Abstract
The proliferation of most primary cells in culture is limited by replicative senescence and crisis, p53-dependent events. However, the regulation of p53 itself has not been defined. We find that deletion of the early growth response 1 (EGR1) transcription factor leads to a striking phenotype, including complete bypass of senescence and apparent immortal growth consistent with loss of a suppressor gene. EGR1-null mouse embryo fibroblasts (MEFs) exhibit decreased expression of p53, p21Cip1/Waf1, and other p53 “marker” proteins. Precrisis WT but not EGR1-null cells exhibit irradiation-induced arrest. WT MEFs that emerge from crisis exhibit a mutated p53 (sequence confirmed), colony formation, and tumorigenicity. In contrast, high-passage EGR1-null MEFs retain the WT p53 sequence but with much reduced expression, remain untransformed, and grow continuously. An EGR1-expressing retrovirus restores p53 expression and sencescence to EGR1-null but not p53-null MEFs or postcrisis WT cells. Taken together, the results establish EGR1 as a major regulator of cell senescence and previously undescribed upstream “gatekeeper” of the p53 tumor suppressor pathway.
Keywords: early growth response 1 gene|cancer|retrovirus|mouse embryo fibroblasts
The proliferative capacity of most primary cells in culture is limited by the induction of senescence. The senescent state depends on a number of pathways that together result in permanent cell-cycle blockade (1). In most rodent cells, induction of the tumor suppressor genes p53 and p19ARF are critical to the induction of senescence, as inactivation of either gene allows “bypass” of replicative senescence, leading to continuous growth (2, 3). The function of p53 is determined in part by p19ARF and by murine double minute-2 protein (MDM2). The study of senescence in cultured cells and its related aspects of lifespan extension and immortialization has become an experimental system of great value for understanding tumorigenesis (4). Senescent populations undergo “crisis” and deteriorate, however, rare primary mouse embryo fibroblasts (MEFs) that acquire “escape” the senescent state and mitotically expand. Most such postcrisis cells are hypotetraploid and contain mutant p53 alleles, whereas others are mutated at p19ARF and remain pseudodiploid (5). The potential importance of MDM2 in senescence is illustrated by amplification of MDM2 in lymphomas (6). Thus, the p53-MDM2-p19ARF pathway is critical for the induction of senescence. However, the upstream regulatory mechanisms controlling this pathway remain unclear.
Another factor that has been discussed as a tumor suppressor is the early growth response 1 (EGR1) transcription factor (7). EGR1 is a member of the immediate early gene family and regulates transcription of target genes through GC-rich elements. EGR1 is involved in the regulation of growth and differentiation (7). However, many human tumor cell lines express little or no EGR1 in contrast to their normal counterparts (8–10). Furthermore, EGR1 has been found to be decreased or undetectable in small cell lung and human breast tumors (11, 12) as well as in human gliomas (13). Taken together, these data suggest a potential role for EGR1 in tumor suppression.
The mechanism of growth suppression by EGR1 as well as EGR1-dependent pathways are incompletely understood. We show here that EGR1 deficiency leads to a complete bypass of replicative senescence and an apparent immortal growth of MEFs. This effect of EGR1 is found to depend on its ability to act as an upstream regulator of the p53 tumor suppressor pathway. Our results thereby establish EGR1 as a previously undescribed gatekeeper of p53-dependent growth regulatory mechanisms in replicative senescence and cell growth.
Materials and Methods
Cells, Cell Culture, and Irradiation Treatment.
MEFs were prepared as described (14) from 15- or 19-day-old embryos from EGR1 WT, EGR1-null, and EGR1 heterozygous (HTZ) mice kindly provided by J. Milbrandt, Washington University, St. Louis (15). The predicted genotype and expression properties of the MEFs derived from EGR1-null and HTZ mice were confirmed by PCR-based analysis of DNA and RNA and by Western analysis of protein expression. Genotyping of MEFs derived from mice generated by Charnay and coworkers (16) was performed as described.
The p53-null MEFs were a gift from P. Puri (The Salk Institute, San Diego) and originally derived by I. Hunton in the laboratory of J. Y. J. Wang (University of California, San Diego).
For growth (proliferation) curve determinations, cells were seeded into six-well tissue culture plates at 20,000 cells per well in DMEM (high glucose) supplemented with 10% FBS and 75 μg/ml hygromycin B. Cell numbers were determined on days 2, 4, 5, and 6 by using a Multisizer II Coulter counter equipped with a channel analyzer for exclusion of noncell counts.
For irradiation experiments (Fig. 4), freshly prepared WT MEFs were isolated and compared to EGR1-null cells. All cells were seeded into six-well tissue culture plates at 70,000 cells per well grown as above and irradiated with 7.5 Gy by using a Cs source. Cell numbers were determined on days 1, 3, and 5, after irradiation or 3 days after reseeding by direct cell counting (Coulter).
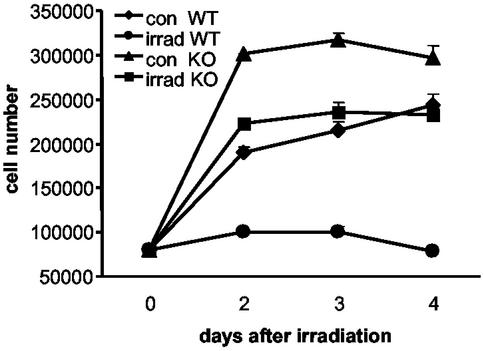
Figure 4.
DNA damage induced by irradiation leads to growth arrest of precrisis WT MEFs as compared to EGR1-null MEFs. Precrisis WT MEFs and EGR1-null MEFs were seeded into six-well tissue culture plates at 70,000 cells per well and were irradiated with 7.5 Gy. Cell numbers were determined by direct cell counting in duplicate on days 2, 3, and 4 after irradiation and 3 days after a subsequent reseeding (data not shown). Error bars indicate SDs (2σ) from the mean, and SDs with the symbol width are not visible. The experiment was replicated in triplicate with independent clones, which yielded very similar results.
Colony Formation Assay.
Cells were seeded into 6-cm diameter tissue culture dishes at 600, 900, or 1,200 cells per dish and grown as above. After 10 days of culture, the colonies were stained with 2% crystal violet, and cell numbers were determined in a parallel experiment.
Tumorigenicity Assay.
Six-week-old female athymic mice (Harlan–Sprague–Dawley) were placed in a pathogen-free environment. At the time of assay, postcrisis WT MEFs at passage 56 or EGR1-null MEFs at passage 62 were trypsinized, counted, washed twice in PBS, and resuspended in PBS 0.1% BSA at 7 × 106 cells/0.1 ml. The same volume of matrigel (Becton Dickinson) was added, and 7 × 106 cells (200 μl) were injected s.c. into each mouse on the dorsal-lateral surface. Mice were monitored for 16 wk for tumor formation.
Retroviral Vector Construction.
A retroviral vector, pLHC-EGR1, was prepared and used as described (10). The titer was monitored by Western analysis of producer cells and supernatants. Forty-eight hours after retrovirus infection, cells were cultured in medium containing 75 μg/ml hygromycin B. After 2 wk of selection, the hygromycin-resistant colonies were used for Western analysis to assess the expression of EGR1.
Quantitative Real-Time PCR (Q-PCR) and Western Analysis.
RNA expression levels were quantified by Q-PCR (Applied Biosystems 7900). Total RNA (0.5 μg) was reverse-transcribed into cDNA by using Superscript II RNase H− Reverse Transcriptase kit from Invitrogen. Q-PCR primer sequences were selected for each cDNA with the aid of primer express software (Applied Biosystems) and are available on request. Q-PCR and quantitative measurements were performed with the SYBR-Green PCR-Master Mix (Applied Biosystems) (Applied Biosystems 7700 user bulletin no. 2). The results were normalized to the relative amounts of GAPDH. For Western analysis, cells were lysed in radioimmunoprecipitation assay buffer with protease inhibitors as described (10), and the membranes were labeled with Abs specific for EGR1 (sc-189, Santa Cruz Biotechnology); p53 (Pab246; sc-100, Santa Cruz Biotechnology); p21Cip1/Waf1 (sc-397, Santa Cruz Biotechnology); EGR2 (sc-190, Santa Cruz Biotechnology); or actin (Sigma).
Immunoprecipitation.
Cells were lysed in 150 mM NaCl/50 mM Tris (pH 8.0)/5 mM sodium EDTA/0.5% Nonidet P-40 supplemented with protease inhibitors mixture and 2 μM lactacystin β-lactone on ice. Protein (400 μg) was precipitated overnight at 4°C by using either a monoclonal mouse-specific and conformation-dependent Ab (Pab 246; sc-100, Santa Cruz Biotechnology) that recognizes WT but not mutant p53 (17) or with a mAb (ab26; Pab 240, Abcam, Cambridge, U.K.) that recognizes many mutant p53s but not WT p53 protein in its native form (18). Precipitates were solubilized in denaturing sample buffer, electrophoretically separated, and transferred to Immobilon P membranes for detection with a polyclonal p53 Ab (sc-6243).
Sequencing.
RNA from pre- and postcrisis WT MEFs and high-passage EGR1-null MEFs was reverse transcribed into cDNA. PCR was performed by using the following primers for p53 (GenBank accession no. K01700): forward position, 420–438, 5′-ggcccctgtcatcttttgt-3′; reverse position, 1,164–1,183, 5′-attcagctcccggaacatct-3′. Sequence reactions of these PCR products were done by BATJ (San Diego).
Results
EGR1-Null MEFs Show Enhanced Cell Growth and Bypass Senescence.
The cell growth of MEFs derived from WT mice, EGR1 HTZ mice, and EGR1-null mice was monitored by cell number counting. In EGR1-null mice, expression of EGR1 is interrupted by the insertion of a neomycin resistance gene cassette upstream of the DNA-binding domain, which introduces in-frame stop codons (15). MEFs derived from the EGR1-null mouse strain established by Milbrandt and coworkers (15) exhibit high aberrant transcript levels corresponding to the altered EGR1 locus; however, no immunoreactive protein product is expressed (Fig. 3D), consistent with findings from Milbrandt and coworkers (15, 19). Normally, MEFs stop dividing and go into crisis after a characteristic number of passages as illustrated in Fig. 1. This so-called “replicative senescence” is p53-dependent (5).
Figure 3.
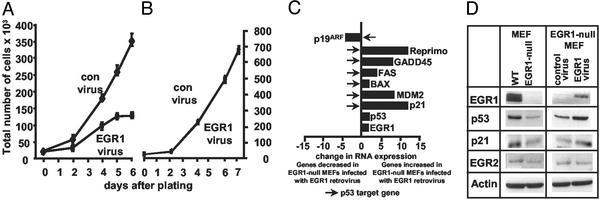
Reexpression of EGR1 in EGR1-null MEFs restores replicative senescence. (A) MEFs from EGR1-null mice generated by Milbrandt and coworkers (15) were infected with an EGR1-expressing or an empty vector control virus and cultured in medium containing 75 μg/ml hygromycin B. After 2 wk of selection, the hygromycin-resistant colonies were counted and then seeded into six-well tissue culture plates at 20,000 cells per well. Cell numbers were determined by direct cell counting on days 2, 4, 5, and 6. Data are the average of three different wells of cells. Error bars indicate SDs (2σ) from the mean. (B) p53-null MEFs were infected with an EGR1-expressing virus or an empty vector control virus and were cultured in medium containing 75 μg/ml hygromycin B. After 2 wk of selection, the hygromycin-resistant colonies were counted and then seeded into six-well tissue culture plates at 20,000 cells per well. Cell numbers were determined by direct cell counting on days 2, 4, 6, and 7. Data are the average of three different wells of cells. Error bars indicate SDs (2σ) from the mean. (C) RNA expression levels were quantified by Q-PCR by using the 7900 Sequence Detection system from Applied Biosystems. Total RNA (0.5 μg) from EGR1-null MEFs infected with an EGR1-expressing virus or an empty vector control virus was reverse-transcribed into cDNA and amplified by using the SYBR-Green PCR-Master Mix and specific primers for each cDNA. Relative amounts of each gene were calculated by reference to standard curves and were then normalized to the relative amounts of GAPDH as detect in the same run. The fold change in RNA expression from EGR1-null MEFs infected with an EGR1-expressing virus as compared to EGR1-null MEFs infected with an empty vector control virus is shown. Arrows indicate known p53 target genes. (D) WT MEFs, EGR1-null MEFs, EGR1-null MEFs infected with an EGR1-expressing virus, or EGR1-null MEFs infected with an empty vector control virus were scored for EGR1 (≈80 kDa), p53, p21inCip1/Waf1, and EGR2 (≈40 kDa) protein expression by Western analysis. Equivalent protein loading was confirmed by reexposing the same membranes to anti-β-actin Abs (≈48 kDa).
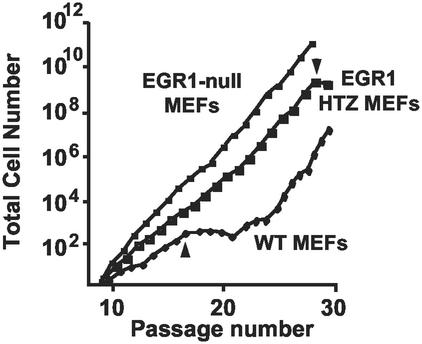
Figure 1.
EGR1-null MEFs bypass replicative senescence. MEFs from WT, EGR1 HTZ, or EGR1-null mice generated by Milbrandt and coworkers (15) were passaged every 3 or 4 days from passage 1 by counting the total number of cells and reseeding 3 × 105 cells per 60-mm dish. Accumulative cell numbers were calculated at each passage. Each growth curve is the average of two independent primary MEF isolates of the indicated genotype. Start of crisis is indicated by arrows; note that postcrisis cells are “WT MEFs” in origin only and exhibit non-WT sequences and phenotype (see text).
In our experiment, passaged WT MEFs initially underwent one population doubling in 3 days to become confluent, at which time they were harvested and reseeded. However, their growth virtually ceased by passage 17–20 (Fig. 1). After this senescent state, postcrisis “survivors” became established as permanent cell lines. In contrast, EGR1-null MEFs grew linearly for >60 passages appearing to bypass senescence. MEFs from EGR1 HTZ mice showed an intermediate growth rate, i.e., paused after 28 passages and resumed rapid growth (Fig. 1). The growth curves in Fig. 1 are the average of two independent primary MEF for each genotype. MEFs prepared from earlier embryos exhibited an essentially identical phenotype (not shown). In addition to the EGR1-null mice generated by Milbrandt and coworkers (15), we prepared and examined the growth of primary MEFs from EGR1-null mice developed by using a different plan by Charney and coworkers (16). In the MEFs derived from these EGR1-null mice, EGR1 RNA and protein levels were undetectable and they showed the same characteristics as the MEFs from Milbrandt and coworkers (not shown). In all, five complete sets of independent isolates were examined and varied only in the passage number of onset of the growth plateau for WT MEF and the prominence of the transiently reduced growth for HTZ MEFs.
These results, therefore, suggest the possibility that EGR1 is required in a gene-dose-dependent manner for the senescence response of WT MEFs observed in culture.
Expression Analysis by Using Q-PCR Reveals Decreased Expression of Several Regulators of Growth and Differentiation Such as p53, p21Cip1/Waf1 as Well as a Number of p53 Marker Proteins in EGR1-Null MEFs.
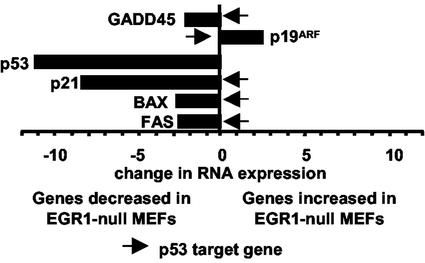
To identify EGR1-regulated genes, which may be responsible for the absence of a senescence state in EGR1-null MEFs, expression analyses were performed by using the mouse Affymetrix Gene Chip (unpublished data). Among the differentially expressed genes were the genes involved in growth differentiation and cell-cycle control. In this regard, transforming growth factor type β1 and p53 mRNA expression was decreased as well as the mRNA expression of a number of known p53 target genes, e.g., p21Cip1/Waf1 (20), GADD45 (21), Bax (22), and Fas (23) (unpublished data). We confirmed the decreased expression of these genes by Q-PCR (Fig. 2). Target gene expression results were supported by using independent RNA preparations from WT MEFs and EGR1-null MEFs and by using different primers and expression analysis methods (e.g., semiquantiative PCR), which included confirmation of the predicted product size by visualization on agarose gels. The sum of results indicate that deletion of EGR1 is associated with decreased expression of a number of established p53-regulated genes.
Figure 2.
Q-PCR reveals expression of genes differentially expressed between WT and EGR1-null MEFs. RNA expression levels were quantified by Q-PCR (ABI 7900). Total RNA (0.5 μg) from WT MEFs or EGR1-null MEFs was reverse-transcribed into cDNA and amplified by using the SYBR-Green PCR-Master Mix and specific primers for each cDNA as described (Materials and Methods). The relative amounts of each gene amplification products were calculated by reference to standard curves and were then normalized to the relative amounts of GAPDH as detected in the same run. The fold change in RNA expression from EGR1-null MEFs as compared to WT MEFs is shown. Known p53 target genes are indicated by arrows.
Reexpression of EGR1 in EGR1-Null MEFs by Retroviral Infection Restores Replicative Senescence.
To verify the EGR1 dependence of the senescence phenotype in murine MEFs, we performed reconstitution experiments by using an EGR1-expressing retrovirus system. When infected with an EGR1-expressing retrovirus, the cells became hygromycin-B resistant and showed increased steady-state protein levels of EGR1 (Fig. 3 C and D). Infection of EGR1-null MEFs with the EGR1-expressing retrovirus completely rescued the WT MEF phenotype (Fig. 3A). EGR1-infected cells were no longer able to bypass senescence and stopped growing 5 days after infection. In contrast, EGR1-null MEFs infected with an “empty vector” control virus became hygromycin-B resistant but did not stop growing and did not senesce (Fig. 3A). Similarly, WT MEFs infected with an EGR1-expressing retrovirus were not retarded in growth compared to MEFs infected with an empty vector control virus (not shown). Q-PCR demonstrated that a number of genes poorly expressed in EGR1-null MEFs became up-regulated after EGR1 virus infection (Fig. 3C). Among these genes is p53 as well as known marker genes of p53 transcriptional activity such as p21Cip1/Waf1 (20), Reprimo (24), GADD45 (21), MDM2 (25), Bax (22), and Fas (23). EGR1 itself was up-regulated twofold. In contrast, p19ARF, known to be negatively regulated by p53 (26), was down-regulated in these cells. The regulation of a variety of p53 target genes by EGR1 reexpression indicated that p53 might play an important role in the EGR1-mediated growth suppression.
To confirm these results, we also studied the effect of retroviral-mediated expression of EGR1 on p53 and p21Cip1/Waf1 protein levels (Fig. 3D). Western analysis demonstrated that p53 and p21Cip1/Waf1 protein expression was down-regulated in EGR1-null MEFs whereas the expression of both proteins was up-regulated in EGR1-null MEFs infected with an EGR1-expressing retrovirus (Fig. 3D). As expected, EGR1 was not detectable in EGR1-null MEFs but was restored in these cells after EGR1 retrovirus infection (Fig. 3D). No changes could be observed in the expression of EGR2. Overall, these results, confirm the observations that reexpression of EGR1 in EGR1-null MEFs leads to increased expression of p53 and p21Cip1/Waf1 at the RNA and protein levels.
Reexpression of EGR1 Does Not Restore Replicative Senescence in p53-Null MEFs.
Our experiments suggest that EGR1 might regulate the induction of the senescent state through the p53 tumor suppressor gene. To test this hypothesis directly, immortalized p53-null MEFs (2) were infected with an EGR1-expressing retrovirus. Stably infected primary, early passage p53-null cells isolated in the presence of hygromycin-B, exhibited 2-fold increased EGR1 protein expression compared to cells infected with an empty vector control virus (data not shown). However, the p53-null MEFs that were productively infected with the EGR1-expressing retrovirus were not able to undergo senescence and showed the same growth curve as the parental p53-null MEFs infected with an empty control virus (Fig. 3B). These results indicate that WT p53 is an important downstream intermediate of EGR1-dependent senescence and growth suppression.
DNA Damage Does Not Arrest EGR1-Null MEFs at a Dosage That Arrests WT MEFs.
To further confirm the observation that EGR1 is an upstream regulator of p53, precrisis MEFs (Materials and Methods) were seeded into six-well tissue culture plates and irradiated with 7.5 Gy to induce DNA damage. The subsequent proliferation of the irradiated cultures as well as nonirradiated controls was monitored by cell number counting in triplicate over the course of 5 days (Fig. 4). Inspection of the plates alone as well as cell counting revealed that WT cells exhibited a striking arrest in growth. These cultures remained very sparse with a high frequency of larger and flatter looking irregular cells. In contrast, the growth of irradiated Egr-1-null cells was brisk leading to a maximum density >3-fold than the irradiated WT cells (Fig. 4). Unirradiated WT cells also exhibited a rapid growth profile similar to irradiated Egr-1-null cells. In contrast, the growth of the irradiated WT cells is significantly reduced (P < 0.01) and the curve defines a broad plateau of little net growth over the 4-day postirradiation period. Moreover, when the cells are harvested and reseeded on day 5 at lower density, a common growth-stimulatory manipulation, irradiated EGR1-null cells resume growth whereas irradiated WT cells remain significantly arrested. Experiments with two independent MEF preparations lead to the same results (not shown). These results indicate that EGR1 is necessary to stimulate growth arrest after DNA damage and therefore further support that EGR1 is an upstream regulator of p53.
Inactivation of p53 Enhances Colony Formation in Postcrisis (High-Passage) WT MEFs Compared to Precrisis WT MEFs and High-Passage EGR1-Null MEFs.
Our results suggest that enhanced unlimited growth of murine MEFs predominantly is due to the absence of intact EGR1 and its effect on the p53 tumor suppressor pathway. However, rare immortal WT MEFs can emerge. These cells invariably exhibit increased growth rate and ability to proliferate at low density because of mutations of the p53-MDM2-p19ARF pathway (5, 27). However, if the role of p53 in promoting senescence in fact depends on EGR1 as indicated here, EGR1-null cells would be expected to be spared any mutations in p53 and to be protected from transformation.
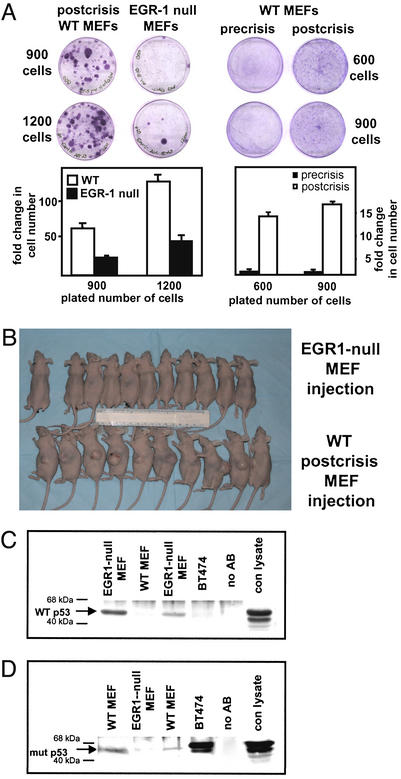
WT MEFs became senescent after ≈17 passages, and postcrisis survivors became established as permanent cell lines (Fig. 1). To determine whether these cells had become transformed, colony formation assays were performed. Precrisis WT MEFs, postcrisis WT MEFs, or EGR1-null MEFs were plated at low density (600, 900, and 1,200 cells per plate) and were grown for 10 days. Staining and colony counting revealed that postcrisis WT MEFs had a greater ability to proliferate at low densities and formed 10-fold more colonies when compared to either precrisis or EGR1-null MEFs (Fig. 5A). Similarly, in replicate experiments (Fig. 5A), colonies were harvested with trypsin, and in parallel cultures the disaggregated cells were counted, which confirmed the large increase in proliferation by the postcrisis MEFs. The t tests indicated significantly increased proliferation for all replicate experiments: P ≥ 0.01. To further assess transformation, 10 athymic mice were s.c. inoculated with postcrisis WT MEFs or EGR1-null MEFs. All mice inoculated with postcrisis cells developed tumors, whereas none of the 10 athymic mice inoculated s.c. with EGR1-null MEFs developed tumors. The difference is significant with P < 0.0001 (χ2) (Fig. 5B). This experiment showed that postcrisis WT MEFs are highly tumorigenic in concordance with the colony formation results whereas EGR1-null cells, which had been in culture considerably longer than postcrisis WT cells, were entirely unable to develop tumors.
Figure 5.
Inactivation of p53 leads to a transformed phenotype of postcrisis (high-passage) WT MEFs as compared to precrisis WT MEFs and high-passage EGR1-null MEFs. (A) For colony formation, precrisis WT MEFs, postcrisis WT MEFs, or EGR1-null MEFs were counted and then seeded into 6-cm diameter tissue culture dishes at 600, 900, or 1,200 cells. After 8 days of incubation at 37°C, the colonies were stained with 2% crystal violet and cell numbers in parallel plates were determined by cell harvesting and direct cell counting. (B) Ten 6-wk-old female athymic mice were injected with postcrisis WT MEFs at passage 56, ten 6-wk-old female athymic mice were injected with EGR1-null MEFs at passage 62, and one mouse was kept without injection. Mice were monitored for 16 wk for tumor formation. (C and D) Lysates from postcrisis WT MEFs and EGR1-null MEFs were analyzed to determine the relative expression of WT and mutant p53 by immunoprecipitation using a conformation-dependent Ab (Pab 246) that recognizes WT (C) but not mutant p53 or an Ab (Pab 240) recognizing mutant p53 (D) but not WT p53 protein in its native form. Precipitates were used for Western analysis with a polyclonal p53 Ab. Lysates from BT474 human breast carcinoma cells, which are known to express mutated p53 (45), were used as a positive control for Pab 240. Mock-immunoprecipitated samples (Ab only omitted) and Western transfers that were not exposed to Ab were used as controls.
Given the importance of p53 in the regulation of cell growth, we analyzed the p53 status in the different cell types by performing immunoprecipitation assays with two Abs that recognize either WT or mutant p53 proteins (17, 18). As shown in Fig. 5C, postcrisis WT MEFs contained no detectable WT p53 protein whereas this protein was readily precipitated from high-passage EGR1-null MEFs. However, cell lysates of postcrisis WT MEFs contained a readily immunoprecipitated mutated form of p53 whereas mutant p53 could not be detected in lysates from high-passage EGR1-null MEFs precipitated with an anti-mutant p53 Ab (Fig. 5D).
Sequencing of a 764-bp region of the p53 gene (GenBank accession no. K01700) in precrisis MEFs, two different clones of postcrisis WT MEFs and high-passage EGR1-null MEFs starting at position 420 to position 1,183, which is homologous to a region in the human p53 gene where most of the mutations occur, revealed point mutations at codon 211 in the postcrisis WT MEFs. One clone of postcrisis WT MEFs showed a C-to-G nucleotide exchange leading to serine-to-arginine change. The other clone of postcrisis WT MEFs showed a G-to-A nucleotide exchange leading to a serine-to-asparagine change. None of these mutations occurred in the precrisis WT MEFs or in the high-passage EGR1-null MEFs. Interestingly, the epitope of the Ab recognizing mutant p53 protein contains the codon 211 in which the mutation occurred (www.abcam.com/index.html?pageconfig=datasheet&intAbID=26), confirming the immunoprecipitation results (Fig. 5 C and D).
Thus, the results confirm that the transformed phenotype of postcrisis WT MEFs is exclusively associated with the absence of WT and presence of mutant p53. In contrast, in the precrisis state p53 appears to be normal. These results indicate that the reduced expression of functional p53 as observed here for the EGR1-null cells results in the unlimited and increased growth when compared to precrisis WT MEFs and indicates that Egr-1 is required for functional p53 expression and senescence.
Discussion
EGR1 Is a Growth Suppressor in Primary MEFs and Is Absolutely Required for Replicative Senescence.
In most human tumors such as breast cancer, fibrosarcoma, and glioblastoma EGR1 is described to be a tumor suppressor gene (8–10). Paradoxically, higher levels of EGR1 were found in prostate cancer (28–31) and are thought to play a role in tumorigenesis. Therefore, it is very important to understand the growth regulation mechanism of EGR1. We investigated the role of EGR1 by use of contrasting genetic backgrounds of primary MEFs from WT and EGR1-null mice (15). Primary MEFs derived from WT mice as observed here exhibited many of the hallmarks attributed to replicative senescence (5, 32), including cessation of growth at low passage and increased expression of p21Cip1/Waf1 followed by a marked decline in cell numbers and a deterioration of morphology. Cultures of WT MEFs that survive replicative senescence commonly exhibit mutations of p53 or, less frequently, genetic alterations of the major regulators of p53, p19ARF, and MDM2 (33). Indeed, mutation of p53 itself or amplification of MDM2 or deletion of p19ARF all tend to inactivate p53-dependent regulation and promote transformation. The significance is shown by the fact that one of these changes occurs in 75% of cancers (33). At passage numbers considerably beyond the passage number characteristic of replicative senescence, we observed that MEFs lacking EGR1 are protected from mutations of p53. These cells retain the WT p53 sequence and therefore do not exhibit characteristics of transformation such as colony formation and tumorigenicity in contrast to cells containing mutant p53 that survive crisis are transformed (Fig. 5 A and B). The reduced expression of p53 in EGR1-null cells results in the unlimited and increased growth, which is not observed in precrisis WT MEFs. The observations presented here are in concordance with recent studies of Sherr and coworkers (5). In the view of Sherr and coworkers, senescence of WT MEFs is a phenotype of the in vitro (experimental tissue culture) environment. This environment promotes DNA damage that activates p53 thereby promoting the growth arrest and replicative senescence. Escape from senescence requires alterations of the p53-MDM2-p19ARF pathway, leading to transformation of the formerly euploid cells (5).
Consistent with a critical role for the p53-MDM2-p19ARF pathway, it was shown recently that the transcriptional repressors BMI-1 and TBX-2 inhibit senescence through down-regulation of p19ARF expression (34, 35). Furthermore, disruption of DMP-1, a positive regulator of p19ARF also leads to the bypass of senescence (36). Similarly p19ARF-null MEFs are not able to undergo senescence (3), MEFs from p16Ink4a-deficient mice do undergo senescence (37). These studies further illustrate the role of the p53-MDM2-p19ARF pathway in the regulation of replicative senescence. In addition, protein levels of p21Cip1/Waf1, an important p53 target gene, are elevated in senescent human fibroblasts (38), and the p21Cip1/Waf1 gene was identified in a screening for senescence-inducing genes (39). Nevertheless, the question of whether p21Cip1/Waf1 is essential has yet to be unambiguously answered (40, 41).
EGR1 Is Required for the Function of p53.
Taken together, our observations indicate that EGR1 is required for senescence by MEFs. These results suggest that EGR1 functions by activating the p53-MDM2-p19ARF pathway. Moreover, p53 is essential for the role of EGR1 in effecting senescence. Therefore, we propose that EGR1 represents a previously undescribed upstream gatekeeper of the p53 tumor suppressor pathway activity and, thereby, has an important impact on cell growth and cell-cycle progression. This function of EGR1 may apply to human tumors as well. EGR1 protein was found to be highly suppressed in 21 of a series of 31 human gliomas when WT p53 was retained but nearly normally expressed in 10 cases with mutant p53, suggesting that expression of EGR1 is not required if p53 is inactivated (13).
The exact mechanism of EGR1-dependent regulation of p53 is unknown. However, it has been observed that EGR1 transactivates the p53 gene promoter (42, 43). Another potential regulatory interaction is suggested by Liu et al. (44) who identified a physical association between EGR1 and p53 in vitro and in vivo. It will be of interest, therefore, to examine whether these events are the basis of the gatekeeper function of EGR1 in cell cycle regulation.
Acknowledgments
We thank J. Milbrandt for EGR1-null mice, N. Mackman for EGR1-null mice generated by P. Charney, P. Puri for p53-null cells, I. Hunton for advice, R. Urcis for help with mouse work, C. Liu for the EGR1-expressing virus, and V. Baron and R. Gjerset for advice and critical reading of the manuscript. This work was supported in part by a fellowship from the Deutscher Akademischer Austauschdienst and a 2002 Scholar-in-Training award from the American Association for Cancer Research (to A.K.-H.), U.S. Public Health Service grants from the National Institutes of Health (CA76173 to D.M. and CA67888 to E.A.), and a Department of Defense California Breast Cancer Research Project grant (DAMD17-01-005 to E.A.).
Abbreviations
- EGR1
early growth response 1
- HTZ
heterozygous
- MEF
mouse embryo fibroblast
- Q-PCR
quantitative-real-time PCR
- MDM2
murine double minute-2 protein
References
- 1.Bringold F, Serrano M. Exp Gerontol. 2000;35:317–329. doi: 10.1016/s0531-5565(00)00083-8. [DOI] [PubMed] [Google Scholar]
- 2.Harvey M, Sands A T, Weiss R S, Hegi M E, Wiseman R W, Pantazis P, Giovanella B C, Tainsky M A, Bradley A, Donehower L A. Oncogene. 1993;8:2457–2467. [PubMed] [Google Scholar]
- 3.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 4.Schmitt C A, Fridman S F, Yang M, Lee S, Baranov E, Hoffman R M, Lowe S W. Cell. 2002;109:335–346. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 5.Sherr C J, DePinho R A. Cell. 2000;102:407–410. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- 6.Eischen C M, Weber J D, Roussel M F, Sherr C J, Cleveland J L. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, Calogero A, Ragona G, Adamson E, Mercola D. Crit Rev Oncog. 1996;7:101–125. [PubMed] [Google Scholar]
- 8.Calogero A, Cuomo L, D'Onofrio M, de Grazia U, Spinsanti P, Mercola D, Faggioni A, Frati L, Adamson E D, Ragona G. Oncogene. 1996;13:2105–2112. [PubMed] [Google Scholar]
- 9.Huang R P, Fan Y, de Belle I, Niemeyer C, Gottardis M M, Mercola D, Adamson E D. Int J Cancer. 1997;72:102–109. doi: 10.1002/(sici)1097-0215(19970703)72:1<102::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 10.Liu C, Yao J, Mercola D, Adamson E. J Biol Chem. 2000;275:20315–20323. doi: 10.1074/jbc.M909046199. [DOI] [PubMed] [Google Scholar]
- 11.Huang R P, Fan Y, Ni Z, Mercola D, Adamson E D. J Cell Biochem. 1997;66:489–499. [PubMed] [Google Scholar]
- 12.Levin W J, Press M F, Gaynor R B, Sukhatme V P, Boone T C, Reissmann P T, Figlin R A, Holmes E C, Souza L M, Slamon D J. Oncogene. 1995;11:1261–1269. [PubMed] [Google Scholar]
- 13.Calogero A, Arcella A, De Gregorio G, Porcellini A, Mercola D, Liu C, Lombari V, Zani M, Giannini G, Gagliardi F M, et al. Clin Cancer Res. 2001;7:2788–2796. [PubMed] [Google Scholar]
- 14.Robertson E J. In: Practical Approach Series. Robertson E J, editor. Oxford: IRL Press; 1987. pp. 71–112. [Google Scholar]
- 15.Lee S L, Tourtellotte L C, Wesselschmidt R L, Milbrandt J. J Biol Chem. 1995;270:9971–9977. doi: 10.1074/jbc.270.17.9971. [DOI] [PubMed] [Google Scholar]
- 16.Topilko P, Levi G, Merlo G, Mantero S, Desmarquet C, Mancardi G, Charnay P. J Neurosci Res. 1997;50:702–712. doi: 10.1002/(SICI)1097-4547(19971201)50:5<702::AID-JNR7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 17.Yewdell J W, Gannon J V, Lane D P. J Virol. 1986;59:444–452. doi: 10.1128/jvi.59.2.444-452.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gannon J V, Greaves R, Iggo R, Lane D P. EMBO J. 1990;9:1595–1602. doi: 10.1002/j.1460-2075.1990.tb08279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S L, Sadovsky Y, Swirnoff A H, Polish J A, Goda P, Gavrilina G, Milbrandt J. Science. 1996;273:1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- 20.el-Deiry W S, Harper J W, O'Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y, et al. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 21.Zhan Q, Fan S, Smith M L, Bae I, Yu K, Alamo I, Jr, O'Connor P M, Fornace A J., Jr DNA Cell Biol. 1996;15:805–815. doi: 10.1089/dna.1996.15.805. [DOI] [PubMed] [Google Scholar]
- 22.Miyashita T, Reed J C. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 23.Munsch D, Watanabe-Fukunaga R, Bourdon J C, Nagata S, May E, Yonish-Rouach E, Reisdorf P. J Biol Chem. 2000;275:3867–3872. doi: 10.1074/jbc.275.6.3867. [DOI] [PubMed] [Google Scholar]
- 24.Ohki R, Nemoto J, Murasawa H, Oda E, Inazawa J, Tanaka N, Taniguchi T. J Biol Chem. 2000;275:22627–22630. doi: 10.1074/jbc.C000235200. [DOI] [PubMed] [Google Scholar]
- 25.Barak Y, Juven T, Haffner R, Oren M. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherr C J. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 27.Todaro G J, Green H. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdulkadir S A, Qu Z, Garabedian E, Song S K, Peters T J, Svaren J, Carbone J M, Naughton C K, Catalona W J, Ackerman J J, et al. Nat Med. 2001;7:101–107. doi: 10.1038/83231. [DOI] [PubMed] [Google Scholar]
- 29.Svaren J, Ehrig T, Abdulkadir S A, Ehrengruber M U, Watson M A, Milbrandt J. J Biol Chem. 2000;275:38524–38531. doi: 10.1074/jbc.M005220200. [DOI] [PubMed] [Google Scholar]
- 30.Eid M A, Kumar M V, Iczkowski K A, Bostwick D G, Tindall D J. Cancer Res. 1998;58:2461–2468. [PubMed] [Google Scholar]
- 31.Thigpen A E, Cala K M, Guileyardo J M, Molberg K H, McConnell J D, Russell D W. J Urol. 1996;155:975–981. [PubMed] [Google Scholar]
- 32.Campisi J. In Vivo. 2000;14:183–188. [PubMed] [Google Scholar]
- 33.Sherr C J. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- 34.Jacobs J J, Kieboom K, Marino S, DePinho R A, van Lohuizen M. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 35.Jacobs J J, Keblusek P, Robanus-Maandag E, Kristel P, Lingbeek M, Nederlof P M, van Welsem T, van de Vijver M J, Koh E Y, Daley G Q, et al. Nat Genet. 2000;26:291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 36.Inoue K, Wen R, Rehg J E, Adachi M, Cleveland J L, Roussel M F, Sherr C J. Genes Dev. 2000;14:1797–1809. [PMC free article] [PubMed] [Google Scholar]
- 37.Krimpenfort P, Quon K C, Mooi W J, Loonstra A, Berns A. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 38.Tahara H, Sato E, Noda A, Ide T. Oncogene. 1995;10:835–840. [PubMed] [Google Scholar]
- 39.Noda A, Ning Y, Venable S F, Pereira-Smith O M, Smith J R. Exp Cell Res. 1994;211:90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 40.Brown J P, Wei W, Sedivy J M. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 41.Pantoja C, Serrano M. Oncogene. 1999;18:4974–4982. doi: 10.1038/sj.onc.1202880. [DOI] [PubMed] [Google Scholar]
- 42.Nair P, Muthukkumar S, Sells S F, Han S S, Sukhatme V P, Rangnekar V M. J Biol Chem. 1997;272:20131–20138. doi: 10.1074/jbc.272.32.20131. [DOI] [PubMed] [Google Scholar]
- 43.Das A, Chendil D, Dey S, Mohiuddin M, Milbrandt J, Rangnekar V M, Ahmed M M. J Biol Chem. 2001;276:3279–3286. doi: 10.1074/jbc.M008454200. [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Grogan L, Nau M M, Allegra C J, Chu E, Wright J J. Int J Oncol. 2001;18:863–870. doi: 10.3892/ijo.18.4.863. [DOI] [PubMed] [Google Scholar]
- 45.Elstner E, Linker-Israeli M, Said J, Umiel T, de Vos S, Shintaku I P, Heber D, Binderup L, Uskokovic M, Koeffler H P. Cancer Res. 1995;55:2822–2830. [PubMed] [Google Scholar]