Abstract
We examined iron nitrosylation of non-heme protein and enzymatic activity of the Fe-S cluster protein, aconitase, in acute cardiac allograft rejection. Heterotopic transplantation of donor hearts was performed in histocompatibility matched (isografts: Lewis → Lewis) and mismatched (allografts: Wistar–Furth → Lewis) rats. On postoperative days (POD) 4–6, Western blot analysis and immunohistochemistry revealed inducible nitric-oxide synthase (iNOS) protein in allografts but not isografts. EPR spectroscopy revealed background signals at g = 2.003 (for semiquinone) and g = 2.02 and g = 1.94 (for Fe-S cluster protein) in isografts and normal hearts. In contrast, in allografts on POD4, a new axial signal at g = 2.04 and g = 2.02 appeared that was attributed to the dinitrosyl–iron complex formed by nitrosylation of non-heme protein. Appearance of this signal occurred at or before significant nitrosylation of heme protein. Iron nitrosylation of non-heme protein was coincidental with decreases in the nonnitrosylated Fe-S cluster signal at g = 1.94. Aconitase enzyme activity was decreased to ≈50% of that observed in isograft controls by POD4. Treatment with cyclosporine blocked the (i) elevation of plasma nitrate + nitrite, (ii) up-regulation of iNOS protein, (iii) decrease in Fe-S cluster EPR signal, (iv) formation of dinitrosyl–iron complexes, and (v) loss of aconitase enzyme activity. Formation of dinitrosyl–iron complexes and loss of aconitase activity within allografts also was inhibited by treatment of recipients with a selective iNOS inhibitor, l-N6-(1-iminoethyl)lysine. This report shows targeting of an important non-heme Fe-S cluster protein in acute solid organ transplant rejection.
Keywords: graft rejection‖aconitase‖dinitrosyl–iron complexes
Increased synthesis of nitric oxide (NO) via the up-regulation of the inducible NO synthase (iNOS) isoform is believed to play a key role in acute rejection after solid organ transplantation. For cardiac transplant rejection, treatment with cyclosporine (to inhibited up-regulation of iNOS) (1–3), NOS inhibitors (to inhibit NO production by NOS) (2–4), or NO scavengers (to inhibit NO function) (5–7) has been shown to prolong graft survival. Deletion of iNOS gene also decreases histological rejection scores in acute cardiac transplant (8).
The precise molecular mechanisms by which these individual interventions prolong graft survival are not known with certainty. In general, one of the more important biological actions of physiological concentrations of NO in various cells is the interaction with iron-containing proteins (9). In contrast, in the setting of excess NO production via iNOS, it is possible that molecular interactions of NO with iron-containing proteins may lead to altered cell function or cell injury. Indeed, nitrosylation of heme protein (notably myoglobin) has been demonstrated by EPR spectroscopy in acute cardiac allograft rejection (10–12). EPR signals for nitrosylated heme protein are found in allograft tissue but not isograft tissue, suggesting that the increase in nitrosylation is due to alloimmune activation rather than secondarily to artifacts of surgery. The cause–effect relationship in nitrosylation of heme protein on graft function during rejection is incompletely understood. Nevertheless, intervention with NOS inhibitors (11, 12) or NO scavengers (5–7) that decreased heme nitrosylation also decreased histological rejection scores and prolonged graft survival.
In addition to heme protein, NO is known to interact with iron in non-heme protein (13–15). Despite, the use of EPR to document nitrosylation of heme protein, examination of the potential interaction of NO with iron in non-heme protein by EPR in acute allograft rejection has received little attention. The importance of this interaction cannot be overstated because several Fe-S cluster proteins located within mitochondria comprise the most abundant non-heme proteins within the cell. Candidate proteins include: aconitase, NADH ubiquinone oxidoreductase (complex I), succinate ubiquinone oxidoreductase (complex II), and ubiquinol cytochrome C oxidoreductase (complex III) (16, 17). Excess NO is known to inhibit cell respiration presumably by targeting mitochondrial Fe-S cluster proteins. Previous studies have shown that, of various potential Fe-S proteins, aconitase was the most susceptible to inactivation by ex vivo exposure of isolated macrophage cells to excess NO concentration followed by complex I and complex II (17, 18).
The purpose of the present study was to examine changes in Fe-S cluster protein signals for nonnitrosylated and nitrosylated species by EPR spectroscopy and aconitase enzyme activity in an in vivo model of acute cardiac allograft rejection. Also, treatment groups were designed to determine the potential species responsible for these changes.
Materials and Methods
Animals and Transplantation of Grafts.
Rats weighing ≈210–230 g were obtained from Harlan–Sprague–Dawley. Lewis (Lew: RT11) and Wistar–Furth (WF: RT1U) rat strains were chosen to represent genetic disparity at both the major and minor histocompatibility loci. Sterile surgery was performed in rats anesthetized with an i.p. injection of 50 mg/kg sodium pentobarbital. Heterotopic cardiac transplantation to the abdominal aorta and vena cava was performed by using established techniques. Graft function was monitored twice daily by standard external palpation. Acute rejection was defined as a loss of palpable contractile activity. Loss of graft function was confirmed on direct inspection after laparotomy.
Donor-to-recipient combinations were Lew→Lew (for isografts) or WF→Lew (for allografts).
Experimental Groups and Biopsy Procedures.
Isograft or allograft experiments were terminated at either postoperative day (POD) 4, POD6, or on the day of rejection. A subset of allograft recipients received 30 μg/ml l-N6-(1-iminoethyl)lysine (L-NIL) added to the drinking water or 2.5 mg/kg i.p. cyclosporine (CsA) beginning the day of surgery until day of tissue harvesting or rejection. These concentrations are within the therapeutic range used in inflammatory diseases (19). Also, concentrations of L-NIL up to 1 mg/ml do not alter systemic blood pressure in normal rats (19, 20), confirming that L-NIL under these conditions does not affect constitutive NOS activity. For POD4 and POD6, grafts were arrested and flushed with cold University of Wisconsin solution, minced, and frozen in liquid nitrogen. Frozen tissue was stored at −80°C (for Western blotting and gel-shift assays). For EPR spectroscopy, tissue samples were frozen in 4.0-mm quartz tubes (EPR quality) under liquid nitrogen. Plasma was obtained from individual rats for determination of total NO by-products, nitrate + nitrite, by using a commercial kit (Cayman Chemical, Ann Arbor, MI).
Histological Rejection Scoring.
Tissue from a portion of grafts were fixed at POD6 in 4% phosphate-buffered formalin, and paraffin-embedded sections were stained with hematoxylin and eosin. Rejection scoring was performed blinded. Scoring was based on criteria established by the International Society for Heart and Lung Transplantation and performed blinded as described (21).
EPR Spectroscopy.
Nitrosylation of myocardial heme protein was detected by using X-band EPR spectroscopy with a liquid-nitrogen finger dewar in a Varian E-109 spectrometer. Samples from each group were analyzed on the same day under similar instrument settings consisting of: 1,000-G scan range; 4-min scan time; 0.25-s time constant; 2-G modulation amplitude; 100-kHz modulation frequency; and 5-mW microwave power. The magnetic field was calibrated with Fremy's salt, giving a g value of 2.0055 ± 0.0001.
Aconitase Enzyme Activity.
Frozen tissue was homogenized in 50 mM Tris (pH 7.4) with 1 mM citrate, 1 mM cysteine, and 0.5 mM MnCl2 followed by ice-cold 0.4% Triton X-100. The sample was incubated on ice for 30 min followed by centrifugation at 4,000 × g for 15 min at 4°C. The supernatant was diluted 1:10 and assayed in 20 mM d,l-isocitrate-Na by following absorbance changes for 2 min at 240 nm on an Agilent 8453 spectrophotometer (Agilent Technologies, Palo Alto, CA) as described (22).
Western Blotting.
Frozen tissue was homogenized in ice-cold PBS with 1% Triton X-100/1 mM PMSF/35 ng/ml pepstatin A/10 ng/ml leupeptin. Homogenates were centrifuged at 10,000 × g for 10 min at 4°C. Protein concentration of the supernatant was determined by using the Bio-Rad DC protein assay, with BSA as a standard. Fifty micrograms of each sample was precipitated with 12.5% trichloroacetic acid containing 0.5 mg/ml deoxycholate. After washing the pellet in ice-cold acetone, the pellet was resuspended in SDS/PAGE loading buffer, neutralized with 1 M Tris, and electrophoresed on 7.5% SDS-polyacrylamide gels (for iNOS). Proteins were transferred to Nytran membranes, and blots were probed with 1:1,000 dilution of rabbit anti-iNOS (Santa Cruz Biotechnology) and visualized by using 1:5,000 dilution of donkey anti-rabbit IgG horseradish peroxidase conjugate and enhanced chemiluminescence. Densitometry was performed on an AlphaImager 2000 image analysis system (Alpha Innotech, San Leandro, CA).
Immunohistochemistry.
A portion of harvested grafts were fixed in formalin and embedded in paraffin for later immunohistochemical analysis. To quench endogenous background, sections were incubated with 1% H2O2 in methanol for 5 min, followed by washing twice in 7.4% buffered saline. Nonspecific binding was blocked at room temperature for 30 min by using commercial blocking serum (Vector Elite ABC kit; Vector Laboratories). Sections were incubated for 45 min with a 1:50 dilution of primary antibody for iNOS (Santa Cruz Biotechnology) followed by washing twice in buffered saline and incubation with biotinylated secondary antibody (Vector Elite kit). After washing in buffered saline, sections were incubated with Vectastain Elite ABC reagent at room temperature for 30 min. Sections then were washed twice in buffered saline, incubated with 0.03% (wt/vol) 3,3′-diaminobenzidine with 0.003% (vol/vol) H2O2, rinsed, and examined. Slides were counterstained with hematoxylin and eosin for viewing.
Data Analysis.
EPR spectra were processed for presentation by using sumspec and grapher programs (Golden Software, Golden, CO). All quantitative data were determined as the mean value ± SEM. Statistical analyses of data were performed by ANOVA for multiple-group means or by Student's t test for comparisons between two group means. Statistical significance was set at the level of P < 0.05.
Results
Histological Evidence for Rejection.
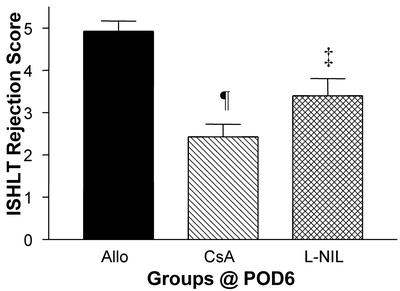
Using a standard International Society for Heart and Lung Transplantation scoring system for graft rejection, we observed elevated rejection scores indicative of inflammatory cell infiltration into the graft in untreated allografts analyzed at POD6 (Fig. 1). The rejections scores were elevated significantly compared with isograft controls or with native hearts of allograft recipients (not shown). Treatment with CsA or L-NIL decreased histological rejection scores relative to untreated allograft recipients.
Figure 1.
International Society for Heart and Lung Transplantation rejection scores in untreated and treated allografts. Results are mean ± SEM (n = 3–5 each). ‡, P < 0.01 vs. allograft (Allo) control; ¶, P < 0.001 vs. allograft.
Heme and Non-Heme Protein Nitrosylation.
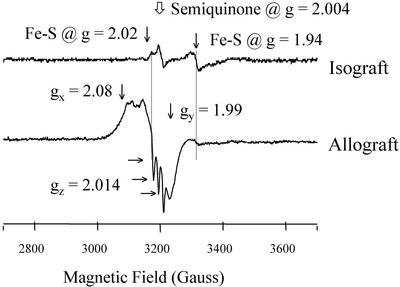
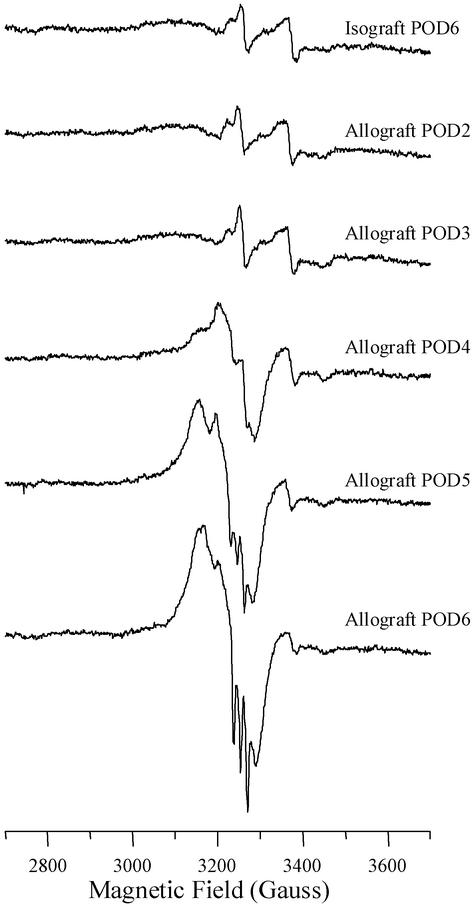
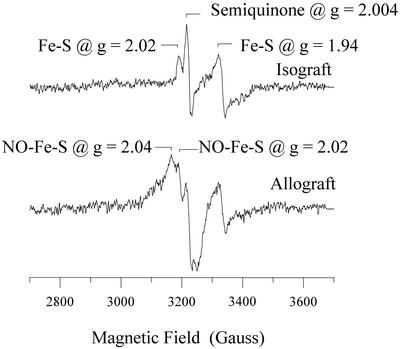
EPR analysis revealed different spectra for isografts vs. rejecting allografts at POD6 (Fig. 2). For isografts, signals were observed at g = 2.004 (denoted as a semiquinone radical) and g = 2.02 and 1.94 (denoted as Fe-S cluster protein) (12). These signals are identical to that seen by our laboratory in normal hearts and denoted as background signals (not shown). In contrast, a new signal was seen in allografts denoted as nitrosylated heme protein. This signal is characterized by a broad signal at g = 2.08, a deflection at g = 1.99, and a triplet feature at g = 2.014. We attribute these to the principle gx, gy, and gz components of nitrosylated purified heme protein (23). The onset of nitrosylation of heme protein was most evident by POD5, with maximum formation by POD6 (Fig. 3). In addition, at POD4, we observed an additional signal that could not be assessed adequately at POD5 and POD6 because of the overriding EPR signal for nitrosylated heme protein. This new signal had a calculated g = 2.04 and a feature at 2.02 (Fig. 4). This is designated as an axial signal of iron nitrosylation of non-heme protein (10, 24). Iron-nitrosylated species for non-heme protein was not observed in any sample examined at POD2 or POD3 (Fig. 3).
Figure 2.
Example showing EPR spectra measured at 77 K for isograft vs. allograft at POD6. Nitrosylation of heme protein is seen in allograft but not isograft. Dotted line to the right indicates the decrease in signal for Fe-S cluster protein at g = 1.94 in allograft vs. that seen in isograft controls. Spectra were collected under identical conditions including spectrometer gain settings.
Figure 3.
Examples of EPR spectra obtained from individual allografts at various periods of time posttransplant relative to an isograft control at POD6. A prominent peak with triplet signal for heme protein nitrosylation is seen by POD5.
Figure 4.
EPR spectra showing iron nitrosylation of non-heme protein in allograft but not isograft controls at POD4 relative to the background signals of native Fe-S protein and semiquinone.
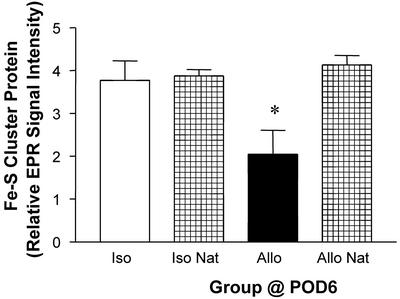
Along with nitrosylation of heme protein, we observed a decrease in the EPR signal for Fe-S cluster protein at g = 1.94 in the samples examined at POD6 (see location designed by the dotted line in Fig. 2). Quantitative analysis of this peak at POD6 revealed a decrease of 50% relative to isografts at POD6 or native hearts of allograft recipients (Fig. 5).
Figure 5.
Decrease in the relative intensity (mean ± SEM; n = 3–5 each) of EPR signal for Fe-S protein (g = 1.94 peak) in cardiac allografts (Allo) compared with isografts (Iso) or native (Nat) hearts of allograft recipients. *, P < 0.05 vs. isografts or native hearts.
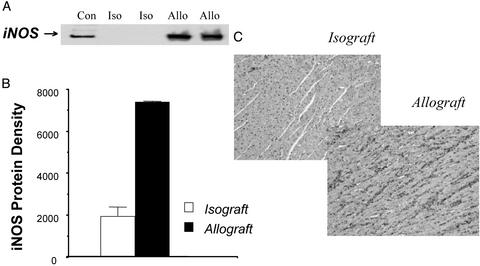
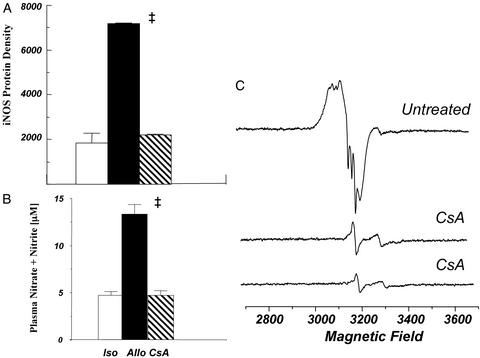
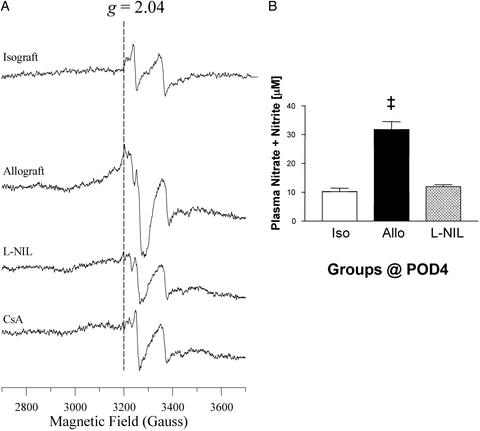
There was up-regulation of iNOS in allografts at POD4 based on Western blot analysis and immunohistochemical analysis by using antibody to iNOS (Fig. 6). Chronic treatment with CsA blocked iNOS protein, the increase in plasma nitrate + nitrite levels, and nitrosylation of both myocardial heme and non-heme protein (Fig. 7). Treatment with the iNOS inhibitor, L-NIL, prevented the formation of plasma NO metabolites and markedly reduced or essentially eliminated heme and non-heme protein nitrosylation (Fig. 8). L-NIL had no significant action to limit iNOS protein levels (not shown).
Figure 6.
Western immunoblot (A), immunoblot densitometry (B), or immunostaining (C) showing up-regulation of iNOS protein in cardiac allografts (Allo) vs. isografts (Iso) at POD4. Con, iNOS-positive control. Densitometry data are expressed as mean ± SEM.
Figure 7.
Treatment with CsA prevents the increase in iNOS protein level by Western blotting (n = 2–3 each) (A), increase in plasma NO by-products, nitrate + nitrite, (n = 5–9 each) (B), and nitrosylation of heme and non-heme protein by EPR spectroscopy (C). Values are expressed as mean ± SEM.
Figure 8.
Chronic treatment of recipients with the iNOS inhibitor, L-NIL, prevents iron nitrosylation of the non-heme Fe-S protein (i.e., EPR signal at g = 2.04) (A) and blocks increased NO by-products (n = 6–9 each) (B). Values are expressed as the mean ± SEM (n = 3–5 each). ‡, P < 0.01.
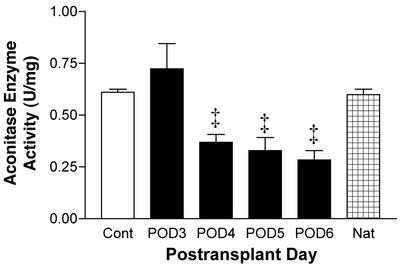
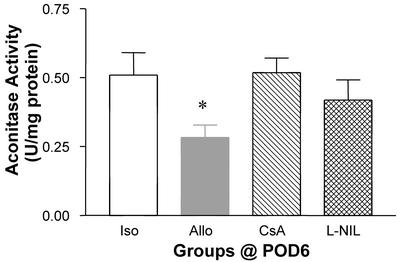
NADH dehydrogenase and succinate dehydrogenase are the most likely Fe-S proteins previously assigned to the EPR signal at g = 1.94 (25). Although this was used to determine changes in Fe-S protein, in general, aconitase does not display signals at g = 1.94 at 77 K. Therefore, decreases in g = 1.94 cannot be ascribed directly to inactivation of aconitase. Thus, we examined changes in aconitase enzyme activity directly. Enzyme activity of aconitase was decreased at POD4 through POD6 but not at POD3 compared with isograft controls (Fig. 9). By POD6, aconitase enzyme activity was decreased by 50% relative to isograft controls and native hearts of allograft recipients (Fig. 9). The decrease in aconitase enzyme activity in allograft recipients at POD6 was inhibited significantly or prevented by treatment with L-NIL or CsA, respectively (Fig. 10).
Figure 9.
Time-dependent decreases in aconitase enzyme activity (mean ± SEM) in cardiac allografts (n = 3–9 each) vs. isograft controls (n = 5) and native (Nat, n = 3) hearts of allograft recipients. ‡, P < 0.01 vs. controls.
Figure 10.
Decrease in aconitase enzyme activity (mean ± SEM) in allografts (n = 9) compared with isografts (n = 5) and prevention of loss of activity by treating allograft recipients with CsA or L-NIL (n = 5 each). *, P < 0.05 vs. isografts.
Discussion
Previously, we (7, 12, 21) and other investigators (10, 24) have detected by EPR spectroscopy the nitrosylation of myocardial heme protein before rejection in untreated cardiac allografts. Two studies described the formation of dinitrosyl–iron complexes of non-heme protein that was characterized by a specific axial EPR signal at g = 2.04 and g = 2.015 in either rat (10) or mouse (23) cardiac transplant models. Despite this description, there has not been any subsequent examination of the functional significance of this g = 2.04 signal and its potential origin of formation in this model. The present study is important in that it examines, in detail, the iron nitrosylation of non-heme protein coupled with determination of the functional enzymatic activity of a Fe-S cluster protein, aconitase, as a potential molecular target of excess NO in cardiac allograft rejection. Treatment with two separate agents that limit histological rejection, CsA and an iNOS inhibitor, inhibited dinitrosyl–iron complex formation and the loss of aconitase enzyme activity.
Aconitase is an enzyme consisting of a [4Fe-4S] cluster protein found in both the cytosol and mitochondria. We have found that the mitochondrial activity of aconitase in normal cardiomyocytes is 100-fold higher than that of cytosolic aconitase (unpublished observations). Thus, the decrease in myocardial aconitase enzyme activity in allografts suggests that mitochondrial dysfunction is a significant consequence of acute rejection.
We found that a temporal increase in iron nitrosylation of non-heme protein (g = 2.04 EPR signal) was associated with a decreased EPR signal for native Fe-S cluster protein (g = 1.94). These findings agree with studies in spinal cords of experimental encephalomyelitis (26) or in cells exposed to Clostridium botulinum (27) in which a decrease in signal for Fe-S protein was associated with an increase in nitrosylated Fe-S protein after treatment with exogenous NO donors. Our data do not corroborate the conclusion of Vanin et al. (28) that short-term (i.e., 5-min) exposure of cultured macrophages to a bolus of NO does not cause a disappearance in native Fe-S protein but, rather, that dinitrosyl–iron complexes arise from iron derived from low-molecular-mass iron pools. This may reflect differences in experimental conditions/models. Also, it should be noted that challenging with bolus NO does not provide reproducible EPR data compared with data derived from longer-term continuous NO exposure (29).
Of the few studies that have used EPR technology in cardiac transplant rejection, changes in the EPR signal for native Fe-S protein at g = 1.94 have not been routinely examined. Bastian et al. (24) showed that the signal at g = 1.94 disappeared in mouse heart allografts both with or without treatment with a nonselective NOS inhibitor, suggesting that the loss of signal was unrelated to NO production. Because they derived data from nonbeating, necrotic tissue and NO can be derived from nonenzymatic sources in ischemic myocardium, it is not possible to compare their results with our studies. In the present study, the coincidental appearance of the axial signal at g = 2.04 representing dinitrosyl–iron complexes of non-heme protein with decreased EPR signal at g = 1.94 representing Fe-S protein is consistent with the loss of the native Fe-S cluster structure upon nitrosylation. Because among various Fe-S proteins aconitase does not display a signal at g = 1.94 at 77 K, effects on this protein were examined by determining changes in enzyme activity. Our additional finding of loss of aconitase enzymatic activity is consistent with and is additional evidence of the significant functional consequence of generalized changes in Fe-S cluster proteins.
The loss of enzymatic activity occurred between POD3 and POD4. This is important because iNOS protein was already up-regulated at POD4. Furthermore, we found no evidence of iron nitrosylation of non-heme protein at POD3, but EPR analysis revealed that nitrosylation began to be observed at POD4. These findings are consistent with the hypothesis that the process of nitrosylation contributes to loss of aconitase enzyme activity. Iron nitrosylation of non-heme protein and loss of aconitase enzyme activity in allografts but not isografts or native hearts of allograft recipients indicates that these changes are specific to alloimmune activity and not due to surgical manipulation per se nor to systemic infection as a consequence to organ transplantation.
Previous studies by Kennedy et al. (29) at our institution have clearly shown that loss of purified aconitase enzyme activity in solution by exposure to a NO donor is associated with conversion of a [4Fe-4S] cluster to a [3Fe-4S] cluster. Furthermore, spin-trapping studies showed definitive evidence that the loss of an atom of Fe in purified aconitase serves as a source of hydroxyl radical formation (30). These findings are interesting considering the previous benefits on graft survival associated with treatment with a hydroxyl radical scavenger, dimethylthiourea, or the metal chelator, pyrrolidine dithiocarbamate (11, 31). These studies indicate that the molecular alteration in activity of the native aconitase enzyme with the loss of a reactive metal ion may have a significant affect on cell respiration and production of cell injury.
In cell-free systems, purified aconitase protein from plant and mammalian sources can be inhibited by oxidants such as H2O2 and superoxide (32, 33). Other early in vitro studies indicated that peroxynitrite, an oxidant derived from superoxide radicals and NO, could inactivate purified aconitase whereas NO could not (33, 34). Subsequent EPR studies from our institution by using purified aconitase provided unequivocal evidence of the loss of an iron atom and loss of aconitase enzyme activity in the presence of an NO donor (30). In contrast to this in vitro data, the species responsible for inactivation in the setting of acute organ rejection is unknown. Currently, there is no available information about potential candidate species in other in vivo models of inflammation. Thus, we designed studies to examine the potential mechanism of inactivation of aconitase in this in vivo model of acute cardiac transplant rejection.
We found that inhibition of aconitase was unique to allografts and not found in isografts or native hearts of allograft recipients. Furthermore, both the iron nitrosylation of non-heme protein and the loss of aconitase activity were blocked by treatment with the immunosuppressant agent, CsA. This suggests that alloimmune activation rather than surgery per se or systemic infection accounts for the loss of aconitase enzyme activity.
Several lines of evidence support a role of NO in the inactivation of aconitase. First, iNOS was up-regulated in untreated allografts at the same time as the onset of loss of enzyme activity. Second, CsA inhibited iNOS up-regulation and also blocked the loss of aconitase enzyme activity. Third, treatment with the highly selective iNOS inhibitor L-NIL protected against formation of dinitrosyl–iron complexes of non-heme protein and the loss of aconitase activity within allografts. Thus, we conclude that NO or a NO-derived species is the predominate entity responsible for loss of aconitase activity.
As stated previously, most of the aconitase enzyme activity in hearts is mitochondrial in origin. Together with recent studies showing tyrosine nitration of mitochondrial proteins in renal allograft rejection (35), our studies support the hypothesis that a variety of mitochondrial proteins are targets of NO in organ rejection. Although our studies do not directly address the potential modification of cytosolic aconitase by NO in this model, it should be recognized that cytosolic aconitase may be important. The cytosolic isoform displays aconitase enzyme activity, whereas the apo-form of the cytosolic protein is an iron-regulatory protein that binds to RNA and regulates genes involved in iron metabolism (36, 37). NO can modulate genes involved in iron homeostasis in isolated macrophage cells in vitro (38). It is not known whether NO regulates iron regulator protein and cytosolic aconitase activity in the setting of transplant rejection in vivo. Our findings suggest that further studies in this area may be warranted.
In conclusion, the present study examines the relationship between iron nitrosylation of non-heme protein and loss of aconitase enzyme activity in acute cardiac allograft rejection. This study suggests that iron nitrosylation and inactivation of non-heme, Fe-S cluster protein may contribute to alloimmune rejection.
Acknowledgments
We acknowledge the assistance of Ms. Karen Hyde in processing EPR data for illustration and Dr. William E. Antholine for discussion of interpretation of EPR signals. This work was supported by National Institutes of Health Grants HL64637 (to G.M.P.) and DK48423 (to O.W.G.) and National Institutes of Health Center Grant RR01008 to the National Biophysics Research Institute.
Abbreviations
- iNOS
inducible NO synthase
- CsA
cyclosporine
- L-NIL
l-N6-(1-iminoethyl)lysine
- POD
postoperative day
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cai B, Roy D K, Sciacca R, Michler R E, Cannon P J. Int J Cardiol. 1995;50:243–251. doi: 10.1016/0167-5273(95)02384-9. [DOI] [PubMed] [Google Scholar]
- 2.Worrall N K, Lazenby W D, Misko T P, Lin T S, Rodi C P, Manning P T, Tilton R G, Williamson J R, Ferguson T B., Jr J Exp Med. 1995;181:63–70. doi: 10.1084/jem.181.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Worrall N K, Misko T P, Sullivan P M, Hui J J, Rodi C P, Ferguson T B., Jr Transplantation. 1996;61:324–328. doi: 10.1097/00007890-199601270-00028. [DOI] [PubMed] [Google Scholar]
- 4.Winlaw D S, Schyvens C G, Smythe G A, Du Z Y, Rainer S P, Lord R S A, Spratt P M, Macdonald P S. Transplantation. 1995;60:77–82. doi: 10.1097/00007890-199507150-00015. [DOI] [PubMed] [Google Scholar]
- 5.Cooper M, Dembny K, Lindholm P, Lai C S, Moore G, Johnson C, Adams M, Pieper G, Roza A. Surg Forum. 1997;48:500–503. [Google Scholar]
- 6.Roza A M, Cooper M, Pieper G, Hilton G, Dembny K, Lai C S, Lindholm P, Komorowski R, Felix C, Johnson C, et al. Transplantation. 2000;69:227–231. doi: 10.1097/00007890-200001270-00006. [DOI] [PubMed] [Google Scholar]
- 7.Pieper G M, Cooper M, Johnson C, Adams M B, Felix C, Roza A M. J Cardiovasc Pharmacol. 2000;35:114–120. doi: 10.1097/00005344-200001000-00015. [DOI] [PubMed] [Google Scholar]
- 8.Koglin J, Glysing-Jensen T, Mudgett J S, Russell M E. Am J Pathol. 1998;153:1371–1376. doi: 10.1016/S0002-9440(10)65723-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper C E. Biochim Biophys Acta. 1999;1411:290–309. doi: 10.1016/s0005-2728(99)00021-3. [DOI] [PubMed] [Google Scholar]
- 10.Lancaster J R, Jr, Langrehr J M, Bergonia H A, Murasse N, Simmons R L, Hoffman R A. J Biol Chem. 1992;267:10994–10998. [PubMed] [Google Scholar]
- 11.Cooper M, Lindholm P, Pieper G, Seibel R, Moore G, Nakanishi A, Dembny K, Komorowski R, Johnson C, Adams M, et al. Transplantation. 1998;66:838–844. doi: 10.1097/00007890-199810150-00005. [DOI] [PubMed] [Google Scholar]
- 12.Nakanishi A L, Roza A M, Adams M B, Seibel R, Moore-Hilton G, Kalyanaraman B, Pieper G M. Free Radical Biol Med. 1998;25:201–207. doi: 10.1016/s0891-5849(98)00051-3. [DOI] [PubMed] [Google Scholar]
- 13.Drapier J C, Hibbs J B., Jr J Clin Invest. 1986;78:790–797. doi: 10.1172/JCI112642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lancaster J R, Jr, Hibbs J B., Jr Proc Natl Acad Sci USA. 1990;87:1223–1227. doi: 10.1073/pnas.87.3.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellat C, Henry Y, Drapier J C. Biochem Biophys Res Commun. 1990;166:119–125. doi: 10.1016/0006-291x(90)91919-j. [DOI] [PubMed] [Google Scholar]
- 16.Henry Y, Lepoivre M, Drapier J C, Ducrocq C, Boucher J L, Guissani A. FASEB J. 1993;7:1124–1134. doi: 10.1096/fasebj.7.12.8397130. [DOI] [PubMed] [Google Scholar]
- 17.Henry Y, Guissani A. Cell Mol Life Sci. 1999;55:1003–1014. doi: 10.1007/s000180050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drapier J C. Methods. 1997;11:319–329. doi: 10.1006/meth.1996.0426. [DOI] [PubMed] [Google Scholar]
- 19.Connor J R, Manning P T, Settle S L, Moore W M, Jerome G M, Webber R K, Tjoeng T S, Currie W M. Eur J Pharmacol. 1995;273:15–24. doi: 10.1016/0014-2999(94)00672-t. [DOI] [PubMed] [Google Scholar]
- 20.Reilly C M, Farrelly L W, Viti D, Redmond S T, Hutchison F, Ruiz P, Manning P, Connor J, Gilkeson G S. Kidney Int. 2002;61:839–846. doi: 10.1046/j.1523-1755.2002.00230.x. [DOI] [PubMed] [Google Scholar]
- 21.Pieper G M, Roza A M, Adams M B, Hilton G, Johnson M, Felix C C, Kampalath B, Darkes M, Wanggui Y, Cameron B, et al. J Cardiovasc Pharmacol. 2002;39:441–448. doi: 10.1097/00005344-200203000-00016. [DOI] [PubMed] [Google Scholar]
- 22.Hausladen C P, Fridovich I. Methods Enzymol. 1996;269:37–40. doi: 10.1016/s0076-6879(96)69007-7. [DOI] [PubMed] [Google Scholar]
- 23.Kobayshi K, Tamura M, Hayashi K. Biochim Biophys Acta. 1982;702:23–29. doi: 10.1016/0167-4838(82)90023-1. [DOI] [PubMed] [Google Scholar]
- 24.Bastian N R, Xu S, Shao X L, Shelby J, Granger D L, Hibbs J B., Jr Biochim Biophys Acta. 1994;1226:225–231. doi: 10.1016/0925-4439(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 25.Moore K P, Holt S G, Patel R P, Svistunenko D A, Zackert W, Goodier D, Reeder B J, Clozel M, Anand R, Cooper C E, et al. J Biol Chem. 1998;273:31731–31737. doi: 10.1074/jbc.273.48.31731. [DOI] [PubMed] [Google Scholar]
- 26.Lin R F, Sin T S, Tilton R G, Cross A H. J Exp Med. 1993;178:643–648. doi: 10.1084/jem.178.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy D, Lancaster J R, Cornforth D P. Science. 1983;221:769–770. doi: 10.1126/science.6308761. [DOI] [PubMed] [Google Scholar]
- 28.Vanin A F, Men'shikov G B, Moroz I A, Mordvintcev P I, Serezhenkov V A, Burbaev Dsh. Biochim Biophys Acta. 1992;1135:275–279. doi: 10.1016/0167-4889(92)90231-y. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy M C, Antholine W E, Beinert H. J Biol Chem. 1997;272:20340–20347. doi: 10.1074/jbc.272.33.20340. [DOI] [PubMed] [Google Scholar]
- 30.Vásquez-Vivar J, Kalyanaraman B, Kennedy M C. J Biol Chem. 2000;275:14064–14069. doi: 10.1074/jbc.275.19.14064. [DOI] [PubMed] [Google Scholar]
- 31.Pieper G M, Olds C, Hilton G, Lindholm P F, Adams M B, Roza A, M. Antioxidants Redox Signal. 2001;3:81–88. doi: 10.1089/152308601750100542. [DOI] [PubMed] [Google Scholar]
- 32.Verniquet F, Gaillard J, Neuburger M, Douce R. Biochem J. 1991;2756:643–648. doi: 10.1042/bj2760643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hausladen A, Fridovich I. J Biol Chem. 1997;269:29405–29408. [PubMed] [Google Scholar]
- 34.Castro L, Rodriguez M, Radi R. J Biol Chem. 1994;269:29409–29415. [PubMed] [Google Scholar]
- 35.MacMillan-Crow L A, Cruthirds D L, Ahki K M, Saunders P W, Thompson J A. Free Radical Biol Med. 2001;31:1603–1608. doi: 10.1016/s0891-5849(01)00750-x. [DOI] [PubMed] [Google Scholar]
- 36.Beinert H, Kiley P J. Curr Opin Chem Biol. 1999;3:152–157. doi: 10.1016/S1367-5931(99)80027-1. [DOI] [PubMed] [Google Scholar]
- 37.Oliveira L, Drapier J C. Proc Natl Acad Sci USA. 2000;97:6550–6555. doi: 10.1073/pnas.120571797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drapier J C, Hirling H, Wietzerbin J, Kaldy P, Kühn L C. EMBO J. 1993;12:3643–3649. doi: 10.1002/j.1460-2075.1993.tb06038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]