Abstract
Type I polyketide synthases (PKSs) are multifunctional enzymes that are organized into modules, each of which minimally contains a β-ketoacyl synthase, an acyltransferase (AT), and an acyl carrier protein. Here we report that the leinamycin (LNM) biosynthetic gene cluster from Streptomyces atroolivaceus S-140 consists of two PKS genes, lnmI and lnmJ, that encode six PKS modules, none of which contain the cognate AT domain. The only AT activity identified within the lnm gene cluster is a discrete AT protein encoded by lnmG. Inactivation of lnmG, lnmI, or lnmJ in vivo abolished LNM biosynthesis. Biochemical characterization of LnmG in vitro showed that it efficiently and specifically loaded malonyl CoA to all six PKS modules. These findings unveiled a previously unknown PKS architecture that is characterized by a discrete, iteratively acting AT protein that loads the extender units in trans to “AT-less” multifunctional type I PKS proteins for polyketide biosynthesis. This PKS structure provides opportunities for PKS engineering as exemplified by overexpressing lnmG to improve LNM production.
Polyketide natural products include many clinically important drugs such as erythromycin (antibacterial), epothilone (anticancer), rapamycin (immunosuppressant), and lovastatin (antihypercholesterolemic) (1). They are biosynthesized from short carboxylic acid precursors by polyketide synthases (PKSs) (2). Three types of bacterial PKSs are known to date. (i) Type I PKSs are multifunctional enzymes that are organized into modules, each of which harbors a set of distinct, noniteratively acting activities responsible for the catalysis of one cycle of polyketide chain elongation (3). (ii) Type II PKSs are multienzyme complexes that carry a single set of iteratively acting activities and minimally consist of the β-ketoacyl synthase (KS) α and β subunits and an acyl carrier protein (ACP) (4). [The KSβ subunit is also known as chain-length factor (5) or chain-initiation factor (6).] (iii) Type III PKSs, also known as chalcone synthase-like PKSs, are distributed predominately in plant and have been characterized from microorganisms only very recently (7, 8). They are essentially condensing enzymes that lack ACP and act directly on acyl CoA substrates.
Among type I PKSs characterized to date, each module minimally contains three domains, β-KS, acyltransferase (AT), and ACP, that select, activate, and catalyze a decarboxylative Claisen condensation between the extender unit and the growing polyketide chain, generating a β-ketoacyl-S-ACP intermediate. Optional domains are found between AT and ACP, which carry out the variable set of reductive modifications of the β-keto group before the next round of chain extension. The order of modules in the PKS enzymes dictates the sequence of biosynthetic events, and the variation of domains within the modules affords the structural diversity observed in the resultant polyketide products (2, 3). This one-to-one correspondence between the modularity of enzyme activities and the structures of the resultant products provides the molecular basis for combinatorial manipulation of type I PKSs for structural diversity (9–12).
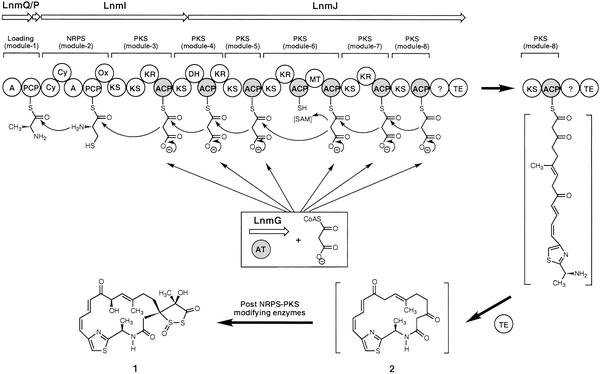
Leinamycin (LNM, 1) (Fig. 1) is a hybrid peptide–polyketide natural product that showed potent antitumor activity, most significantly against tumor cell lines that were resistant to clinically important anticancer drugs (13–16). We recently cloned and localized the lnm biosynthesis gene cluster to a 172-kb DNA region from Streptomyces atroolivaceus S-140, and DNA sequence analysis of the four overlapping cosmids (pBS3004, pBS3005, pBS3006, and pBS3007) revealed 72 ORFs (17). Sequential inactivation of ORFs from both ends of the sequenced region led to the assignment of the lnm gene cluster to consist of 27 ORFs, of which two (lnmQ and lnmP) encode nonribosomal peptide synthetase (NRPS), one (lnmI) encodes a hybrid NRPS–PKS enzyme, and two (lnmJ and lnmG) encode PKS. Surprisingly, all six PKS modules encoded by lnmI and lnmJ lack their cognate AT domain, and the only AT activity identified within the lnm gene cluster is a discrete AT protein encoded by lnmG (Fig. 1).
Figure 1.
Hypothesis for LNM (1) biosynthesis and modular organization of the Lnm hybrid NRPS–PKS megasynthetase with the discrete LnmG AT enzyme loading the malonyl CoA extender to all six PKS modules. The structures in brackets are hypothetical. It is not known whether one or both ACPs in module 6 are loaded with the malonyl group in vivo, although LnmG prefers ACP6-2 in vitro. Cy, condensation/cyclization; DH, dehydratase; MT, methyl transferase; Ox, oxidation; ?, domain of unknown function; TE, thioesterase.
Because a PKS module cannot be functional unless its ACP domain is loaded with the extender unit (2, 3), we proposed that LnmG provides the missing AT activity in trans to the LnmI and LnmJ PKS enzymes. To test this hypothesis, we first inactivated lnmI, lnmJ, and lnmG, confirming that they are essential for 1 biosynthesis. We then overexpressed lnmG and the ACP domains from all six PKS modules either as an individual domain or multidomain PKS module, along with lnmP, which encodes a peptidyl carrier protein (PCP), as a negative control. We finally showed that LnmG efficiently and specifically loaded malonyl CoA to the ACP domains from all six PKS modules but not to LnmP PCP. Our findings unveiled a previously unknown PKS architecture that provides opportunities for PKS engineering as exemplified by overexpressing lnmG to improve 1 production.
Materials and Methods
Sequence and in Vivo Analysis of the lnm Gene Cluster.
DNA sequencing, bioinformatic analyses of the sequenced 135,638-bp contiguous DNA, and functional assignments of the deduced gene products were summarized in GenBank (accession no. AF484556) (17). The boundaries of the lnm gene cluster were identified by gene replacement of orf(−13), orf(−11), orf(−2), orf(−1), and lnmA for the upstream boundary and of orfZ′, orf(+1), orf(+2), orf(+3), orf(+4), and orf(+6) for the downstream boundary, respectively. Inactivation of genes within the lnm gene cluster abolished 1 production, whereas that of genes outside the lnm gene cluster had no effect on 1 production.
LNM Production, Isolation, and Analysis.
LNM production and isolation from both the wild-type and recombinant S. atroolivaceus strains were carried out as reported (17). HPLC analysis was carried out on a Microsorb-MV C-18 column (5 μm, 100 Å, 250 × 4.6 mm, Varian) eluted with a gradient from 100% buffer A (20% CH3CN, pH 3.6 with HOAc) to 68% buffer B (80% CH3CN, pH 3.6 with HOAc) in 40 min at a flow rate of 1 ml/min and UV detection at 320 nm.
Inactivation by Gene Replacement.
To inactivate lnmI, lnmJ, and lnmG, internal fragments (a 5,906-bp SphI–XhoI fragment that harbors the condensation/cyclization, adenylation, PCP, and oxidation domains of NRPS module 2 and the first KS domain of PKS module 1 for lnmI, a 4,571-bp EcoRI–NotI fragment that harbors the ketoreductase (KR) and ACP domain of PKS module 7 and the KS domain of PKS module 8 for lnmJ, and a 388-bp SalI–BamHI fragment for lnmG) were replaced with the aac(3)IV apramycin-resistance gene (18), and the mutated lnmI, lnmJ, and lnmG genes were cloned into pSET151 (19) to yield pBS3017, pBS3018, and pBS3019, respectively. These constructs were introduced into S. atroolivaceus S-140 by conjugation (17) and selected for apramycin resistance and thiostrepton-sensitive phenotype to isolate the desired double-crossover mutant strains SB3002 (ΔlnmI), SB3003 (ΔlnmJ), and SB3004 (ΔlnmG), respectively. The genotypes of these mutants were confirmed by Southern analysis.
Expression of lnm Gene in Escherichia coli and Purification of the Resultant Recombinant Proteins.
The lnmG gene, the seven ACP domains from lnmI and lnmJ, the tridomain PKS module 4 of lnmJ-(DH-ACP-KR), and the lnmP gene all were amplified by PCR using the following primers: for lnmG, 5′-CG GAA TTC CAT ATG GTG GCA CTG GTT TTC CCG-3′ and 5′-CG GCC AAG CTT GCG GCG GGC GAG GAC GTC-3′; for lnmP, 5′-CG GAA TTC CAT ATG TGG GAC CAC AAG TTC GAG-3′ and 5′-CG CGC AAG CTT TCG GCC GGC TCC GTC GAG-3′; for lnm-ACP3, 5′-CG GAA TTC CAT ATG TCA GTC ACC GGG CCG CCC-3′ and 5′-CG CGC AAG CTT CCC GAG GTC CGC CAG ATG-3′; for lnmJ-ACP4, 5′-CG GAA TTC CAT ATG GGG CCG GAC GCG GTG CGC-3′ and 5′-CG CGC AAG CTT GAA CTC GGC GTA CAG GTG-3′; for lnmJ-ACP5, 5′-CG GAA TTC CAT ATG GAC CCG CAG GAG GTG CTG-3′ and 5′-CG CGC AAG CTT GTG CAG TTC CCT GAC GTG-3′; for lnmJ-ACP6-1, 5′-CG GAA TTC CAT ATG TCG GCC GAG GCC GTG CGG-3′ and 5′-CG CGC AAG CTT GTG TTC CTG GCG GAA GTA CC-3′; for lnmJ-ACP6-2, 5′-CG GAA TTC CAT ATG TCG CCC GAG TCC GTG CGG-3′ and 5′-CG CGC AAG CTT GTG CTC GGC GCT CAG GTA C-3′; for lnmJ-ACP7, 5′-CG GAA TTC CAT ATG CTG CGG GAG CTC GTG GAG-3′ and 5′-CG CGC AAG CTT ATG GTG CTG CGT CAG GTA CT-3′; for lnmJ-ACP8, 5′-CG GAA TTC CAT ATG GCC GCC TCC ACC GTC GTC-3′ and 5′-CG CGC AAG CTT GAC CAG CGG CGC GAC GAA C-3′; and for lnmJ-(DH-ACP4-KR), 5′-A TGA ATT CAT ATG AAC GTG CCC TCC GCA C-3′ and 5′-AT AAG CTT GCC GTC CGG GGA GTC AGG-3′. The numbers after each ACP refer to PKS modules from which they are derived with 6-1 and 6-2 to indicate the first and second ACP, respectively, for PKS module 6, and the restriction sites CAT ATG for NdeI and AAG CTT for HindIII designed in primers are underlined. The resultant products were sequenced to confirm PCR fidelity and cloned as NdeI–HindIII fragments into the same sites of pET28a (Novagen), yielding expression constructs pBS3020–pBS3029 for lnmG, lnmP, lnmJ-ACP3, lnmJ-ACP4, lnmJ-ACP5, lnmJ-ACP6-1, lnmJ-ACP6-2, lnmJ-ACP7, lnmJ-ACP8, and lnmJ-(DH-ACP4-KR), respectively. Introduction of pBS3020–pBS3029 into E. coli BL-21 (DE-3) resulted in the overproduction of these gene products as His6-tagged fusion proteins. The latter were purified by affinity chromatography on Ni-nitrilotriacetic acid resin (Qiagen, Valencia, CA), dialyzed against 25 mM Tris⋅HCl, pH 7.0 (for LnmG) or pH 8.0 (for ACPs or LnmP), 25 mM NaCl/10% glycerol/2 mM DTT, and stored at −80°C.
In Vitro Assay of LnmG with ACPs or PCP and [2-14C]Malonyl CoA.
LnmG-catalyzed loading of the malonyl group from malonyl CoA to ACPs or PCP was assayed in a two-step reaction. First, apo-ACPs or PCP was phosphopantetheinylated by Streptomyces verticillus phosphopatetheinyl transferase (Svp) with CoA (20, 21). A typical reaction of 75 μl contained 100 mM Tris·HCl, pH 7.5, 12.5 mM MgCl2, 2.5 mM DTT, 33.3 μM CoA, 10 μM ACP or PCP, and 2 μM Svp incubated at 25°C for 60 min. Second, a mixture of 2 μM LnmG and 10 μl of [2-14C]malonyl CoA [200 μM, 51 mCi/mmol (1 Ci = 37 GBq), Perkin–Elmer] in a 15-μl volume was added to each reaction. The reaction was incubated at 25°C and subsequently quenched by the addition of 900 μl of acetone at various time points. Proteins were precipitated by centrifugation at 4°C for 30 min after being frozen at −80°C for at least 1 h. The protein pellet was redissolved in 1× sampler buffer and separated on 4–15% SDS/PAGE gels (Bio-Rad). The resolved gels were visualized by Coomassie blue staining and phosphorimaging (LE phosphor screen, Amersham Pharmacia).
HPLC and Electrospray Ionization MS (ESI-MS) Analyses of Apo-, Holo-, and Malonyl-ACPs.
For HPLC preparation of apo-, holo-, and malonyl-ACPs, the phosphopantetheinylation reaction or complete loading reaction was scaled up three times, and cold malonyl CoA was used instead. The second step of loading the malonyl group to holo-ACP proceeded for 10 min at 25°C. HPLC analysis was carried out on a Jupiter C-18 column (5 μm, 300 Å, 250 × 4.6 mm, Phenomenex, Belmont, CA), eluted with a gradient from 85% buffer A (H2O + 0.1% CF3CO2H) to 90% buffer B (CH3CN + 0.1% CF3CO2H) in 25 min at a flow rate of 1 ml/min, and UV detection was at 220 nm to separate the proteins in the reaction mixture. Individual protein peaks were collected, lyophilized, and redissolved in H2O for ESI-MS analyses. The latter was performed on an Agilent (Palo Alto, CA) 1000 HPLC-MSD SL instrument.
LNM Yield Improvement by Overexpressing lnmG in S. atroolivaceus SB3004.
To construct the lnmG overexpression constructs, a 450-bp EcoRI–SacI fragment that harbors the ErmE* fragment (18) and a 2,883-bp SacI–BglII fragment of the lnmG gene were cloned into pBS3030 [a low-copy-number vector derived from the SCP2* origin of replicon (18)] and pBS3031 [a high-copy-number vector derived from the pIJ101 origin of replicon (18)] vectors, respectively, to yield pBS3032 and pBS3033. The latter was introduced into S. atroolivaceus SB3004 by conjugation, and the resultant SB3005 and SB3006 strains that harbor pBS3032 and pBS3033, respectively, were cultured and analyzed by HPLC for LNM production with the S. atroolivaceus S-140 wild-type strain as a control (17).
Results and Discussion
LnmI and LnmJ are “AT-less” PKSs That Are Essential for LNM Biosynthesis.
We recently cloned and sequenced the lnm biosynthetic gene cluster from S. atroolivaceus S-140 and confirmed its involvement in LNM biosynthesis by genetic and biochemical characterization of the lnmGHI locus (17). We subsequently established the lnm gene cluster boundaries by sequential inactivation of ORFs from both ends of the sequenced 172-kb DNA region, defining the lnm gene cluster to be comprised of 27 ORFs (unpublished data). Among the genes identified from the lnm cluster are lnmG, lnmI, lnmJ, lnmQ, and lnmP that together encode the Lnm hybrid NRPS–PKS megasynthetase (Fig. 1).
LnmQ and LnmP are NRPS adenylation enzyme (22) and PCP (23), respectively, constituting the loading module. LnmI contains the previously characterized thiazole-forming NRPS module (17) as well as PKS module 3 and the KS domain of PKS module 4. LnmJ harbors PKS modules 4–8 plus a thioesterase domain. The Lnm megasynthetase-templated synthesis of 1 could be envisaged to begin at LnmQ and end with the cyclization of the full-length linear peptide–polyketide intermediate by the thioesterase domain of LnmJ to yield a macrolactam intermediate such as 2 (Fig. 1). Although it remains unclear what the origin of the 1,3-dioxo-1,2-dithiolane is and how it is spirofused to the 18-membered macrolactam ring, subsequent modification of 2 by the action of post-PKS enzymes could be envisaged to furnish 1 (Fig. 1).
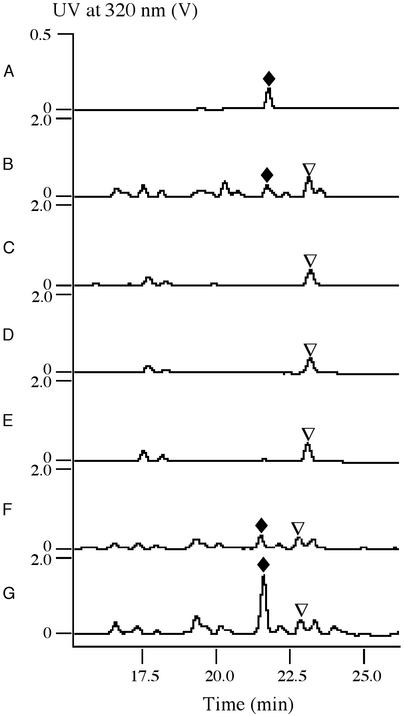
The deduced Lnm NRPS and PKS functions are consistent with what would be required for the biosynthesis of 1 from the amino acid and acyl CoA precursors. However, the Lnm hybrid NRPS–PKS megasynthetase is characterized by several intriguing features, and most strikingly, it lacks the cognate AT domain from all six PKS modules, i.e., the LnmJ and LnmI are AT-less type I PKSs (Fig. 1). To investigate the role of lnmI and lnmJ in vivo, we replaced lnmI and lnmJ with mutant copies in which domains of NRPS module 2 and PKS module 3 (for lnmI) and of PKS modules 7 and 8 (for lnmJ) were substituted with the apramycin-resistance gene, aac(3)IV, respectively. The resultant S. atroolivaceus SB3002 and SB3003 mutant strains lost its ability to produce 1 (Fig. 2 C and D), confirming that lnmI and lnmJ are essential for 1 production.
Figure 2.
HPLC analysis of LNM production by S. atroolivaceus wild-type and recombinant strains. (A) LNM standard. (B) S-140. (C) SB3002 (ΔlnmI). (D) SB3003 (ΔlnmJ). (E) SB3004 (ΔlnmG). (F) SB3005 (SB3004 harboring the lnmG overexpression plasmid of pBS3032). (G) SB3006 (SB3004 harboring the lnmG overexpression plasmid of pBS3033). ♦, LNM; ▿, an unknown metabolite with production that is independent to LNM biosynthesis.
LnmG Is a Discrete AT Protein That Is Essential for LNM Biosynthesis.
Lack of AT domain in all six PKS modules raises the question of how the LnmI and LnmJ PKSs are charged with the extender unit malonyl CoA for the biosynthesis of 1. To search for the missing AT activity, we reexamined genes within the lnm cluster and identified lnmG, the deduced product of which (the N-terminal half) showed high sequence homology to AT domains (3). We inactivated lnmG by replacing it with a mutant copy in which lnmG was disrupted by aac(3)IV. The resultant S. atroolivaceus SB3004 mutant lost its ability to produce 1, confirming that lnmG is essential for 1 production (Fig. 2E). These results inspired us to propose that LnmG provides the AT activity in trans to LnmI and LnmJ and loads the malonyl CoA extender unit to all ACP domains of the six PKS modules for 1 biosynthesis (Fig. 1).
LnmG Loads Malonyl CoA in Trans to All Six PKS Modules of LnmI and LnmJ.
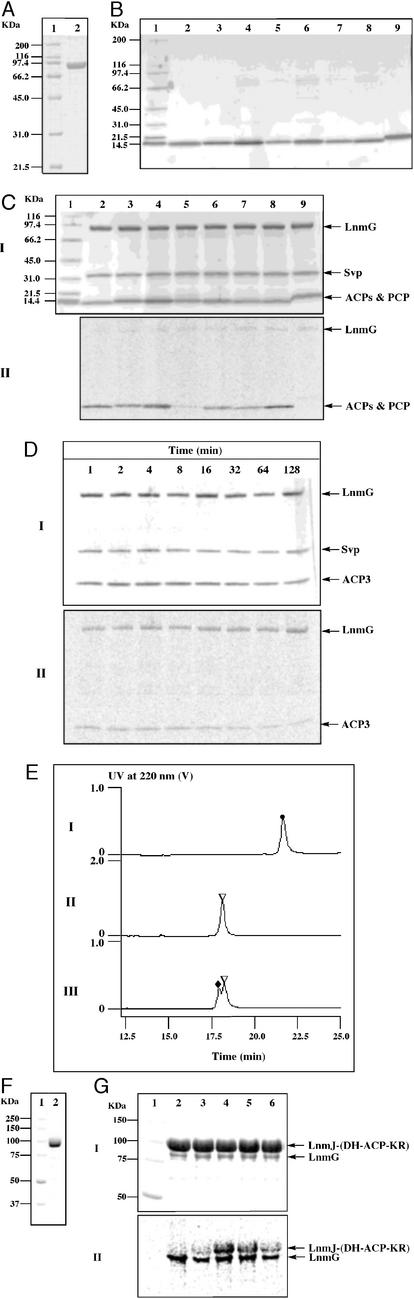
To validate this hypothesis, we expressed both lnmG and the seven ACP domains from the six PKS modules encoded by lnmI and lnmJ as well as lnmP in E. coli and purified the resultant LnmG (Fig. 3A), ACPs, and LnmP PCP (Fig. 3B) as His6-tagged fusion proteins to homogeneity. [We included LnmP, a discrete PCP identified within the lnm gene cluster (17), as a negative control to demonstrate that LnmG discriminates the LnmI and LnmJ PKS ACPs from other carrier proteins.] Because most ACPs or PCPs overproduced in E. coli are in the nonfunctional apo forms, we incubated them with CoA and the Svp phosphopantetheinyl transferase to ensure that all carrier proteins are converted into the functional holo forms (20, 21). We incubated the holo-carrier proteins with [2-14C]malonyl CoA and LnmG to directly test malonyl CoA extender-unit loading. The reaction mixtures were subjected to (i) SDS/PAGE and phosphorimaging to detect specific loading of the [2-14C]malonyl group to the phosphopantetheinyl group of ACPs (Fig. 3 C and D) and (ii) HPLC (Fig. 3E) and ESI-MS analyses to confirm the predicted molecular species (Table 1).
Figure 3.
In vitro assays of LnmG-catalyzed loading of malonyl CoA to individual LnmIJ PKS ACP domains and the LnmJ-(DH-ACP-KR) tridomain protein. (A) Purified LnmG on 4–15% SDS/PAGE. Lane 1, molecular mass standards; lane 2, LnmG. (B) Purified LnmIJ ACPs and LnmP on 4–15% SDS/PAGE. Lane 1, molecular mass standards; lane 2, ACP3; lane 3, ACP4; lane 4, ACP5; lane 5, ACP6-1; lane 6, ACP6-2; lane 7, ACP7; lane 8, ACP8; lane 9, LnmP. The numbers after the ACPs refer to the PKS modules from which they are derived with 6-1 and 6-2 to indicate the first and second ACPs, respectively, for PKS module 6. (C) Incubation of holo-ACPs or PCP with [2-14C]malonyl CoA and LnmG as visualized on 4–15% SDS/PAGE (I) and by phosphorimaging (II). Lane 1, molecular mass standards; lanes 2–8; ACP3–ACP8; lane 9, LnmP. (D) Time course of LnmG-catalyzed loading of [2-14C]malonyl CoA to ACP3 as visualized on 4–15% SDS/PAGE (I) and by phosphorimaging (II). (E) HPLC analysis of LnmG-catalyzed loading of malonyl CoA to ACP3. I, a negative control in the absence of Svp; II, a negative control in the absence of LnmG; III, complete assay. ●, apo-ACP3; ▿, holo-ACP3; ♦, malonyl-S-ACP3. (F) Purified LnmJ-(DH-ACP-KR) on 9% SDS/PAGE. Lane 1, molecular mass standards; lane 2, LnmJ-(DH-ACP-KR). (G) Incubation of holo-LnmJ-(DH-ACP-KR) with [2-14C]malonyl CoA and LnmG as visualized on 9% SDS/PAGE (I) and by phosphorimaging (II). Lane 1, molecular mass standards; lane 2, a negative control in the absence of Svp; lanes 3–6, complete assay with incubation times of 2, 5, 15, and 60 min, respectively.
Table 1.
ESI-MS analysis of apo-, holo-, and malonyl-S-ACPs
| ACPs* | apo-ACP [M + H]+
|
holo-ACP [M + H]+
|
Malonyl-S-ACP [M + H]+
|
|||
|---|---|---|---|---|---|---|
| Calcd. | Found | Calcd. | Found | Calcd. | Found | |
| LnmI-ACP3 | 11,702 | 11,700 | 12,042 | 12,040 | 12,128 | 12,126 |
| LnmJ-ACP4 | 12,245 | 12,241 | 12,585 | 12,582 | 12,671 | 12,669 |
| LnmJ-ACP5 | 12,520 | 12,517 | 12,860 | 12,857 | 12,946 | 12,943 |
| LnmJ-ACP6-1 | 12,209 | 12,206 | 12,549 | 12,546 | 12,635 | 12,632 |
| LnmJ-ACP6-2 | 12,151 | 12,147 | 12,491 | 12,486 | 12,577 | 12,572 |
| LnmJ-ACP-7 | 12,322 | 12,318 | 12,662 | 12,665 | 12,748 | 12,751 |
| LnmJ-ACP-8 | 12,090 | 12,087 | 12,430 | 12,427 | 12,516 | 12,512 |
The numbers after ACPs refer to the PKS modules from which they are derived with 6-1 and 6-2 to indicate the first and second ACPs, respectively, for PKS module 6.
LnmG specifically and efficiently catalyzes the loading of the malonyl CoA extender unit to the LnmI and LnmJ PKS ACPs, and no loading was observed in the absence of LnmG. After 5 min of incubation in the presence of LnmG and [2-14C]malonyl CoA, six of the seven ACPs were loaded efficiently with the malonyl group (Fig. 3C, lanes 2–4 and 6–8) with ACP6-1 being loaded less efficiently (Fig. 3C, lane 5), and no loading was observed for the LnmP PCP (Fig. 3C, lane 9). [The observation that ACP6-1 is loaded less efficiently is consistent with the finding that PKS module 6 has two ACP domains, one of which may be the preferred site for malonyl CoA loading (Fig. 1).] LnmG was also labeled by [2-14C]malonyl CoA (Fig. 3 C, D, and G). ATs are known to form acyl-O-enzyme intermediates at their active-site Ser residues before transferring the acyl groups from their CoA substrates to the nucleophilic recipients such as an ACP, and labeling of ATs by extender units has been confirmed for both the AT domain of type I PKS (24) and the malonyl CoA–ACP transacylase of type II PKS (25). Interestingly, the extent of ACP labeling decreased with longer incubation time, reaching a maximum in the first 5 min and falling to <10% after ≈2 h as exemplified by LnmI ACP3 (Fig. 3D). The malonyl-S-ACP product is apparently not stable under the assay condition, the malonyl group of which undergoes hydrolysis in the absence of chain elongation. Analogous results have been found for both the 6-deoxyerythronolide B synthase (24) and fatty acid synthase (26). In contrast, LnmG labeling appeared to be constant (Fig. 3D), suggesting that the malonyl-O-LnmG species is stable under the assay condition. To exclude any ambiguity associated with these assays, we subsequently purified the apo-, holo-, and malonyl-S-ACP species from the assay mixtures by HPLC, as exemplified by the LnmI–ACP3 (Fig. 3E), and established their identities by ESI-MS analysis. As summarized in Table 1, distinct [M + H]+ ions at m/z values exact or near the calculated values of the corresponding ACP species were observed for all samples analyzed.
Finally, we expressed the PKS module 4 as a tridomain protein, LnmJ-(DH-ACP-KR), in E. coli and purified it as a His6-tagged fusion protein (Fig. 3F). LnmJ-(DH-ACP-KR) was similarly assayed in the presence of LnmG and [2-14C]malonyl CoA to confirm that LnmG can load the malonyl group to a multidomain PKS module as efficiently as to individual ACP domains. As summarized in Fig. 3G, LnmG efficiently loaded the malonyl group to holo-LnmJ-(DH-ACP-KR) (lanes 3–6); the extent of [2-14C]malonyl labeling reached a maximum in 5 min (lane 4) and decreased with longer incubation time as a result of hydrolysis (lanes 4–6), and no loading was observed with the apo-LnmJ-(DH-ACP-KR) protein (lane 2).
The Interaction Among LnmG, LnmI, and LnmJ Represents a Previously Uncharacterized PKS Architecture.
Among type I PKSs characterized to date, each module minimally contains the KS, AT, and ACP domains (2, 3). Limited structural studies suggested that deoxyerythronolide B synthase, the archetype of type I PKSs, forms a parallel homodimer, possibly a helical structure (27, 28). At the core of the helix is a tetrahedron formed by the KS and AT domains of each module with the ACP domain brought into close juxtaposition with the KS domain of the opposite subunit. The optional reductive domains form loops that protrude out from the central core while remaining within range of the phosphopantetheine arm of the adjacent ACP. This model, consistent with the recently solved crystal structure of the homodimer of the thioesterase domain of deoxyerythronolide B synthase (29), has important general implications for both the mechanism of enzyme catalysis and genetic engineering of type I PKSs.
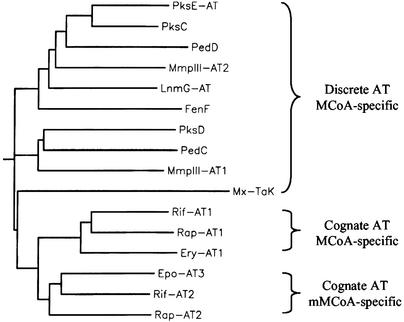
We now report that the LnmI and LnmJ are AT-less type I PKSs consisting minimally of the KS and ACP domains and that LnmG is a discrete AT enzyme that interacts with LnmI and LnmJ to form functional type I PKS modules by providing the AT activity in trans. These findings suggest an alternative model for type I PKS in which the KS and ACP domains of each module could minimally constitute the core structure. Although AT-less PKS is very rare and the recently reported putative pederin cluster was the only other example to our knowledge that lacks the cognate AT domain from all modules of a type I PKS (30), individual modules that lack the AT domain have been noted in a few other type I PKS or hybrid NRPS–PKS systems (31–35). The AT activity in all these AT-less type I PKS clusters remains unknown. We propose that these AT-less PKS clusters represent a previously unknown architecture for type I PKSs, which is characterized by discrete, iteratively acting AT(s), that loads the extender units in trans to AT-less type I PKS(s). Inspired by this model, we performed sequence analysis of all AT-less type I PKS clusters known to date to search for the discrete AT enzymes. Preliminary phylogenetic analysis indeed has led to the identification of LnmG homologs from these clusters (Fig. 4), which, in a mechanistic analogy to LnmG, could provide the AT activity in trans to the AT-less PKS modules for polyketide biosynthesis.
Figure 4.
Phylogenetic analysis of LnmG AT and its homologs from other AT-less type I PKS clusters and their relationships to cognate ATs from type I PKS clusters. The LnmG AT and its homologs fall into distinct groups that differ from cognate ATs of type I PKSs. Multiple sequence alignment and phylogenetic analysis were performed by the GCG program. We predict that PksC/PksD/PksE-AT [for the unknown polyketide in Bacillus subtilis (33)], PedC/PedD [for pederin in a bacterial symbiont of Paederus beetles (30)], MmpIII-AT1/AT2 [for mupirocin in Pseudomonas fluorescens (GenBank accession no. AF318063)], FenF [for mycosubtilin in B. subtilis ATCC6633 (31)], and Mx-TaK [for Ta1 in Myxococcus xanthus (32)], acting in a mechanistic analogy to LnmG, load the malonyl CoA extender unit onto the AT-less PKS modules in trans for polyketide biosynthesis in these clusters. For cognate ATs from rifamycin (Rif), rapamycin (Rap), erythromycin (Ery), and epothilone (Epo) clusters, protein GenBank accession numbers are given after the protein names: Rif-AT4 and Rif-AT2, AF04570; Rap-AT1 and Rap-AT2, X86780; Ery-AT1, Q03131; Epo-AT3, AF217189. MCoA, malonyl CoA; mMCoA, methyl malonyl CoA.
The AT-Less Type I PKS Architecture Provides Opportunities for PKS Engineering.
The model of AT-less type I PKS reported here provides opportunities for PKS engineering. For example, given the mechanism that LnmG is responsible for the loading of the malonyl CoA extender unit to all six PKS modules of the Lnm hybrid NRPS–PKS megasynthetase, we reasoned that LnmG could be a rate-limiting factor for 1 biosynthesis. We therefore explored yield improvement for 1 by overexpressing lnmG under the constitutive ErmE* promoter in both low- and high-copy-number vectors in S. atroolivaceus SB3004. The resultant recombinant strains SB3005, harboring the low-copy-number expression construct pBS3032, and SB3006, harboring the high-copy-number expression construct pBS3033, produce 3- to 5-fold more 1 (Fig. 2 F and G) than the wild-type S-140 strain (Fig. 2B) as determined by HPLC analysis. We also envisage applying lnmG and its homologs to either type I PKSs or AT-less PKSs to alter the extender-unit specificity by combinatorial biosynthesis methods, further expanding the size and diversity of polyketide natural products.
Acknowledgments
We thank Kyowa Hakko Kogyo (Tokyo) for an authentic sample of LNM, the wild-type S. atroolivaceus S-140 strain, and assistance in sequencing the lnm gene cluster. B.S. is a recipient of National Science Foundation Career Award MCB9733938 and National Institutes of Health Independent Scientist Award AI51689. This work was supported in part by University of California BioSTAR Program Grant Bi099-10045 and KOSAN Biosciences (Hayward, CA).
Abbreviations
- PKS
polyketide synthase
- KS
β-ketoacyl synthase
- ACP
acyl carrier protein
- AT
acyltransferase
- LNM
leinamycin
- NRPS
nonribosomal peptide synthetase
- PCP
peptidyl carrier protein
- KR
ketoreductase
- Svp
Streptomyces verticillus phosphopantetheinyl transferase
- ESI-MS
electrospray ionization MS
Footnotes
References
- 1.O'Hagan D. Polyketides. Chichester, U.K.: Horwood; 1991. [Google Scholar]
- 2.Hopwood D A. Chem Rev (Washington, DC) 1997;97:2465–2497. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 3.Staunton J, Weissman K J. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 4.Shen B. Top Curr Chem. 2000;209:1–51. [Google Scholar]
- 5.McDaniel R, Ebert-Khosla S, Hopwood D A, Khosla C. Science. 1993;262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- 6.Bisang C, Long P F, Cortes J, Westcott J, Crosby J, Matharu A, Cox R J, Simpson T J, Staunton J, Leadlay P F. Nature. 1999;401:502–505. doi: 10.1038/46829. [DOI] [PubMed] [Google Scholar]
- 7.Funa N, Ohnishi Y, Fujii I, Shibuya M, Ebizuka Y, Horinouchi S. Nature. 1999;400:897–899. doi: 10.1038/23748. [DOI] [PubMed] [Google Scholar]
- 8.Moore B S, Hopke J N. Chembiochem. 2001;2:35–38. doi: 10.1002/1439-7633(20010105)2:1<35::AID-CBIC35>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 9.Cane D E, Walsh C T, Khosla C. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 10.Cane D E, Walsh C T. Chem Biol. 1999;6:R319–R325. doi: 10.1016/s1074-5521(00)80001-0. [DOI] [PubMed] [Google Scholar]
- 11.Du L, Shen B. Curr Opin Drug Discov Dev. 2001;4:215–228. [PubMed] [Google Scholar]
- 12.Walsh C T. Chembiochem. 2002;3:124–134. doi: 10.1002/1439-7633(20020301)3:2/3<124::AID-CBIC124>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 13.Hara M, Asano K, Kawamoto I, Takiguchi T, Katsumata S, Takahashi K, Nakano H. J Antibiot. 1989;42:1768–1774. doi: 10.7164/antibiotics.42.1768. [DOI] [PubMed] [Google Scholar]
- 14. Hirayama, N. & Matsuzawa, E. S. (1993) Chem. Lett., 1957–1958.
- 15.Kanda Y, Fukuyama T. J Am Chem Soc. 1993;115:8451–8452. [Google Scholar]
- 16.Nakano H, Tamaoki T. In: Harnessing Biotechnology for the 21st Century: Proceedings of the Ninth International Biotechnology Symposium and Exposition, Crystal City, Virginia. Ladisch M R, Bose A, editors. Washington, DC: Am. Chem. Soc.; 1992. pp. 72–75. [Google Scholar]
- 17.Cheng Y-Q, Tang G-L, Shen B. J Bacteriol. 2002;184:7013–7024. doi: 10.1128/JB.184.24.7013-7024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieser T, Bibb M J, Buttner M J, Chater K F, Hopwood D A. Practical Streptomyces Genetics. Norwich, U.K.: The John Innes Foundation; 2000. [Google Scholar]
- 19.Bierman M, Logan R, O'Brien K, Seno E T, Nagaraja R, Schoner B E. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 20.Walsh C T, Gehring A M, Weinreb P H, Quadri L E N, Flugel R S. Curr Opin Chem Biol. 1997;1:309–315. doi: 10.1016/s1367-5931(97)80067-1. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez C, Du L, Edwards D J, Toney M D, Shen B. Chem Biol. 2001;8:725–738. doi: 10.1016/s1074-5521(01)00047-3. [DOI] [PubMed] [Google Scholar]
- 22.Conti E, Stachelhaus T, Marahiel M A, Brick P. EMBO J. 1997;16:4147–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber T, Baumgartner R, Renner C, Marahiel M A, Holak T A. Structure Fold Des. 2000;8:407–418. doi: 10.1016/s0969-2126(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 24.Marsden A F A, Caffrey P, Aparicio J F, Loughran M S, Staunton J, Leadlay P F. Science. 1994;263:378–380. doi: 10.1126/science.8278811. [DOI] [PubMed] [Google Scholar]
- 25.Carreras C W, Khosla C. Biochemistry. 1998;37:2084–2088. doi: 10.1021/bi972919+. [DOI] [PubMed] [Google Scholar]
- 26.Wakli S J. Biochemistry. 1989;28:4523–4530. doi: 10.1021/bi00437a001. [DOI] [PubMed] [Google Scholar]
- 27.Staunton J, Caffrey P, Aparicio J F, Roberts G, Bethell S, Leadlay P F. Nat Struct Biol. 1996;3:188–192. doi: 10.1038/nsb0296-188. [DOI] [PubMed] [Google Scholar]
- 28.Gokhale R S, Lau J, Cane D E, Khosla C. Biochemistry. 1998;37:2524–2528. doi: 10.1021/bi971887n. [DOI] [PubMed] [Google Scholar]
- 29.Tsai S-C, Miercke L J W, Krucinski J, Gokhale R, Chen J C-H, Foster P G, Cane D E, Khosla C, Stroud R M. Proc Natl Acad Sci USA. 2001;99:14002–14007. doi: 10.1073/pnas.011399198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piel J. Proc Natl Acad Sci USA. 2002;98:14808–14813. [Google Scholar]
- 31.Duitman E H, Hamoen L W, Rembold M, Venema G, Seitz H, Saenger W, Bernhard F, Reinhardt R, Schmidt M, Ullrich C, et al. Proc Natl Acad Sci USA. 1999;96:13294–13299. doi: 10.1073/pnas.96.23.13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paitan Y, Alon G, Orr E, Ron E Z, Rosenberg E. J Mol Biol. 1999;286:465–474. doi: 10.1006/jmbi.1998.2478. [DOI] [PubMed] [Google Scholar]
- 33.Albertini A M, Caramori T, Scoffone F, Scotti C, Galizzi A. Microbiology. 1995;141:299–309. doi: 10.1099/13500872-141-2-299. [DOI] [PubMed] [Google Scholar]
- 34.Huang G, Zhang L, Birch R G. Microbiology. 2001;147:631–642. doi: 10.1099/00221287-147-3-631. [DOI] [PubMed] [Google Scholar]
- 35.Zhu G, LaGier M J, Stejskal F, Millership J J, Cai X, Keithly J S. Gene. 2002;298:79–89. doi: 10.1016/s0378-1119(02)00931-9. [DOI] [PubMed] [Google Scholar]