Abstract
It has been proposed that the topoisomerase II (TOP2)β–DNA covalent complex arrests transcription and triggers 26S proteasome-mediated degradation of TOP2β. It is unclear whether the initial trigger for proteasomal degradation is due to DNA damage or transcriptional arrest. In the current study we show that the TOP2 catalytic inhibitor 4,4-(2,3-butanediyl)-bis(2,6-piperazinedione) (ICRF-193), which traps TOP2 into a circular clamp rather than the TOP2–DNA covalent complex, can also arrest transcription. Arrest of transcription, which is TOP2β-dependent, is accompanied by proteasomal degradation of TOP2β. Different from TOP2 poisons and other DNA-damaging agents, ICRF-193 did not induce proteasomal degradation of the large subunit of RNA polymerase II. These results suggest that proteasomal degradation of TOP2β induced by the TOP2–DNA covalent complex or the TOP2 circular clamp is due to transcriptional arrest but not DNA damage. By contrast, degradation of the large subunit of RNA polymerase II is due to a DNA-damage signal.
DNA topoisomerase (TOP)-mediated DNA damage (i.e., TOP–DNA covalent complexes) is an important form of DNA lesion that can be induced by xenobiotics (e.g., antibiotics and anticancer drugs), DNA structural perturbations (e.g., UV and carcinogen–DNA adducts), and physiological stresses (e.g., thiol stress and acidic pH) (1, 2). This form of DNA lesion is due to accumulation of the covalent intermediate of the TOP reaction, often referred to as the TOP cleavable or cleavage complex (1, 2). TOP-mediated DNA damage induced by anticancer drugs has been shown to be highly efficient in inducing tumor-cell death (3). Despite the importance of TOP-mediated DNA damage, the molecular mechanism(s) for repair of TOP-mediated DNA damage has been unclear. Recent studies have demonstrated that TOP–DNA covalent complexes induced by either the TOP1-specific poison camptothecin (CPT) or TOP2-specific poison 4′-demethylepipodophyllotoxin thenylidene-β-d-glucoside (VM-26) undergo 26S proteasome-mediated degradation referred to as TOP down-regulation (4, 5). It has been suggested that this down-regulation represents a cellular repair/stress response to TOP-mediated DNA damage.
Both TOP1 and TOP2 down-regulation induced by drugs that trap TOP covalent complexes depend on transcription but not new protein synthesis (4, 5). It has been suggested that TOP–DNA covalent complexes can arrest transcription and trigger TOP down-regulation (4, 5).
Surprisingly, transcription-dependent TOP2 degradation targets predominantly the TOP2β isozyme (4). Two TOP2 isozymes, TOP2α and TOP2β, are present in human cells and share ≈72% sequence identity (6–8). However, they are regulated very differently. The TOP2α protein level peaks at the G2/M phase and is elevated greatly in proliferating and tumor cells (9). TOP2α has been considered a cell-proliferating marker (10). Recently, chicken TOP2α, but not TOP2β, has been colocalized with sites of DNA replication (11). By contrast, the TOP2β protein level is not changed significantly during the cell cycle and is often present in both proliferating and differentiated cells (9). TOP2β has been mapped within the transcribed region of the rRNA-encoding DNA and shown to affect expression of certain neuronal genes during neuronal differentiation (12–14). It seems that TOP2α may play a more important role in DNA replication and chromosome condensation/segregation, whereas TOP2β may be more important for transcription. Preferential degradation of TOP2β induced by TOP2 poisons thus may be related to the role of TOP2β in transcription.
The initial trigger for TOP2β down-regulation is still unclear. One possibility is that TOP2β–DNA covalent complexes may induce a DNA-damage signal. Alternatively, arrest of the RNA polymerase (Pol) may signal a 26S proteasome pathway. To distinguish between these two possibilities, the TOP2 catalytic inhibitor 4,4-(2,3-butanediyl)-bis(2,6-piperazinedione) (ICRF-193), which does not trap TOP2–DNA covalent complexes but stabilizes ATP-bound TOP2 in the closed-clamp conformation (15), was used in the current study. We show that ICRF-193 can also arrest transcription and triggers proteasomal degradation of TOP2β. However, unlike TOP2 poisons and other DNA-damaging agents, ICRF-193 does not induce proteasomal degradation of the large subunit of Pol II.
Materials and Methods
Materials.
ICRF-193 was purchased from ICN. VM-26 was a gift from Bristol-Myers Squibb. 5,6-Dichlorobenzimidazole riboside (DRB), demethylepipodophyllotoxin ethylidene-β-d-glucoside (etoposide), CPT, aphidicolin, and cycloheximide were purchased from Sigma. Staphylococcus aureus S7 nuclease was purchased from Roche Molecular Biochemicals. The caspase inhibitor Z-Asp(OCH3)-Glu(OCH3)-Val-Asp(OCH3)-fluoromethyl ketone was purchased from Calbiochem. Rabbit sera against human (h)TOP2α and hTOP2β were prepared as described (4). Anti-actin and anti-p53 antibodies were purchased from Oncogene. ARNA-3 antibodies (Research Diagnostics, Flanders, NJ) directed against a non-carboxyl-terminal domain, unphosphorylated epitope (amino acids 806–820) were used to detect both Pol IIa (hypophosphorylated form) and Pol II0 (hyperphosphorylated form). hTOP2α and hTOP2β isozymes were purified from yeast by using the published procedure (16).
Cell Culture.
HeLa and human breast cancer ZR75-1 cells were cultured in a humidified atmosphere of 5% CO2 at 37°C in RPMI medium 1640 containing penicillin-streptomycin and 10% FBS. The HL-60 cell line and its mitoxantrone-resistant (TOP2-deficient) variant, HL-60/MX2, were obtained from the American Type Culture Collection (17). The simian virus 40-transformed TOP2β knockout mouse embryo fibroblast cell line TOP2β(−/−) and its parental simian virus 40-transformed TOP2β(+/+) cell line were cultured under the same condition (18).
Immunoblotting Analysis.
Treated cells were lysed either directly with SDS sample buffer or by an alkaline solution as described (4). For alkaline lysis, 100 μl of an alkaline lysis solution (200 mM NaOH/2 mM EDTA) was added to each sample. The lysate was neutralized by 8 μl of 1.2 M Tris (pH 8.0) and 8 μl of 2 M HCl. The neutralized lysate was mixed with 13.2 μl of 10× S7 nuclease reaction buffer (50 mM MgCl2/50 mM CaCl2/5 mM DTT/1 mM EDTA/50 μg/ml leupeptin/50 μg/ml aprotinin/50 μg/ml pepstatin A/1 mM PMSF) and 60 units of staphylococcal S7 nuclease (the staphylococcal S7 nuclease treatment was omitted in the band-depletion assay, which is used to monitor the amount of TOP–DNA covalent complexes) (19). After 20 min of ice nuclease digestion, 60 μl of 3× SDS sample buffer (150 mM Tris⋅HCl, pH 6.8/45% sucrose/6 mM EDTA/9% SDS/10% 2-mercaptoethanol/0.03% bromophenol blue) was added to each sample. The samples were subjected to SDS/PAGE and immunoblotted with anti-TOP antibodies.
Uridine Incorporation.
Cells were treated with different drugs for 30 min. 3H-labeled uridine (1 μCi/ml; 1 Ci = 37 GBq) then was added to the medium and cultured for 15 min. Cells were lysed with 100 μl of a lysis buffer (4 M guanidine isothiocyanate/0.5% sodium lauryl sarcosine/2 mM sodium acetate/0.1 M 2-mercaptoethanol). Twenty-microliter aliquots were spotted onto DE-81 paper. The dried paper was washed three times with 0.3 M ammonium formate (pH 7.8) and once with 90% ethanol. The dried paper was placed in a vial containing 4 ml of a scintillation fluid (EcoLite, ICN) and counted in a scintillation counter.
Nucleosomal DNA-Fragmentation Assay.
The nucleosomal DNA-fragmentation assay was performed as described (20).
In Vivo Complex of Enzyme (ICE) Assay.
The ICE assay was used to monitor the amount of covalent protein–DNA complexes in cells (21). Briefly, 1 × 107 HL-60 cells per sample were treated with different drugs for 30 min. Cells then were collected and lysed with 1 ml of 1% Sarkosyl or 6 M guanidine hydrochloride (Gdn⋅HCl). The lysates were passed through a 26-gauge needle five times. Two milliliters of each CsCl solution (1.82, 1.72, 1.50, and 1.37 g/ml) was layered successively in a polyallomer tube to generate the step CsCl gradient. The volume of the lysate was adjusted with 1% Sarkosyl or 6 M Gdn⋅HCl to 2 ml and layered gently on top of the gradient. The samples were centrifuged at 20°C in a Beckman SW41 rotor at 31,000 rpm for 24 h. Approximately 1 ml of each fraction was collected from the bottom of the tube. One hundred microliters of each fraction was diluted with 100 μl of 25 mM sodium-phosphate buffer (pH 6.5) and then spotted in a slot-blot apparatus onto the nitrocellulose membrane presoaked in sodium-phosphate buffer for 30 min. The membrane was washed with sodium-phosphate buffer and immunoblotted with anti-TOP2 antibodies.
Results
The TOP2 Catalytic Inhibitor ICRF-193 Induces Down-Regulation of hTOP2β.
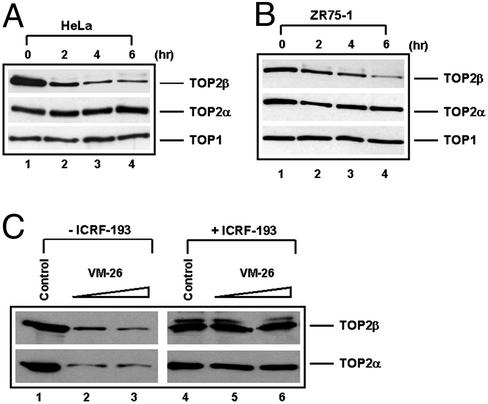
Earlier studies have demonstrated that the TOP2 poison VM-26, which traps TOP2–DNA covalent complexes, induces proteasomal degradation of TOP2 (4). To test whether TOP2 degradation is in response to a DNA-damage signal induced by VM-26, a TOP2 catalytic inhibitor, ICRF-193, that does not trap TOP2–DNA covalent complexes, was used in the current study. ICRF-193 traps TOP2 in its ATP-bound closed-clamp form by interfering with the ATPase activity of TOP2 and the reopening of the closed-clamp form (15). Treatment of HeLa cells (Fig. 1A) and the breast cancer ZR75-1 cells (Fig. 1B) with 100 μM ICRF-193 resulted in a time-dependent reduction of the total cellular TOP2β protein level. In both cell lines, 50% reduction of the TOP2β protein level occurred within 2 h. Degradation of TOP2β was shown to be specific because no significant reduction of the overall protein was observed (data not shown). In addition, the TOP1 protein level was essentially unchanged over the 6-h period (Fig. 1 A and B). The reduction of TOP2β in ICRF-193-treated cells was not related to covalent TOP2β–DNA complex formation because the lysates were treated with staphylococcal S7 nuclease to remove DNA. S7 nuclease treatment has been shown to completely release covalently bound TOP2 from covalent TOP2–DNA complexes (4). The reduction of TOP2β thus is likely to be the result of specific proteolytic degradation of TOP2β.
Figure 1.
ICRF-193 induces time-dependent reduction in the hTOP2β protein level in mammalian cells. HeLa (A) and ZR75-1 (B) cells were treated with 100 μM ICRF-193 for different times (0, 2, 4, and 6 h). Cells then were lysed with the alkaline lysis procedure with staphylococcal S7 nuclease treatment as described in Materials and Methods. Cell lysates were analyzed by immunoblotting with anti-hTOP2α, anti-hTOP2β, and anti-hTOP1 antibodies, respectively. (C) HL-60 cells were treated with VM-26 (25 and 50 μM) in the presence or absence of ICRF-193 (100 μM) for 30 min. Cells then were lysed with the alkaline lysis solution without staphylococcal S7 nuclease treatment. Cell lysates were analyzed by immunoblotting with anti-hTOP2α and anti-hTOP2β antibodies, respectively.
There was no significant change of the TOP2α protein level in either cell line treated with ICRF-193 (Fig. 1 A and B). The preferential degradation of TOP2β over TOP2α has been observed previously in cells treated with the TOP2 poison VM-26 (4). To test whether ICRF-193 preferentially inhibits TOP2β over TOP2α, the effect of ICRF-193 on TOP2–DNA covalent complexes was determined by the band-depletion assay. The antagonistic effect of ICRF-193 on TOP2 poisons-induced TOP2–DNA covalent complexes has been noted before (22). As shown in Fig. 1C, VM-26 trapped an equal amount of TOP2α and TOP2β covalent complexes as evidenced by depletion of the TOP2α and TOP2β protein bands, respectively. ICRF-193 (100 μM) was shown to reduce VM-26-induced TOP2α and TOP2β covalent complexes with about the same efficiency (Fig. 1C). This result suggests that preferential degradation of TOP2β over TOP2α is not due to preferential inhibition of TOP2β over TOP2α by ICRF-193.
26S Proteasome Is Involved in hTOP2β Down-Regulation.
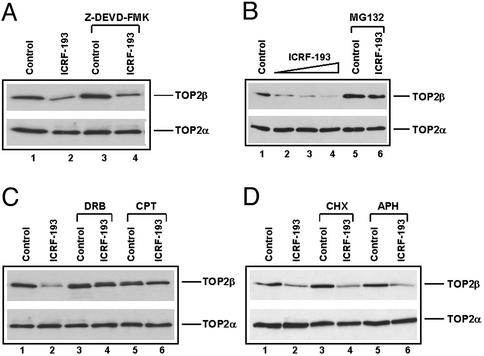
ICRF-193 is known to induce apoptotic cell death (23). Degradation of TOP2β could be due to the indirect result of cell death. To test this possibility, the caspase inhibitor Z-Asp(OCH3)-Glu(OCH3)-Val-Asp(OCH3)-fluoromethyl ketone (50 μM) was used to inhibit apoptosis. As shown in Fig. 2A, treatment with the caspase inhibitor had no effect on ICRF-193-induced TOP2β degradation, suggesting that apoptosis is not responsible for TOP2β degradation. However, treatment with the 26S proteasome inhibitor MG132 substantially abrogated ICRF-193-induced TOP2β degradation in HL-60 cells, suggesting the involvement of 26S proteasome in TOP2β degradation. It should be noted that ICRF-193 is highly efficient and specific in inducing TOP2β degradation in HL-60 cells. More than 50% TOP2β was degraded with as low as 1 μM ICRF-193 in 4 h, whereas TOP2α was essentially unchanged even with 100 μM ICRF-193 (Fig. 2B).
Figure 2.
Effect of caspase inhibitor and metabolic inhibitors on ICRF-193-induced down-regulation of hTOP2β. (A) HL-60 cells were treated with ICRF-193 (100 μM) in the presence or absence of the caspase inhibitor Z-Asp(OCH3)-Glu(OCH3)-Val-Asp(OCH3)-fluoromethyl ketone (Z-DEVD-FMK, 50 μM) for 4 h. (B) HL-60 cells were treated with ICRF-193 (0, 1, 10, and 100 μM, lanes 1–4, respectively), MG132 (10 μM, lane 5), and MG132 (10 μM) plus ICRF-193 (100 μM, lane 6) for 4 h. (C) HL-60 cells were treated with 100 μM ICRF-193 for 4 h in the presence and absence of 150 μM DRB or 25 μM CPT. (D) HL-60 cells were treated with 100 μM ICRF-193 for 4 h in the presence or absence of 50 μM cycloheximide (CHX) or 10 μM aphidicolin (APH). Cells were lysed by the alkaline lysis method, and cell lysates were analyzed by immunoblotting with anti-hTOP2α and anti-hTOP2β antibodies, respectively.
Transcription Is Involved in hTOP2β Down-Regulation.
Previous studies have demonstrated that VM-26-induced degradation of TOP2–DNA covalent complexes depends on transcription but not DNA or protein synthesis (4). It has been suggested that blockage of transcription is primarily responsible for proteasomal degradation of TOP2. To test whether ICRF-193-induced degradation of TOP2β also involves transcription, various metabolic inhibitors were used. As shown in Fig. 2C, inhibition of transcription by DRB (150 μM) substantially blocked TOP2β degradation. This result is confirmed with another potent transcription inhibitor, CPT (25 μM; see Fig. 2C; ref. 24). On the other hand, the DNA replication inhibitor aphidicolin did not affect ICRF-193-induced TOP2β degradation (Fig. 2D). Most interestingly, the protein-synthesis inhibitor cycloheximide had no effect on ICRF-193-induced TOP2β degradation, suggesting that the effect of transcription is not due to the expression of a gene product(s).
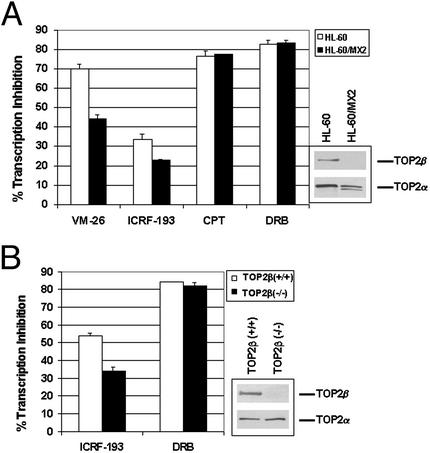
One possible explanation for the involvement of transcription in ICRF-193-induced TOP2β degradation is that the TOP2β circular protein clamp, similar to TOP2–DNA covalent complexes, blocks transcription elongation. To test this possibility, the effect of ICRF-193 on transcription was measured by 3H-labeled uridine incorporation in HL-60 and its TOP2-deficient HL-60/MX2 cells. HL-60/MX2 cells are known to express reduced nuclear TOP2α and undetectable TOP2β (ref. 17; also see Fig. 3A Inset). ICRF-193 (200 μM), similar to VM-26, inhibited uridine incorporation in HL-60 cells (Fig. 3A). Inhibition on transcription was significantly less in TOP2-deficient HL-60/MX2 cells (Fig. 3A), suggesting the involvement of TOP2 in transcription inhibition by ICRF-193 and VM-26. We also tested the effect of ICRF-193 on transcription in simian virus 40-transformed TOP2β(−/−) and TOP2β(+/+) cells. Again, ICRF-193 inhibited uridine incorporation more in TOP2β(+/+) (57.3 ± 1.7%) than in TOP2β(−/−) cells (34.0 ± 1.2%) (Fig. 3B), suggesting the involvement of TOP2β in transcription inhibition by ICRF-193. The reduced inhibitory effect of ICRF-193 on transcription in TOP2-deficient cell lines is not due to a nonspecific effect, because other transcription inhibitors such as DRB and CPT inhibited uridine incorporation to the same extent in both TOP2-deficient and -proficient cell lines (Fig. 3). These results are consistent with the notion that transcription inhibition is due to blockage of the Pol elongation complex by the TOP2 circular clamp (in the case of ICRF-193) or the TOP2–DNA covalent complex (in the case of VM-26). We noticed that ICRF-193 also inhibited transcription in TOP2β(−/−) cells, although to a lesser extent than that in TOP2β(+/+) cells, suggesting that some TOP2α circular clamps may also block transcription in TOP2β(−/−) cells. This surprising result could be explained, because TOP2α was found to be degraded in TOP2β(−/−) cells treated with ICRF-193 (data not shown). One possible explanation for this result is that TOP2α may partially substitute for TOP2β function in transcription in TOP2β(−/−) cells.
Figure 3.
ICRF-193 inhibits transcription. Transcription was monitored by 3H-labeled uridine incorporation. HL-60 and HL-60/MX2 cells (A) or mouse embryo fibroblast TOP2β(+/+) and TOP2β(−/−) cells (B) were treated with VM-26 (25 μM), ICRF-193 (200 μM), CPT (25 μM), or DRB (200 μM) for 30 min followed by a 15-min incubation with 3H-labeled uridine. The amount of uridine incorporated into RNA was determined by counting in a scintillation counter. The percentage of transcription inhibition was normalized to cells without drug treatment.
ICRF-193 Up-Regulates p53 and Induces Apoptosis.
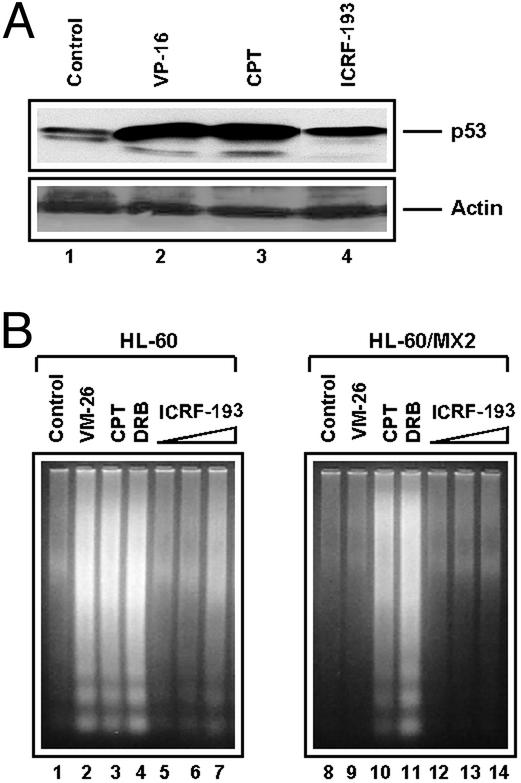
Transcriptional arrest caused by DNA adducts and other agents (e.g., DRB and α-amanitin) have been shown to up-regulate p53 and induce apoptosis (25, 26). As shown in Fig. 4A, ICRF-193, similar to demethylepipodophyllotoxin ethylidene-β-d-glucoside and CPT, up-regulated p53 in the breast cancer ZR75-1 cells. In addition, ICRF-193 also induced apoptosis in HL-60 cells as evidenced by the formation of nucleosomal DNA fragments (Fig. 4B Left). ICRF-193-induced apoptosis in HL-60 cells was shown to depend on the presence of TOP2, because no nucleosomal DNA fragments were observed in the TOP2-deficient HL-60/MX2 cells (Fig. 4B Right). VM-26 was used as a positive control, which induced apoptosis in HL-60 but not HL-60/MX2 cells. CPT and DRB were used as negative controls, which induced apoptosis in both cell lines (Fig. 4B). It is noted that although ICRF-193 is as efficient as VM-26 in inducing TOP2β down-regulation when compared at the same concentration, ICRF-193 is significantly less potent in up-regulation of p53 and induction of apoptosis.
Figure 4.
ICRF-193 up-regulates p53 and induces TOP2-dependent apoptosis. (A) The breast cancer ZR75-1 cells were treated with demethylepipodophyllotoxin ethylidene-β-d-glucoside (25 μM), CPT (25 μM), or ICRF-193 (100 μM) for 1 h. Treated cells were lysed directly with SDS sample buffer and prepared for immunoblotting with anti-p53 and anti-actin antibodies, respectively. (B) HL-60 cells and HL-60/MX2 cells (TOP2-deficient mutant cells) were treated with VM-26 (2.5 μM), CPT (2.5 μM), DRB (100 μM), or increasing concentrations of ICRF-193 (25, 100, and 400 μM) for 4 h. Cells then were lysed and processed for detection of the nucleosomal DNA ladders.
ICRF-193 Does Not Trap TOP2–DNA Covalent Complexes.
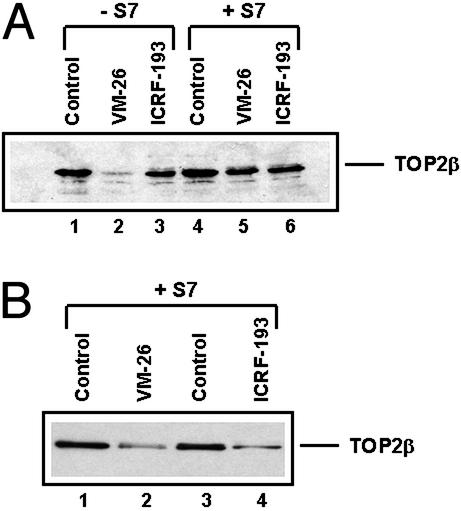
Earlier studies have demonstrated that ICRF-193 induces TOP2 circular clamps and does not trap a significant amount of TOP2–DNA covalent complexes (27). However, a more recent study has suggested that ICRF-193 can trap a substantial amount of covalent TOP2β (but not TOP2α)–DNA complexes if Gdn⋅HCl rather than SDS is used to terminate the reaction (28). Their results could explain why ICRF-193 induces preferential degradation of TOP2β, because only TOP2β can be trapped efficiently by ICRF-193 into covalent TOP2β–DNA complexes. We thus initiated the following studies to ascertain the effect of ICRF-193 on trapping of covalent TOP2–DNA complexes. As shown in Fig. 5A, ICRF-193 (200 μM, 30 min) did not trap any detectable amount of TOP2–DNA covalent complexes in HL-60 cells by using the alkaline lysis procedure (compare lanes 3 and 6). By contrast, VM-26 (100 μM, 30 min) trapped a significant amount (>60%) of TOP2β–DNA covalent complexes (Fig. 5A, compare lanes 2 and 5). The recovery of TOP2β in S7 nuclease-treated samples was ≈60% (Fig. 5A, compare lanes 5 and 6 with 4), most likely reflecting covalent modification of TOP2β by ubiquitin and other related proteins (4, 29). For comparison, TOP2β degradation in HL-60 cells treated with 100 μM VM-26 and 100 μM ICRF-193 for 4 h was shown to be approximately the same (Fig. 5B). These results suggest that covalent TOP2β–DNA complexes are unlikely to be the cause of ICRF-193-induced degradation of TOP2β.
Figure 5.
ICRF-193 does not induce TOP2–DNA covalent cleavable complexes in HL-60 cells. (A) HL-60 cells were treated with ICRF-193 (200 μM) or VM-26 (100 μM) for 30 min and lysed with the alkaline lysis procedure as described in Materials and Methods. Cell lysates were treated with (+S7) or without (−S7) staphylococcal nuclease S7. Treatment with the nuclease releases hTOP2β from covalent TOP2–DNA complexes, which migrate more slowly (not shown in the gel) than free hTOP2β. (B) Cells were treated with ICRF-193 (100 μM) or VM-26 (100 μM) for 4 h. Cells were lysed and processed for immunoblotting with anti-hTOPβ antibodies as described in the Fig. 2 legend.
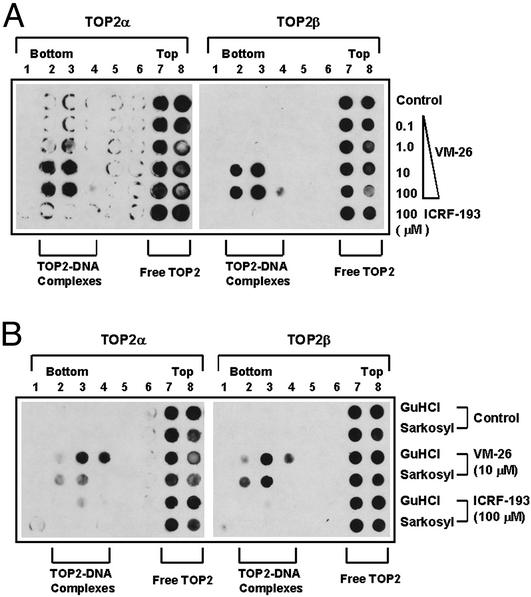
We have also measured the amount of covalent TOP2–DNA complexes using the ICE assay (21). As shown in Fig. 6A (bottom row), ICRF-193 (100 μM) did not induce any detectable amount of TOP2–DNA complexes when treated cells were lysed with 1% Sarkosyl. By contrast, VM-26 induced a dose-dependent increase in the amount of both covalent TOP2α–DNA and TOP2β–DNA complexes by using the same lysis procedure (Fig. 6A).
Figure 6.
ICRF-193 does not induce detectable amounts of covalent TOP2–DNA complexes as determined by the ICE assay. (A) HL-60 cells were treated with ICRF-193 (100 μM) and different concentrations of VM-26 for 30 min. Cells were collected and lysed with 1% Sarkosyl and loaded onto a preformed CsCl step gradient. Centrifugation and immunoblotting of the fractions were performed as described in Materials and Methods. Covalent TOP2–DNA complexes sedimented near the bottom of the gradient (see fractions marked “TOP2–DNA complexes”), whereas free TOP2 enzymes sedimented near the top of the gradient (see fractions marked “Free TOP2”). (B) HL-60 cells were treated with ICRF-193 (100 μM) and VM-26 (100 μM) for 30 min and lysed with either 1% Sarkosyl or 6 M Gdn⋅HCl (GuHCl). The ICE assay was performed as described for A.
In addition, we tested whether ICRF-193 could trap covalent TOP2β–DNA complexes in HL-60 cells by using 6 M Gdn⋅HCl as the termination/lysis solution. Using the ICE assay, we showed that ICRF-193 (100 μM) did not trap any detectable amount of covalent TOP2β–DNA complexes in HL-60 cells by using either Sarkosyl (1%) or Gdn⋅HCl (6 M) as the termination/lysis solution (Fig. 6B, bottom two rows). By contrast, a substantial amount of covalent TOP2α–DNA and TOP2β–DNA complexes was trapped by VM-26 (10 μM; Fig. 6B). We also tested the effect of Gdn⋅HCl on the trapping of ICRF-193-induced TOP2–DNA covalent complexes in the purified system using linear DNA and purified hTOP2 isozymes (30). Compared with SDS (1%) as the terminating agent, Gdn⋅HCl (0.4 M) was ineffective in trapping either TOP2 isozyme into covalent complexes (data not shown).
VM-26 but Not ICRF-193 Induces Proteasomal Degradation of Pol II.
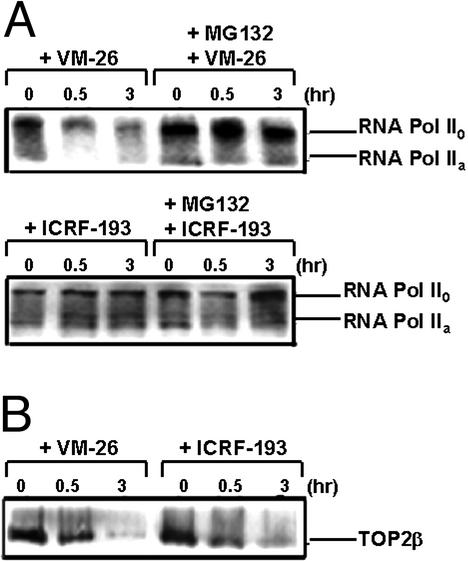
Many DNA-damaging agents have been shown to arrest transcription and trigger degradation of the large subunit of Pol II through the ubiquitin/26S proteasome pathway (31, 32). As shown in Fig. 7A, VM-26 (100 μM) induced degradation of the large subunit of Pol II in breast cancer ZR75-1 cells. Degradation of the large subunit of Pol II was shown to be inhibited by MG132, suggesting the involvement of 26S proteasome (Fig. 7A). By contrast, ICRF-193 (100 μM) had no effect on the large subunit of Pol II (Fig. 7A). On the other hand, both VM-26 and ICRF-193 were effective in inducing TOP2β degradation (Fig. 7B). We have also tested the effect of VM-26 and ICRF-193 on Pol II in Chinese hamster V79 cells, and the same results were obtained (data not shown). These results suggest that TOP2β degradation, unlike degradation of the large subunit of Pol II, is not the result of DNA damage.
Figure 7.
VM-26 but not ICRF-193 induces degradation of Pol II. Breast cancer ZR75-1 cells (106 cells per sample) were treated with ICRF-193 (100 μM) or VM-26 (100 μM) for 30 min and 3 h in the presence or absence of the 26S proteasome inhibitor MG132 (1 μM). Cells were then lysed with the alkaline lysis procedure and immunoblotted as described in Materials and Methods. (A) Degradation of the large subunit of Pol II. Pol IIa and Pol II0 were detected by immunoblotting with ARNA-3 antibodies. (B) Degradation of TOP2β. Cell lysates were analyzed by immunoblotting with anti-TOP2β antibodies.
Discussion
Using the prototypic TOP2 poison VM-26, we have shown that TOP2–DNA covalent complexes are degraded by 26S proteasome (4). VM-26-induced proteasomal degradation is transcription-dependent and predominantly targets TOP2β (4). One possible explanation is that the TOP2-concealed strand breaks may trigger TOP2 degradation (4). Results from our current study with the TOP2 catalytic inhibitor ICRF-193 argue against this possibility. ICRF-193 belongs to a different class of TOP2 inhibitors than TOP2 poisons. ICRF-193 interferes with ATP hydrolysis and traps TOP2 in the ATP-bound closed-clamp form (33). The fact that the TOP2β circular clamp induced by ICRF-193 can trigger transcription-dependent proteasomal degradation of TOP2β suggests that DNA strand breaks are not likely to be responsible for TOP2β degradation induced by either ICRF-193 or VM-26.
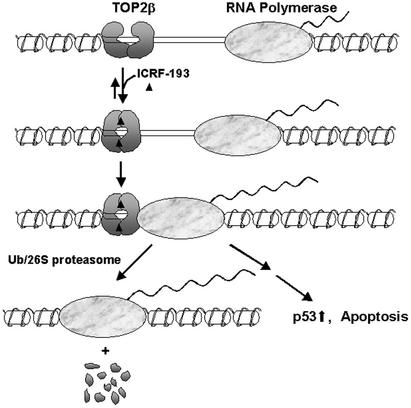
Another possibility is that the arrest of the Pol elongation complex could trigger TOP2β degradation. Our previous studies have shown that TOP2β down-regulation induced by VM-26 depends on transcription but not synthesis of new proteins (4). Our current studies with ICRF-193 have revealed a similar dependence of TOP2β degradation on transcription. In addition, we have shown that transcription inhibition by ICRF-193 is reduced significantly in TOP2β(−/−) cells. It seems that TOP2β down-regulation depends on both the process of transcription and the presence of the TOP2β circular clamp. A transcription collision model has been proposed previously for the interaction between the TOP1–DNA covalent complex and the Pol elongation complex (5, 34). In this model, TOP1–DNA covalent complexes on the template strands block Pol elongation complexes, which results in displacement (melting) of the 5′ hydroxyl-containing broken DNA strands from the cleavage sites and hence the formation of nonreversible TOP1-linked DNA breaks (5, 34). A similar transcription collision model has been proposed for the interaction between TOP2–DNA covalent complexes and the Pol elongation complex (4). It has been suggested that the transcriptional arrest by the TOP2–DNA covalent complexes signals proteasomal degradation of TOP2β (4). It seems plausible that the same transcription collision model could be applied to explain ICRF-193-induced TOP2β degradation (see Fig. 8 for a model). In this model, the TOP2 circular clamp, similar to the TOP2–DNA covalent complex, acts as a roadblock for the Pol elongation complex. Transcriptional arrest then signals the 26S proteasome pathway to degrade TOP2 (Fig. 8). In this case, blockage of the Pol elongation complex is due to the TOP2 circular clamp but not the covalent protein–DNA adduct.
Figure 8.
A proposed model for ICRF-193-induced degradation of TOP2β. In the presence of ICRF-193 and ATP, TOP2β is trapped as a closed circular clamp on DNA. This clamp blocks the movement of the transcription-elongation complex. Arrest of transcription triggers 26S proteasome-dependent degradation of TOP2β.
It is unclear how the TOP2 circular clamp can block transcription. The TOP2 circular clamp formed in the presence of DNA could be immobile, which, similar to TOP2 covalent complexes, can also function as a roadblock for the Pol. Alternatively, the TOP2β circular clamp may be a sliding clamp (30), which may impede the ability of Pol to negotiate with the nucleosome (35, 36). In either case, degradation of the TOP2β circular clamp could be the solution for recovery of the arrested Pol complex (see Fig. 8).
It is still unclear what the signal is for TOP2 degradation when the Pol elongation complex is arrested by the roadblock. It has been shown that the 19S regulatory subunit of 26S proteasome is involved in transcription elongation and is associated with the Pol elongation complex (37). The 19S regulatory subunit is known to recognize the polyubiquitin chain and contains multiple ATPases that can unfold proteins (38). The 19S regulatory subunit of the 26S proteasome could function to remove protein roadblocks for the Pol elongation complex.
We have noted that VM-26 but not ICRF-193 can also trigger proteasomal degradation of the large subunit of Pol II (Fig. 7). Proteasomal degradation of the large subunit of Pol II has been shown to occur in cells treated with various DNA-damaging agents (31, 32). It has been suggested that proteasomal degradation of the large subunit of Pol II is the result of transcriptional arrest of the Pol elongation complex by DNA adducts (31, 32). The fact that ICRF-193 arrests transcription without inducing degradation of the large subunit of Pol II suggests that transcriptional arrest, which signals TOP2β degradation, may not be sufficient by itself to signal degradation of the large subunit of Pol II. It seems plausible that a DNA-damage signal accompanied by transcriptional arrest at the site of DNA damage may signal for degradation of the large subunit of Pol II. The nature of this DNA-damage signal at the arrested sites remains to be identified. Regardless of the molecular basis for degradation of the large subunit of Pol II, the failure of ICRF-193 to induce its degradation is consistent with the notion that ICRF-193, unlike VM-26, does not induce DNA damage (i.e., TOP2–DNA covalent complexes) in cells.
The preferential degradation of TOP2β over TOP2α occurs in cells treated with either the TOP2 poison VM-26 or TOP2 inhibitor ICRF-193 (4). The mechanism for preferential TOP2β degradation is still unclear. There are at least three possibilities. One is that ICRF-193 preferentially inhibits TOP2β over TOP2α. This seems very unlikely, because we have shown that ICRF-193 inhibits the formation of both covalent TOP2α–DNA and TOP2β–DNA complexes to a similar extent in cells (Fig. 1C). In addition, both VM-26 and ICRF-193 induce preferential degradation of TOP2β. VM-26, similar to ICRF-193, does not preferentially inhibit TOP2β over TOP2α (4). Another possibility is that the ubiquitin-conjugating enzyme/ubiquitin-ligating enzyme for TOP2β is more active than the ubiquitin-conjugating enzyme/ubiquitin-ligating enzyme for TOP2α. Currently, this possibility cannot be ruled out. The third possibility is that TOP2β and TOP2α are differentially located within the cell and perform different functions. This possibility seems reasonable, because TOP2β has been located within the rRNA genes and has been implicated in neuronal differentiation by affecting the transcription of certain genes (12–14). Trapping of TOP2β within the transcribed regions by either VM-26 or ICRF-193 would result in blockage of the Pol elongation complex and thus activation of proteasomal degradation of TOP2β (Fig. 8).
ICRF-193 is known to kill yeast cells expressing hTOP2 and induces apoptosis in mammalian cells (23, 39). Studies in yeast have demonstrated that ICRF-193-induced killing is unrelated to inhibition of the catalytic activity of TOP2 but is consistent with trapping of TOP2 into potentially lethal complexes (39). Consistent with this notion, ICRF-193-induced apoptosis is much reduced in TOP2-deficient HL-60/MX2 cells as compared with HL-60 cells (Fig. 4B). We have noted that although ICRF-193 is as efficient as VM-26 in effecting TOP2β down-regulation, ICRF-193 is much less effective than VM-26 in inducing apoptosis. It seems plausible that apoptosis induced by VM-26 is primarily due to exposed DNA double-strand breaks (through TOPβ degradation), whereas that by ICRF-193 is due to transcriptional arrest (see Fig. 8). The two signaling pathways may be quite different. Although the signaling pathway associated with DNA double-strand breaks has been studied extensively, little is known about the signaling pathway associated with transcriptional arrest. In this regard, it is interesting to note that ICRF-193 also activates G2 checkpoint through ATM and Rad3-related/polo-like kinase 1 (40). It remains to be determined whether transcriptional arrest by the TOP2 circular clamp can activate ATR.
Transcriptional arrest by various DNA adducts is a frequent biological event. It has been shown that transcriptional arrest by DNA adducts or other agents signals p53 up-regulation and apoptosis (25, 26). However, DNA adducts can signal through both transcription arrest and DNA damage. ICRF-193 being a non-DNA-damaging agent may offer a unique tool for studying the signaling mechanism associated with transcription arrest.
Acknowledgments
This work was supported by National Institutes of Health Grants GM 27731 and CA 39662, National Science Council of Taiwan Grant NSC 89-2320-B001-075, and Academia Sinica.
Abbreviations
- TOP
topoisomerase
- CPT
camptothecin
- VM-26
4′-demethylepipodophyllotoxin thenylidene-β-d-glucoside (teniposide)
- Pol
RNA polymerase
- ICRF-193
4,4-(2,3-butanediyl)-bis(2,6-piperazinedione)
- DRB
5,6-dichlorobenzimidazole riboside
- h
human
- ICE
in vivo complex of enzyme
- Gdn⋅HCl
guanidine hydrochloride
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.D'Arpa P, Liu L F. Biochim Biophys Acta. 1989;989:163–177. doi: 10.1016/0304-419x(89)90041-3. [DOI] [PubMed] [Google Scholar]
- 2.Li T K, Liu L F. Annu Rev Pharmacol Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 3.Kaufmann S H. Biochim Biophys Acta. 1998;1400:195–211. doi: 10.1016/s0167-4781(98)00136-5. [DOI] [PubMed] [Google Scholar]
- 4.Mao Y, Desai S D, Ting C Y, Hwang J, Liu L F. J Biol Chem. 2001;276:40652–40658. doi: 10.1074/jbc.M104009200. [DOI] [PubMed] [Google Scholar]
- 5.Desai S D, Li T K, Rodriguez-Bauman A, Rubin E H, Liu L F. Cancer Res. 2001;61:5926–5932. [PubMed] [Google Scholar]
- 6.Tan K B, Dorman T E, Falls K M, Chung T D, Mirabelli C K, Crooke S T, Mao J. Cancer Res. 1992;52:231–234. [PubMed] [Google Scholar]
- 7.Drake F H, Hofmann G A, Bartus H F, Mattern M R, Crooke S T, Mirabelli C K. Biochemistry. 1989;28:8154–8160. doi: 10.1021/bi00446a029. [DOI] [PubMed] [Google Scholar]
- 8.Jenkins J R, Ayton P, Jones T, Davies S L, Simmons D L, Harris A L, Sheer D, Hickson I D. Nucleic Acids Res. 1992;20:5587–5592. doi: 10.1093/nar/20.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woessner R D, Mattern M R, Mirabelli C K, Johnson R K, Drake F H. Cell Growth Differ. 1991;2:209–214. [PubMed] [Google Scholar]
- 10.Bauman M E, Holden J A, Brown K A, Harker W G, Perkins S L. Mod Pathol. 1997;10:168–175. [PubMed] [Google Scholar]
- 11.Niimi A, Suka N, Harata M, Kikuchi A, Mizuno S. Chromosoma. 2001;110:102–114. doi: 10.1007/s004120100140. [DOI] [PubMed] [Google Scholar]
- 12.Govoni M, Neri S, Labella T, Sylvester J E, Novello F, Pession A. Biochem Biophys Res Commun. 1995;213:282–288. doi: 10.1006/bbrc.1995.2127. [DOI] [PubMed] [Google Scholar]
- 13.Yang X, Li W, Prescott E D, Burden S J, Wang J C. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 14.Tsutsui K, Tsutsui K, Sano K, Kikuchi A, Tokunaga A. J Biol Chem. 2001;276:5769–5778. doi: 10.1074/jbc.M008517200. [DOI] [PubMed] [Google Scholar]
- 15.Roca J, Ishida R, Berger J M, Andoh T, Wang J C. Proc Natl Acad Sci USA. 1994;91:1781–1785. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wasserman R A, Austin C A, Fisher L M, Wang J C. Cancer Res. 1993;53:3591–3596. [PubMed] [Google Scholar]
- 17.Harker W G, Slade D L, Parr R L, Feldhoff P W, Sullivan D M, Holguin M H. Cancer Res. 1995;55:1707–1716. [PubMed] [Google Scholar]
- 18.Errington F, Willmore E, Tilby M J, Li L, Li G, Li W, Baguley B C, Austin C A. Mol Pharmacol. 1999;56:1309–1316. doi: 10.1124/mol.56.6.1309. [DOI] [PubMed] [Google Scholar]
- 19.Desai S D, Liu L F, Vazquez-Abad D, D'Arpa P. J Biol Chem. 1997;272:24159–24164. doi: 10.1074/jbc.272.39.24159. [DOI] [PubMed] [Google Scholar]
- 20.Khelifa T, Beck W T. Biochem Pharmacol. 1999;58:1247–1257. doi: 10.1016/s0006-2952(99)00213-0. [DOI] [PubMed] [Google Scholar]
- 21.Subramanian D, Kraut E, Staubus A, Young D C, Muller M T. Cancer Res. 1995;55:2097–2103. [PubMed] [Google Scholar]
- 22.Andoh T, Ishida R. Biochim Biophys Acta. 1998;1400:155–171. doi: 10.1016/s0167-4781(98)00133-x. [DOI] [PubMed] [Google Scholar]
- 23.Hasinoff B B, Abram M E, Barnabe N, Khelifa T, Allan W P, Yalowich J C. Mol Pharmacol. 2001;59:453–461. doi: 10.1124/mol.59.3.453. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Wang J C, Liu L F. Proc Natl Acad Sci USA. 1988;85:1060–1064. doi: 10.1073/pnas.85.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ljungman M, Zhang F, Chen F, Rainbow A J, McKay B C. Oncogene. 1999;18:583–592. doi: 10.1038/sj.onc.1202356. [DOI] [PubMed] [Google Scholar]
- 26.te Poele R H, Okorokov A L, Joel S P. Oncogene. 1999;18:5765–5772. doi: 10.1038/sj.onc.1202961. [DOI] [PubMed] [Google Scholar]
- 27.Ishida R, Hamatake M, Wasserman R A, Nitiss J L, Wang J C, Andoh T. Cancer Res. 1995;55:2299–2303. [PubMed] [Google Scholar]
- 28.Huang K C, Gao H, Yamasaki E F, Grabowski D R, Liu S, Shen L L, Chan K K, Ganapathi R, Snapka R M. J Biol Chem. 2001;276:44488–44494. doi: 10.1074/jbc.M104383200. [DOI] [PubMed] [Google Scholar]
- 29.Mao Y, Desai S D, Liu L F. J Biol Chem. 2000;275:26066–26073. doi: 10.1074/jbc.M001831200. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Mao Y, Zhou N, Hu T, Hsieh T S, Liu L F. J Biol Chem. 2001;276:15990–15995. doi: 10.1074/jbc.M011143200. [DOI] [PubMed] [Google Scholar]
- 31.Lee K B, Wang D, Lippard S J, Sharp P A. Proc Natl Acad Sci USA. 2002;99:4239–4244. doi: 10.1073/pnas.072068399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratner J N, Balasubramanian B, Corden J, Warren S L, Bregman D B. J Biol Chem. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 33.Morris S K, Baird C L, Lindsley J E. J Biol Chem. 2000;275:2613–2618. doi: 10.1074/jbc.275.4.2613. [DOI] [PubMed] [Google Scholar]
- 34. Desai, S. D., Zhang, H., Rodriguez-Bauman, A., Yang, J. M., Wu, X., Gounder, M. K., Rubin, E. H. & Liu, L. F. (2003) Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 35.Hartzog G, Speer J, Lindstrom D. Biochim Biophys Acta. 2002;1577:276–286. doi: 10.1016/s0167-4781(02)00458-x. [DOI] [PubMed] [Google Scholar]
- 36.Shilatifard A. FASEB J. 1998;12:1437–1446. doi: 10.1096/fasebj.12.14.1437. [DOI] [PubMed] [Google Scholar]
- 37.Ferdous A, Gonzalez F, Sun L, Kodadek T, Johnston S A. Mol Cell. 2001;7:981–991. doi: 10.1016/s1097-2765(01)00250-7. [DOI] [PubMed] [Google Scholar]
- 38.Kierszenbaum A L. Mol Reprod Dev. 2000;57:109–110. doi: 10.1002/1098-2795(200010)57:2<109::AID-MRD1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 39.Jensen L H, Nitiss K C, Rose A, Dong J, Zhou J, Hu T, Osheroff N, Jensen P B, Sehested M, Nitiss J L. J Biol Chem. 2000;275:2137–2146. doi: 10.1074/jbc.275.3.2137. [DOI] [PubMed] [Google Scholar]
- 40.Deming P B, Flores K G, Downes C S, Paules R S, Kaufmann W K. J Biol Chem. 2002;277:36832–36838. doi: 10.1074/jbc.M206109200. [DOI] [PubMed] [Google Scholar]