proliferation (3–5). Although these effects are well documented, the relationship between the steroid hormone receptors and the cell-cycle regulatory proteins remains unclear (6).
The glucocorticoid receptor (GR) belongs to a superfamily of transcription factors that includes receptors for steroid and thyroid hormones, retinoic acid, and vitamin D3 (7). GR is normally localized in the cytoplasm, in a nonactive state, in a complex with Hsp90 and other factors. On hormone binding, GR changes conformation, is released from the complex, and migrates to the nucleus. Once in the nucleus, the receptor can induce or repress transcription by binding to specific DNA sequences on target genes.
Progression from the G1 to S phase of the cell cycle is regulated in part by the activation of cyclin-dependent kinases (cdk4 and cdk6) by cyclin D1, resulting in phosphorylation of the retinoblastoma protein (pRb), and consequent release of E2F transcription factors and expression of genes required for the S phase (8). Cyclin–cdk complexes are regulated by a family of kinase inhibitors that can prevent phosphorylation of the corresponding substrate (9). p16INK4a (here referred to as p16) specifically inhibits cyclin D-dependent kinases, preventing them from phosphorylating and inactivating pRb (10). p16 is often mutated or inactivated in primary tumors, including leukemias, melanomas, gliomas, lung carcinomas, osteosarcomas, and fibrosarcomas (FSs), and in many cancer cell lines (11–14). Mice carrying a targeted deletion of the INK4a locus develop spontaneous tumors, particularly lymphomas and FSs, at an early age (15). This phenotype might be the result of the absence of p19ARF and not p16; however, most of the human mutations target p16 and not p14ARF (16, 17).
The conversion of a normal cell to a neoplastic one is a multistep process (18), and one approach to studying this process has used transgenic mice (19). Mice carrying the bovine papillomavirus (BPV) type 1 genome develop dermal FSs in a process that involves distinct proliferative stages. These are the normal dermal fibroblasts, and two histological grades of hyperplasia that arise are mild fibromatosis (MF) and aggressive fibromatosis (AF). Finally, at lower frequency, dermal FSs develop. Cells cultured from each of these stages appear to retain characteristics of the lesions from which they were derived (20). GR displays only modest transcriptional activity in cells derived from the nontumor stages, but is highly active in FS cells (21). On inoculation into mice, the AF cells progress to tumor cells with high GR activity, indicating that the increased GR transcriptional activity correlates with the cellular transition to the tumor stage (21).
Glucocorticoids are used as part of anticancer therapy for some lymphatic leukemias and lymphomas (22). In addition, glucocorticoids have been shown to inhibit in vitro growth of malignant melanoma cells (ref. 23 and references therein) and also to inhibit the growth of carcinogen-induced pulmonary adenoma (24) and FSs (25). To investigate the molecular basis of the potential antiproliferative role of glucocorticoids in tumor development, we took advantage of the multistep tumorigenic pathway that dermal FS development provides and examined the alterations of the components involved in the G1 phase of cell-cycle progression and their consequences on GR transactivation activity.
Materials and Methods
DNA Plasmid Constructs.
The luciferase reporter TAT3-Luc and the Rous sarcoma virus (RSV)-β-gal construct to monitor transfection efficiency have been described (21). The mouse p16, p18INK4c, and p19INK4d expression vectors were a generous gift from Charles J. Sherr (St. Jude Children's Research Hospital, Memphis, TN). The expression vectors encoding p21Cip/Waf1 and p27Kip1 were kindly provided by Sibylle Mittnacht (Institute of Cancer Research).
Cell Culture and Transient Transfections.
Cultures were established from skin and tumor tissues as reported (21), and maintained in growth medium (DMEM supplemented with 8% FCS (Autogen). Mouse embryo fibroblasts (MEFs) were generously provided by Manuel Serrano (National Centre of Biotechnology, Madrid) and maintained in growth medium (DMEM plus 10% FCS). For experiments involving steroid hormone treatments, growth medium was replaced by medium containing charcoal-stripped serum (26) 24 h after plating, and cells were treated with the carrier ethanol, as a negative control, or 100 nM dexamethasone (Sigma) for the required amount of time. Cells were transiently transfected by using the DEAE-dextran method, as described (21). After exposure to the DNA/DEAE-dextran mixture, the cells were incubated for 36 h in fresh medium containing charcoal-stripped serum, with or without 100 nM dexamethasone. Luciferase activity was measured according to the instructions of the manufacturer (Promega) and normalized for β-galactosidase expression.
p16 Expression.
First-strand cDNA was synthesized from 0.5 μg of total RNA primed with poly(dT) using Superscript II reverse transcriptase (Life Technologies). Amplification products for p16 (145 bp) and GAPDH (200 bp, used as control for RNA integrity) were generated from the cDNA template. After cDNA synthesis, 10% of the product from each sample was used for PCR analysis. Amplifications were carried out in 25-μl reaction volumes for 25 cycles, each consisting of denaturation at 95°C for 1 min, annealing at 60°C for 45 sec, and extension at 72°C for 1 min. Primers were as follows: for p16, 5′-AAGCGAACTCGAGGAGAGC-3′ (sense) and 5′-GTACGACCGAAAGAGTTCG-3′ (antisense); and for GAPDH, 5′-TTGTTGCCATCAACGACC-3′ (sense) and 5′-GACATCATACTTGGCAGG-3′ (antisense). Water blank, in which no template was added, was included, and no PCR product was detected in this control.
For analysis of p16 expression after 5-aza-2′-deoxycytidine (5-Aza-dC) treatment, nested PCR was performed. The oligonucleotides used for the primary PCR to amplify the whole gene were as follows: 5′-TCACACGACTGGGCGATTGG-3′ (sense) and 5′-GCCATTATTCCCTTCGCCGC-3′ (antisense). Conditions were the same as above, except for extension for 2 min. For the nested PCR, 1 μl from the primary reaction was used with the oligonucleotides amplifying the exon 2 (see below). Conditions were as for the RT-PCR described above.
Genomic DNA Analysis.
Genomic DNA was prepared by using the Wizard Genomic DNA kit (Promega) according to the instructions of the manufacturer. It was amplified with primer sets designed to recognize sequences from the INK4α promoter region and exon 1α: 5′-TCACACGACTGGGCGATTGG-3′ (sense) and 5′-CACCTGAATCGGGGTACGAC-3′ (antisense); from exon 2: 5′-GGGCAACGTTCACGTAGCAG-3′ (sense) and 5′-GGCGTGCTTGAGCTGAAGCT-3′ (antisense); and from exon 3: 5′-CTGGAACTTCGCGGCCAAT-3′ (sense) and 5′-GCCATTATTCCCTTCGCCGC-3′ (antisense); and for GAPDH as described above. All amplifications were carried out for 35 cycles using Taq polymerase (Promega) and the following conditions: an initial 1-min denaturation at 94°C, annealing at 53°C for 30 sec, and extension at 72°C for 45 sec, in a thermal cycler (PCR System 9700, Applied Biosystems). Control water blanks were introduced in each experiment. PCR products were visualized on 2% agarose gels containing ethidium bromide.
5-Aza-dC Treatment.
Cells were grown in the presence of 5 μM 5-Aza-dC for 96 h. Fresh 5-Aza-dC was added every 24 h and the medium was changed every 48 h. For the transient transfection experiments, cells were split into six-well plates 48 h after initial treatment, transfected by using the DEAE/dextran method (in the absence of 5-Aza-dC), and continued with the experiment as above for the last 36 h of incubation in the absence or presence of dexamethasone.
Preparation and Analysis of Cell Extracts.
Cells were harvested and whole cell extracts were prepared as described (21). Equal amounts of protein (30 μg) from each cell stage were separated by electrophoresis on an SDS/15% polyacrylamide gel and transferred to an Immobilon membrane (Millipore). Blocking, washing, and incubation of the membrane with antibodies were carried out in Tris-buffered saline (10 mM Tris⋅HCl, pH 7.6/150 mM NaCl) containing 4% nonfat dried milk and 0.05% Tween 20. The primary antibodies used were directed against cyclin D1 (72-13G), cdk4 (C-22), p16 (M-156), E2F1 (C-20), cyclin E (M-20), p18 (N-20), p19 (M-167), p21 (F-5), p27 (F-8) (all from Santa Cruz Biotechnology), pRb (14001A, PharMingen), and β-tubulin (Sigma). Proteins were detected by using horseradish peroxidase-conjugated goat anti-rabbit (1:3,000, DAKO), or goat anti-mouse (1:3,000, Bio-Rad). Protein–antibody complexes were visualized by an enhanced chemiluminescence immunoblotting detection system according to the recommendations of the manufacturer (Amersham Pharmacia).
Results
Dexamethasone Reduces Cell Proliferation at All Stages of FS Progression.
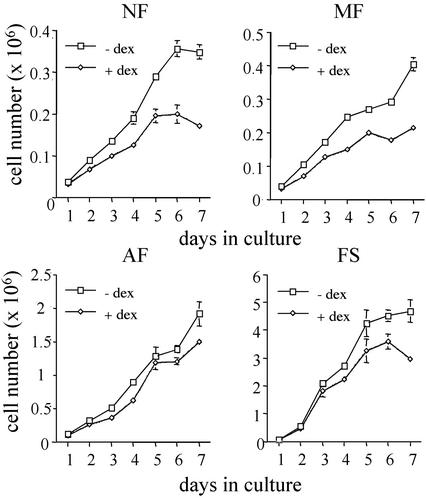
To assess the changes in proliferation during FS progression we monitored cell growth for 1 wk at 24-h intervals, in the absence and presence of dexamethasone. The cells from the earlier stages of the progression proliferated at a low rate, particularly the NF and MF cells, as reported (20), whereas there was a very rapid increase in the number of FS cells. Dexamethasone treatment resulted in a decrease in the proliferation of the cells at all stages of FS progression (Fig. 1), likely as a consequence of an accumulation of cells at the G1 phase of the cell cycle (data not shown). Despite the low levels of GR transcriptional activity in cells from the earlier stages of FS progression, the inhibition of cell growth by dexamethasone was stronger at these stages (between 30% and 40% in NF and MF cells) than at the FS stage (11%), suggesting that FS cells have partially overcome the growth-inhibitory pathways mediated by GR. In conclusion, as documented in many other cell types, dexamethasone reduces cell proliferation during FS development.
Figure 1.
Effect of dexamethasone (dex) on proliferation of cell lines from different stages of FS progression. Cell-growth kinetics of NF (NF 40950, MF 14249, and AF BPV3) and FS cells (FS BPV1). Cells from each stage were plated on day 0 onto six-well plates, and cultured in the absence (□) or presence (⋄) of 100 nM dexamethasone. Total numbers of viable cells were determined on the indicated days by the trypan blue exclusion method. The graph represents one of at least three independent experiments, which were done in triplicate.
p16 Protein Is Absent at the Tumor Stage of FS Progression.
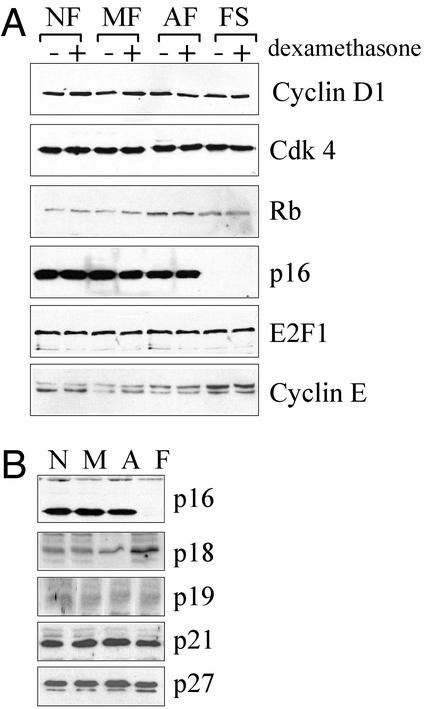
To further investigate the effects of dexamethasone on cell proliferation, we examined the expression of various proteins involved in cell-cycle regulation by immunoblot analysis. The expression levels of cyclin D1, cdk4, and E2F1 were not affected by FS progression or dexamethasone treatment (Fig. 2A). pRb mobility was slightly decreased at the tumor stage, probably reflecting the increased pRb phosphorylation that accompanies increased cell cycling, but pRb mobility was not affected by dexamethasone. There was a small increase in cyclin E levels in FS cells, possibly as a consequence of the increased level of pRb phosphorylation at the tumor stage of FS development. The most striking observation during this analysis was that the expression of p16 protein was completely lost during the transition from the AF to the FS stage. However, the expression of p16 was not influenced by dexamethasone treatment at any of the preneoplastic stages (Fig. 2A). Expression of p16 protein was also found in a second set of clones that were tested corresponding to NF, MF, and AF stages (Fig. 2B), independent of their passage number, but was not observed in any of the four clones that were examined corresponding to the FS stage (Fig. 2B and data not shown). This result indicates that the loss of p16 protein at the tumor stage is inherent to the transition from the AF to the FS stage during FS progression. In contrast, the expression levels of other cdk inhibitors, including members of the INK4 family (p18INK4c and p19INK4d), and members of the Cip/Kip family (p21Cip/Waf1 and p27Kip1), were unchanged during FS development (Fig. 2B).
Figure 2.
Expression of cell-cycle regulatory proteins during FS progression. (A) Cells from each stage (clones as indicated in Fig. 1) were cultured in the absence (−) or presence (+) of 100 nM dexamethasone for 48 h. Whole-cell extracts were probed with antibodies against cyclin D1, cdk4, pRb, p16, E2F1, and cyclin E (antibodies were used as described in Materials and Methods). Each immunoblot is representative of at least three independent experiments. (B) Intracellular levels of various cdk inhibitors during FS progression. A second set of clones, different from the set used above, and representative of each stage of FS progression (NF non-BPV, MF 39614, AF BPV7, and FS BPV22, represented by N, M, A, and F, respectively) were used in this experiment. Each blot was reprobed for β-tubulin as a control for equal loading (data not shown). Note that the apparent increase of p18 expression in FS cells was insignificant after correction with tubulin levels.
p16 Coding Region Is Present in FS Cells.
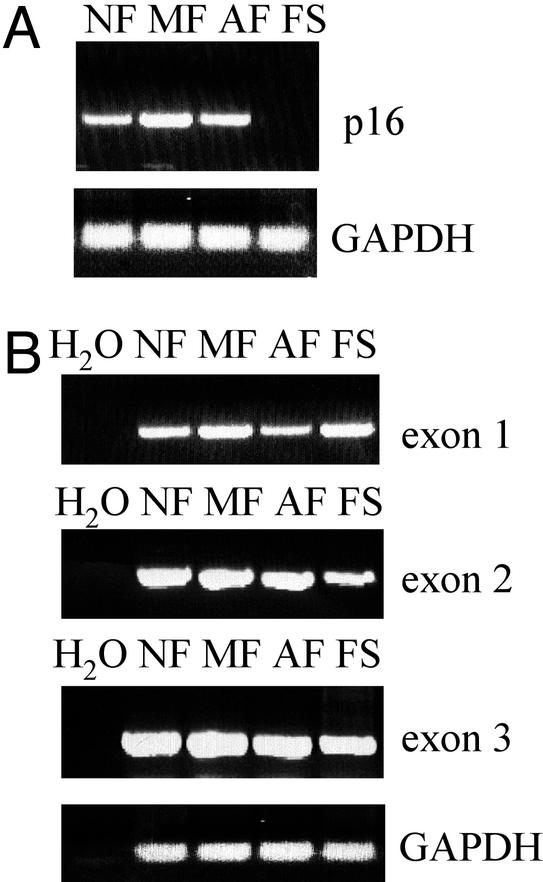
To investigate whether the lack of p16 protein coincides with the absence of mRNA expression, we performed RT-PCR analysis using mRNA from all stages of FS progression. p16 mRNA was present in NF, MF, and AF cells, but not in FS cells (Fig. 3A), which was found to be similar to p16 protein expression levels during FS development. The lack of p16 protein and mRNA expression in FS cells raised the possibility that p16 DNA might be lost, mutated, or inactivated. Homozygous deletions represent a mechanism of p16 inactivation in various types of tumors (11, 27). We used PCR analysis to examine the genomic DNA from cells representing all stages of FS progression to determine whether any of the exons were selectively deleted in FS cells. By using oligonucleotides that hybridized with regions of exons 1, 2, and 3 (28), specific 200-, 350-, and 250-bp (respectively) products were amplified in all samples obtained from each stage of FS development (Fig. 3B). These products represent the whole coding region of mouse p16, and, therefore, they demonstrate that the DNA encoding p16 is not deleted in FS cells.
Figure 3.
RT-PCR and PCR analysis of p16 expression throughout FS progression. (A) Detection of p16 mRNA by RT-PCR analysis in samples obtained from NF, MF, AF, and FS cells. (B) PCR analysis of genomic DNA from NF, MF, AF, and FS cells. The oligonucleotides were designed to hybridize with sequences from each exon of the mouse p16 coding region (28). A PCR without template (H2O) was incorporated in the analysis as a negative control.
Methylation Modulates GR Transactivation Activity.
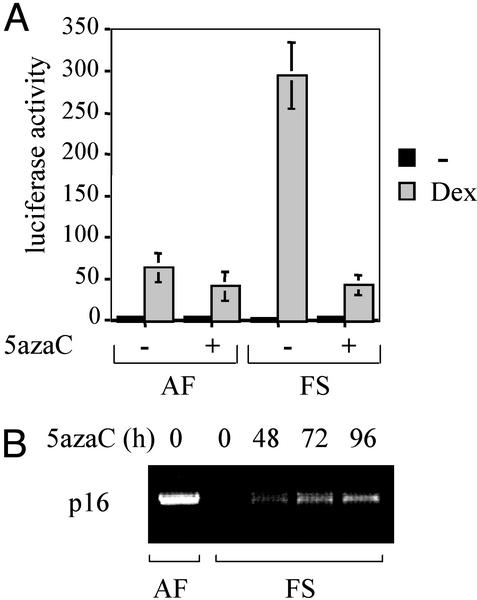
In addition to homozygous deletion and rare inactivating mutations, several recent studies have identified inappropriate methylation of exon 1α and the 5′ promoter region of p16 in cell lines and tumors derived from both humans and mice (29, 30). One possibility of this finding is that the p16 gene is inactivated by methylation at the FS stage, as does occur in many other tumor cell types, including FSs (31, 32). Because the only documented molecular change in the progression from AF to FS is the increased transactivation activity of GR (21), we sought to determine whether there was a link between GR transcriptional activity and p16 inactivation predicted to result from methylation. To this end, we treated the cells with the DNA methyltransferase inhibitor 5-Aza-dC, which has been reported to demethylate the p16 gene promoter, leading to reexpression of p16 (12, 30, 33, 34). AF (expressing p16) and FS (not expressing p16) cells were grown in the presence or absence of 5-Aza-dC treatment and were transfected with the luciferase reporter construct TAT3-Luc, which contains three copies of a simple glucocorticoid response element (21). The results from these transfection experiments (Fig. 4A) revealed that 5-Aza-dC has no significant effect on the hormone responsiveness of AF cells. In contrast, treatment with 5-Aza-dC strongly reduced GR transcriptional activity in FS cells, compared with the results observed in AF cells.
Figure 4.
Methylation modulates GR-mediated transcriptional activity in FS cells. (A) AF and FS cells were grown in the absence (−) or presence (+) of 5 μM 5-Aza-dC for 96 h, and the expression from the TAT3-Luc reporter was determined in the absence or presence of 100 nM dexamethasone. Luciferase activity was normalized against a cotransfected β-galactosidase reporter. The values presented are the average of at least three independent experiments, which were done in triplicate. (B) Detection of p16 mRNA by RT-PCR analysis in AF cells in the absence of 5-Aza-dC treatment, but not in FS cells. Treatment of FS cells with 5 μM 5-Aza-dC during an increasing amount of time (h) results in the reexpression of p16 mRNA.
To directly address whether treatment with the demethylating agent 5-Aza-dC resulted in the reexpression of p16 in FS cells, we performed RT-PCR analysis. Expression of p16 mRNA was restored in FS cells (Fig. 4B) on treatment with 5-Aza-dC, suggesting that the observed effect on GR activity (Fig. 4A) is because of p16 reactivation. These observations support a model in which inactivation of p16 by methylation results in increased GR transcriptional activity in FS cells.
p16 Affects GR Transcriptional Activity and Cell Sensitivity to Dexamethasone.
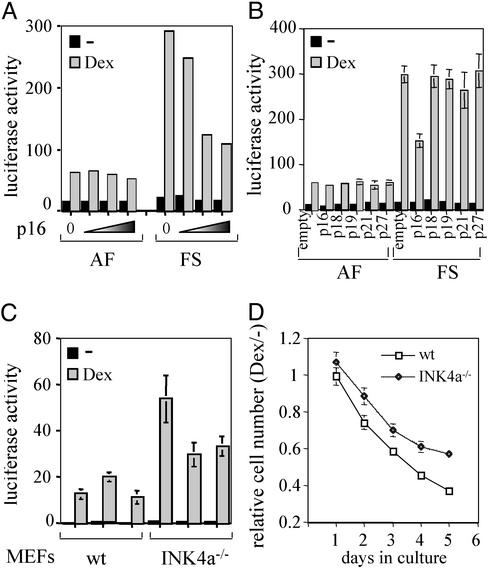
To assess whether expression of p16 protein is sufficient to reduce GR transcriptional activity, we cotransfected a p16 expression vector together with the simple glucocorticoid response element reporter, TAT3-Luc, into AF and FS cells. Over a wide range of cotransfected p16 expression vector, no statistically significant difference in GR activity was detected in AF cells (Fig. 5A). In contrast, cotransfected p16 clearly reduced the magnitude of the dexamethasone response in the FS cells. Importantly, the same results were obtained with three different AF and FS clones we tested, indicating that the reduction of GR transcriptional activity by exogenously expressed p16 is a general phenomenon in FS cells. Notably, cotransfected RSV-β-gal produced comparable β-galactosidase levels throughout the experiment (data not shown), indicating similar transfection efficiencies. In conclusion, ectopic expression of p16 is sufficient to repress GR transcriptional activity. In contrast, cotransfection of expression vectors encoding other cdk inhibitors, including members of the INK4 family (p18INK4c and p19INK4d), and members of the Cip/Kip family (p21Cip/Waf1 and p27Kip1), did not significantly affect GR transcriptional activity in AF or FS cells (Fig. 5B).
Figure 5.
Effects of p16 expression on GR transcriptional activity and the antiproliferative effects of dexamethasone. (A) Effects of cotransfected p16 on hormone responsiveness of AF and FS cells. Expression from the TAT3-Luc reporter in AF and FS cells in the absence (0) or presence of exogenous p16. A wide range of p16 plasmid was tested (0, 0.1, 0.75, and 1.5 μg are shown). Transfected cells were cultured for 36 h in the absence (solid bars) or presence (shaded bars) of 100 nM dexamethasone. Luciferase activity was normalized against a cotransfected β-galactosidase reporter. The values presented are the average of at least three independent experiments, which were done in triplicate. (B) Effects of cotransfected p18INK4c, p19INK4d, p21Cip/Waf1, and p27Kip1 on hormone responsiveness of AF and FS cells. A wide range of concentrations was tested and the cotransfection of 0.75 μg of each expression vector is shown as representative example. Cotransfection of the expression vector without insertion of the sequences encoding the different cdk inhibitors is represented as “empty.” The conditions were as in A. (C) GR transcriptional activity in MEFs derived from WT or INK4a−/− mice. The TAT3-Luc was transfected into three different cell lines derived from WT (clones 41, 42, and 44) and INK4a−/− (clones 35, 36, and 37) mice. The cells were untreated (solid bars) or treated with dexamethasone (shaded bars). Conditions were as in A. (D) Relative cell number of WT (□) and INK4a−/− (◊) MEFs treated with dexamethasone versus those that were untreated. The conditions were as in Fig. 1.
To determine whether endogenous p16 protein affects GR transcriptional activity, we compared the hormone responsiveness of WT and INK4a−/− MEFs. We tested three different clones for each type and, interestingly, all INK4a−/− MEFs displayed higher GR transcriptional activity compared with WT MEFs (Fig. 5C). Taken together, these findings imply that p16 inhibits GR activity and that loss of p16 in FS cells results in elevated GR transcriptional activity.
Finally, we wished to investigate whether there is a relationship between GR transcriptional activity and the antiproliferative effects of dexamethasone that depends on the presence of p16. We were unable to generate FS lines stably expressing p16. This result is, perhaps, not surprising, taking into account the expected growth arrest imposed by overexpression of p16. To determine the effects of dexamethasone specifically caused by the expression of p16, we examined the effects of dexamethasone the cell growth of MEFs, both WT and INK4a−/−. Interestingly, WT MEFs were more sensitive (between 15% and 20%) to the growth inhibitory effects of dexamethasone than were MEFs lacking p16 (Fig. 5D). This finding resembles the higher sensitivity to dexamethasone of NF, MF, and AF cells with respect to FS cells (see Fig. 1). These results suggest that loss of p16 influences the cellular response to the antiproliferative effects of dexamethasone.
Discussion
Control of cell proliferation is deregulated in the majority of tumors (for a review, see ref. 35). Proliferation increases during FS development in vivo (36), and in cultured cells (20), particularly at the FS stage. We observed that dexamethasone reduces cell proliferation throughout the progression; however, this reduction is more apparent in the earlier stages of tumorigenesis than at the FS stage. At the FS stage, duplication of chromosome 8 and/or loss of chromosome 14 are observed (37), and it is therefore possible that some factor lies in the duplicated or deleted regions that might overcome the effects of dexamethasone on cell-cycle regulation. The BPV1 genome acts as a tissue-specific oncogene in transgenic mice, and its transforming properties reside mainly in two genes, E5 and E6, whose products are detectable in AF and FS cells to similar levels, suggesting that increased levels of the oncoproteins are not responsible for the step from AF to FS (20). Another possibility is that in FS cells the reduction in apoptosis in response to dexamethasone (D. Gascoyne and M.d.M.V., unpublished observations), to those levels found in cells from earlier stages of tumorigenesis, mitigates the effects on proliferation. Thus, the final step to FS development is accompanied by an altered proliferation/apoptosis ratio in favor of proliferation. Importantly, our studies of cell-growth inhibition by dexamethasone using WT and INK4a−/− MEFs identifies the loss of p16 as a molecular change that renders cells less sensitive to the antiproliferative effects of dexamethasone.
p16 expression was lost in four different FS lines tested, whereas p16 was detected in all of the cell lines examined from each stage of the tumorigenic pathway, independent of the passage number (note that the cells were used only at low passage numbers, 1–30). This finding suggests that inactivation of p16 is a late event in FS progression and coincides with the high increase in proliferation of FS cells and the transition in transcriptional activation by GR at the tumor stage of FS development (21). In some cases, inactivation of p16 has been found to be an early event during tumorigenesis (12, 38, 39); although in brain tumors (40), and lung carcinomas (41), later stage tumors present higher frequencies of p16 abnormalities, suggesting that different tumor types present different progression events to malignancy.
FS cells do not express p16 mRNA, suggesting that a nontranscriptional mechanism is responsible for the loss of p16 in FS cells. Because p16 genomic DNA can be detected at all stages of FS progression, it is unlikely that deletions in the p16 coding region have occurred in FS cells. These observations lead us to favor the possibility that the loss of p16 in FS cells involves methylation of the p16 promoter. Indeed, methylation of the 5′ CpG island in the p16 promoter is a frequent mechanism of p16 inactivation in many tumor types (42), and, importantly, in FSs (31, 32). Interestingly, and consistent with this hypothesis, inhibition of DNA methylation by treatment with 5-Aza-dC restores p16 expression in FS cells and reduces GR transcriptional activity in FS cells, but not in AF cells. Thus, a reduction in methylation results in decreased GR transcriptional activity.
We carried out two sets of experiments that specifically addressed whether the level of p16 expression influences GR transcriptional activity and obtained the following results. First, ectopic expression of p16 specifically inhibits GR activity in FS cells. Second, MEFs in which the p16 gene has been deleted exhibit increased GR activity relative to their normal counterparts, further supporting a model in which endogenous p16 normally represses GR transcriptional activity, and loss of p16 leads to GR activation.
Conditional expression of p16 in a lymphoblastic leukemia cell line has recently been reported to be associated with enhanced glucocorticoid-mediated transcriptional activity as a result of increased GR protein levels (43). In contrast, during FS development, the level of GR protein expression remains constant, whereas there is an increase in the transcriptional activity of GR at the tumor stage (21); and GR protein expression levels are clearly not affected by the absence (FS cells) or presence (NF, MF, and AF) of p16. This discrepancy highlights how p16 effects on GR might be different, depending on the cell type investigated. In this context, the role of glucocorticoids in inducing apoptosis in leukemic cells is well established (44). However, they inhibit apoptosis in other systems (45–47), including FS progression (D. Gascoyne and M.d.M.V., unpublished observations). The contrasting effects of glucocorticoids on apoptosis in the lymphoblastic leukemia cells and FS cells parallel those of p16 expression on GR transcriptional activity in these two cellular contexts. This finding might reflect the variety of alterations that give rise to cancers in different tissues.
p16 specifically binds to cdk4 and inhibits cdk4-dependent phosphorylation of pRb and progression to the S phase of the cell cycle (48). The analysis of mutations found in human tumors suggests that cyclin D1, cdk4, pRb, and p16 are critical components of a cell-cycle regulatory pathway that is altered in most tumor cells (reviewed in ref. 49). Differences among cell types have been reported for cell-cycle proteins involved in GR-mediated cell-cycle arrest. For example, cdk4 and E2F protein levels are repressed by GR in U2OS cells, but not in SAOS cells (50). We observed a constant level of expression of cyclin D1 and cdk4 during FS progression. The dramatic loss of p16 expression from AF to FS is reflected by a change in pRb mobility, suggesting increased phosphorylation and, therefore, increased proliferation. Interestingly, pRb potentiates GR transcriptional activity through the interaction of pRb's pocket domain with the transcriptional coactivator hBRM (51), whereas hBRM can potentiate the transcriptional activity of GR (52). It remains to be determined whether increased phosphorylation of endogenous pRb (through lack of p16) increases GR transcriptional activity. Interestingly, other cdk inhibitor proteins, including members of the INK4 family (p18INK4c and p19INK4d), and members of the Cip/Kip family (p21Cip/Waf1 and p27Kip1), were expressed at similar levels throughout FS development and, therefore, it is not surprising that their overexpression did not significantly affect GR transcriptional activity in AF or FS cells. This result suggests that the effects of p16 on GR activity are specific.
The identification of the differential transcriptional activity displayed by GR at the critical FS stage was the first molecular parameter to distinguish AF from FS (21). The loss of p16 expression provides a second molecular parameter. Although it is presently unclear how the loss of p16 leads to GR activation, this observation might have important implications for current cancer therapies. For example, treatment of malignant lymphoproliferative disorders by using dexamethasone (53) may have different clinical outcomes, depending on whether the cancer cells express p16. The correlation between activation of GR, sensitivity to the antiproliferative effects of dexamethasone, and the loss of p16 highlights a functional relationship between steroid hormone receptors and cell-cycle inhibitors.
Acknowledgments
We thank Dr. Manuel Serrano for providing the MEFs (derived from WT and INK4a−/− mice); Dr. Charles J. Sherr, and Dr. Sibylle Mittnacht for providing the expression vectors encoding the various cdk inhibitors tested; Gail O'Connell for technical assistance; Dr. Sibylle Mittnacht and Dr. Duncan Gascoyne for helpful discussions; and Prof. Alan Ashworth for critically reading the manuscript. This work was supported by the Leopold Muller Trust and the Institute of Cancer Research.
Abbreviations
- GR
glucocorticoid receptor
- p16
p16INK4a
- NF
normal fibroblast
- MF
mild fibromatosis
- AF
aggressive fibromatosis
- FS
fibrosarcoma
- 5-Aza-dC
5-aza-2′-deoxycytidine
- cdk
cyclin-dependent kinase
- pRb
retinoblastoma protein
- BPV
bovine papillomavirus
- MEF
mouse embryo fibroblast
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cole T J, Blendy J A, Monaghan A P, Krieglstein K, Schmid W, Aguzzi A, Fantuzzi G, Hummler E, Unsicker K, Schutz G. Genes Dev. 1995;9:1608–1621. doi: 10.1101/gad.9.13.1608. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto K R. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- 3.Goya L, Maiyar A C, Ge Y, Firestone G L. Mol Endocrinol. 1993;7:1121–1132. doi: 10.1210/mend.7.9.8247014. [DOI] [PubMed] [Google Scholar]
- 4.Frost G H, Rhee K, Ma T, Thompson E A. J Steroid Biochem Mol Biol. 1994;50:109–119. doi: 10.1016/0960-0760(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez I, Goya L, Vallerga A K, Firestone G L. Cell Growth Differ. 1993;4:215–225. [PubMed] [Google Scholar]
- 6.Jenkins B D, Pullen C B, Darimont B D. Trends Endocrinol Metab. 2001;12:122–126. doi: 10.1016/s1043-2760(00)00357-x. [DOI] [PubMed] [Google Scholar]
- 7.Beato M, Herrlich P, Schutz G. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 8.Harbour J W, Dean D C. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 9.Sherr C J, Roberts J M. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 10.Serrano M, Hannon G J, Beach D. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 11.Maelandsmo G M, Berner J M, Florenes V A, Forus A, Hovig E, Fodstad O, Myklebost O. Br J Cancer. 1995;72:393–398. doi: 10.1038/bjc.1995.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merlo A, Herman J G, Mao L, Lee D J, Gabrielson E, Burger P C, Baylin S B, Sidransky D. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 13.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J P, Davidson N E, Sidransky D, Baylin S B. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 14.Gonzalez-Zulueta M, Bender C M, Yang A S, Nguyen T, Beart R W, Van Tornout J M, Jones P A. Cancer Res. 1995;55:4531–4535. [PubMed] [Google Scholar]
- 15.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 16.Krimpenfort P, Quon K C, Mooi W J, Loonstra A, Berns A. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 17.Sharpless N E, Bardeesy N, Lee K H, Carrasco D, Castrillon D H, Aguirre A J, Wu E A, Horner J W, DePinho R A. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D, Weinberg R A. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D. Annu Rev Genet. 1988;22:479–519. doi: 10.1146/annurev.ge.22.120188.002403. [DOI] [PubMed] [Google Scholar]
- 20.Sippola-Thiele M, Hanahan D, Howley P M. Mol Cell Biol. 1989;9:925–934. doi: 10.1128/mcb.9.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vivanco M d M, Johnson R, Galante P E, Hanahan D, Yamamoto K R. EMBO J. 1995;14:2217–2228. doi: 10.1002/j.1460-2075.1995.tb07216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Distelhorst C W. Cell Death Differ. 2002;9:6–19. doi: 10.1038/sj.cdd.4400969. [DOI] [PubMed] [Google Scholar]
- 23.Landi M T, Baccarelli A, Calista D, Fears T R, Landi G. Int J Cancer. 2001;94:302–303. doi: 10.1002/ijc.1468. [DOI] [PubMed] [Google Scholar]
- 24.Wattenberg L W, Estensen R D. Cancer Res. 1996;56:5132–5135. [PubMed] [Google Scholar]
- 25.Steffen M, Scherdin U, Duvigneau C, Holzel F. Cancer Res. 1988;48:7212–7218. [PubMed] [Google Scholar]
- 26.Miesfeld R, Godowski P J, Maler B A, Yamamoto K R. Science. 1987;236:423–427. doi: 10.1126/science.3563519. [DOI] [PubMed] [Google Scholar]
- 27.Cairns P, Polascik T J, Eby Y, Tokino K, Califano J, Merlo A, Mao L, Herath J, Jenkins R, Westra W, et al. Nat Genet. 1995;11:210–212. doi: 10.1038/ng1095-210. [DOI] [PubMed] [Google Scholar]
- 28.Quelle D E, Ashmun R A, Hannon G J, Rehberger P A, Trono D, Richter K H, Walker C, Beach D, Sherr C J, Serrano M. Oncogene. 1995;11:635–645. [PubMed] [Google Scholar]
- 29.Jones P A, Laird P W. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 30.Malumbres M, Perez de Castro I, Santos J, Melendez B, Mangues R, Serrano M, Pellicer A, Fernandez-Piqueras J. Oncogene. 1997;14:1361–1370. doi: 10.1038/sj.onc.1200969. [DOI] [PubMed] [Google Scholar]
- 31.Rocco J W, Sidransky D. Exp Cell Res. 2001;264:42–55. doi: 10.1006/excr.2000.5149. [DOI] [PubMed] [Google Scholar]
- 32.Orlow I, Drobnjak M, Zhang Z F, Lewis J, Woodruff J M, Brennan M F, Cordon-Cardo C. J Natl Cancer Inst. 1999;91:73–79. doi: 10.1093/jnci/91.1.73. [DOI] [PubMed] [Google Scholar]
- 33.Herman J G, Jen J, Merlo A, Baylin S B. Cancer Res. 1996;56:722–727. [PubMed] [Google Scholar]
- 34.Loughran O, Malliri A, Owens D, Gallimore P H, Stanley M A, Ozanne B, Frame M C, Parkinson E K. Oncogene. 1996;13:561–568. [PubMed] [Google Scholar]
- 35.Adams P D, Kaelin W G., Jr Curr Opin Cell Biol. 1998;10:791–797. doi: 10.1016/s0955-0674(98)80123-3. [DOI] [PubMed] [Google Scholar]
- 36.Lacey M, Alpert S, Hanahan D. Nature. 1986;322:609–612. doi: 10.1038/322609a0. [DOI] [PubMed] [Google Scholar]
- 37.Lindgren V, Sippola-Thiele M, Skowronski J, Wetzel E, Howley P M, Hanahan D. Proc Natl Acad Sci USA. 1989;86:5025–5029. doi: 10.1073/pnas.86.13.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, Corio R, Lee D, Greenberg B, Koch W, Sidransky D. Cancer Res. 1996;56:2488–2492. [PubMed] [Google Scholar]
- 39.Yamano M, Fujii H, Takagaki T, Kadowaki N, Watanabe H, Shirai T. Am J Pathol. 2000;156:2123–2133. doi: 10.1016/S0002-9440(10)65083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker D G, Duan W, Popovic E A, Kaye A H, Tomlinson F H, Lavin M. Cancer Res. 1995;55:20–23. [PubMed] [Google Scholar]
- 41.Okamoto A, Hussain S P, Hagiwara K, Spillare E A, Rusin M R, Demetrick D J, Serrano M, Hannon G J, Shiseki M, Zariwala M, et al. Cancer Res. 1995;55:1448–1451. [PubMed] [Google Scholar]
- 42.Esteller M, Corn P G, Baylin S B, Herman J G. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 43.Ausserlechner M J, Obexer P, Wiegers G J, Hartmann B L, Geley S, Kofler R. J Biol Chem. 2001;276:10984–10989. doi: 10.1074/jbc.M008188200. [DOI] [PubMed] [Google Scholar]
- 44.Smets L A, van den Berg J D. Leuk Lymphoma. 1996;20:199–205. doi: 10.3109/10428199609051608. [DOI] [PubMed] [Google Scholar]
- 45.Feng Z, Marti A, Jehn B, Altermatt H J, Chicaiza G, Jaggi R. J Cell Biol. 1995;131:1095–1103. doi: 10.1083/jcb.131.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans-Storms R B, Cidlowski J A. Endocrinology. 2000;141:1854–1862. doi: 10.1210/endo.141.5.7466. [DOI] [PubMed] [Google Scholar]
- 47.Gorman A M, Hirt U A, Orrenius S, Ceccatelli S. Neuroscience. 2000;96:417–425. doi: 10.1016/s0306-4522(99)00565-5. [DOI] [PubMed] [Google Scholar]
- 48.Lukas J, Parry D, Aagaard L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 49.Sherr C J. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 50.Rogatsky I, Trowbridge J M, Garabedian M J. Mol Cell Biol. 1997;17:3181–3193. doi: 10.1128/mcb.17.6.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh P, Coe J, Hong W. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 52.Muchardt C, Yaniv M. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smets L A, Salomons G, van den Berg J. Adv Exp Med Biol. 1999;457:607–614. doi: 10.1007/978-1-4615-4811-9_67. [DOI] [PubMed] [Google Scholar]