Abstract
Pitx2 is a bicoid-related homeodomain factor that is required for effective cell type-specific proliferation directly activating a specific growth-regulating gene cyclin D2. Here, we report that Pitx2, in response to the Wnt/β-catenin pathway and growth signals, also can regulate c-Myc and cyclin D1. Investigation of molecular mechanisms required for Pitx2-dependent proliferation, in these cases, further supports a nuclear role for β-catenin in preventing the histone deacetylase 1-dependent inhibitory functions of several DNA-binding transcriptional repressors, potentially including E2F4/p130 pocket protein inhibitory complex, as well as lymphoid enhancer factor 1 and Pitx2, by dismissal of histone deacetylase 1 and loss of its enzymatic activity. Thus, β-catenin plays a signal-integrating role in Wnt- and growth factor-dependent proliferation events in mammalian development by both derepressing several classes of repressors and by activating Pitx2, regulating the activity of several growth control genes.
The Wnt pathway induces various cellular responses from cell proliferation to cell fate determination and terminal differentiation (1–3). The binding of Wnt ligands to receptors activates intracellular Dishevelled (Dsh in Drosophila, Dvl in vertebrates), which, in turn, modulates the activity of the GSK-3β signals. In the absence of Wnt signaling, β-catenin (armadillo in Drosophila) is found in a multiprotein complex containing adenomatous polyposis coli and axin, targeting β-catenin for degradation. Activation of Dsh/Dvl leads to the stabilization and accumulation of β-catenin in the cytoplasm. On translocation, β-catenin interacts with members of the T cell factor (TCF)/lymphoid enhancer factor (LEF) family of DNA-binding molecules to influence target gene expression (4, 5), although recent evidence in Drosophila indicates that β-catenin can regulate gene transcription by selective nuclear export of regulatory proteins (6).
β-Catenin is proposed to bind to the TCF/LEF family of transcription factors, changing them from repressors to activators of transcription (7, 8). In part, because the expression patterns of the known TCF/LEFs probably are not broad enough to explain all of the activities of the Wnts expressed in the development, we investigated whether β-catenin may bind to other tissue-restricted transcription factors to modulate specific aspects of Wnt signaling.
Recently, we reported that a Wnt/β-catenin → Pitx2 pathway operates in several specific tissues to control proliferation by regulating expression of cyclin D2 gene in G1 (9). The Wnt pathway directly induces Pitx2 and, with additional growth factor-dependent signaling, dismisses Pitx2-associated corepressors and mediates a temporally specific, sequential recruitment of specific coactivator complexes, which includes a factor Ldb1/NLI/CLIM, previously identified as a coactivator of LIM homeodomain factors (10, 11), and Tip60, a member of the MYST family of coactivators (12, 13), required for activation of the E2F-independent G1 growth regulatory target gene Cyclin D2. Pitx2 is a member of a subfamily of bicoid-related factors (14, 15). Pitx2, expressed in several tissues, initially was identified as one of the genes responsible for the human Reiger syndrome as well as occasional abnormal cardiac and pituitary development (16).
Here, we report that Pitx2 can regulate additional G1 cell cycle control genes including c-Myc and cyclin D1. We find that β-catenin not only reverses the repressive function of LEF1 and Pitx2 but also can overcome the repressive actions of the E2F4-related pocket protein p130 and histone deacetylase 1 (HDAC1), potentially providing an additional component in mediating Wnt-dependent growth and developmental programs. Together, these results suggest that the nuclear response to Wnt-dependent signaling events in target tissues involves a specific antirepressive role for β-catenin in integrating the transcriptional response as well as an activation role of Pitx2.
Materials and Methods
Antibodies.
Anti-Pitx2 IgG was generated from guinea pigs (9). The following antibodies were obtained from Santa Cruz Biotechnology: anti-β-catenin, HDAC1, HDAC2, Tip60, TCFs, and LEF-1. Antiacetylated histone H3 and H4 antibodies were from Upstate Biotechnology (Lake Placid, NY), and anti-RNA polymerase II antibody was from Berkeley Antibody (Richmond, CA).
Immunoprecipitations and HDAC Enzymatic Assays.
Coimmunoprecipitation and other protein-interaction studies were performed as described (12). Fluorogenic histone deacetylase substrate [Boc-Lys(Ac)-AMC] was purchased from Calbiochem. From immunoprecipitates, HDAC enzymatic assay was done by using fluorogenic HDAC substrate as described (17).
Single-Cell Nuclear Microinjection Assays.
Microinjection assays were carried out as described (9). Affinity-purified αPitx2 IgG was used. Each experiment was performed on three independent cover slips consisting of 1,000 cells. Where no experimental antibody was used, preimmune IgGs were coinjected, allowing the unambiguous identification of injected cells in addition to serving as a negative control.
Chromatin Immunoprecipitation (ChIP) Assays.
For the ChIP assay, αT3-1, a murine pituitary cell line, and C2C12 myoblast cell line were used, and LiCl (10 mM) was added for 1 h before harvest. Cells were washed twice with PBS and cross-linked with 1% formaldehyde for 10 min at room temperature. Cross-linked cells were treated as described (12). Cells then were resuspended in 0.3 ml of lysis buffer (50 mM Tris⋅HCl, pH 8.1/1% SDS/10 mM EDTA/protease inhibitors) and sonicated three times for 10 sec followed by centrifugation for 10 min. The average size of sheared fragments was expected to be ≈300–1,000 bp. Immunoprecipitates were eluted three times with 1% SDS/0.1 M NaHCO3. Eluates were pooled and heated at 65°C for 6 h to reverse the formaldehyde cross-linking. DNA fragments were purified with a QIAquick spin kit (Qiagen, Chatsworth, CA). For PCR, 1 μl from a 50-μl DNA extraction and 25 cycles of amplification were used with the following promoter-specific primers: c-Myc forward, 5′-GCTTGG CGGGAAAAAGAAGGG-3′; reverse, 5′-AGAGCTGCCTTCTTAGGTCG-3′; Cyclin D1 forward, 5′-GTTCCTGGAAGGGCGACTAA-3′; reverse, 5′-GGGGTGGGATCTGAGATTTG-3′.
Results
Pitx2 Modulates a Subset of G1 Cell Cycle Control Gene.
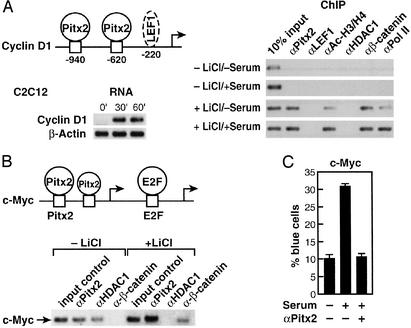
Pitx2 serves as a transcriptional regulator in early to late G1 in cells and can regulate cyclin D2 (9). Because it is likely that several cell cycle control genes are modulated by Pitx2, as well as cyclin D2, we investigated Pitx2 on several growth regulatory promoters. A second potential G1 regulatory target was considered. In C2C12 cells, Wnt pathway activation and serum stimulation caused a marked increase in cyclin D1 mRNA levels by 30–60 min, correlating with the recruitment of Pitx2 to the cyclin D1 promoter (Fig. 1A). Although there is a potential 3′ LEF1 site further in the murine Cyclin D1 promoter (18), no detectable LEF1 recruitment was observed in C2C12 or αT3-1 cells (Fig. 1A). Also, several Pitx2-binding sites were found in the c-Myc promoter, one of which corresponds precisely to the consensus CTAATCC bicoid recognition sequence (19) (Fig. 1B). c-Myc is an example of a cell cycle control gene that is regulated positively or negatively based on combinatorial interactions involving E2F transcription factors and other classes of transcriptional activators or repressors (20). For example, in proliferating myeloid cells where the c-Myc gene is expressed, the c-Myc promoter is occupied by the activators E2F1 and Ets2 (21). In contrast, in terminally differentiated cells in which the c-Myc gene is repressed, the c-Myc promoter instead is occupied by the E2F repressor E2F4, the cell type-restricted ets repressor METS, and the E2F4-associated pocket proteins p107 and p130 that function as cell cycle-dependent corepressors (22, 23). Furthermore, previous studies of the human c-Myc promoter suggested that it is a target of the Wnt pathway, and it contains upstream functional TCF/LEF-binding sites (24).
Figure 1.
Pitx2 modulates a subset of G1 cell cycle control gene. (A) Recruitment of Pitx2, but not LEF1, to the murine Cyclin D1 promoter in C2C12 cells. A schematic of response elements is shown, with a potential (not proven) LEF1 site. Serum + LiCl caused rapid induction of Cyclin D1 transcripts by RT-PCR in C2C12 cells. A ChIP analysis reveals recruitment of Pitx2, but not LEF1, in response to LiCl treatment, even when we used primers out to −2 kb. (B) Pitx2 and β-catenin on the c-Myc promoter. Murine C2C12 myoblast cells were treated with lithium for 1 h in the presence of serum. ChIP assay was performed by using αPitx2, αHDAC1, and αβ-catenin-specific IgGs. In the absence of lithium, Pitx2 and HDAC1 were bound on the c-Myc promoter in C2C12 cells, whereas, after induction with lithium for 1 h, Pitx2 binding was stronger and release of HDAC1 and the presence of β-catenin were noted. PCR analysis was performed by using oligonucleotide primers flanking the Pitx2- and E2F-binding sites on the murine c-Myc promoter. Schematic shows the location of Pitx2 (−212; −188) and E2F (−154) elements. RT-PCR analysis was performed to check message level after treatment with lithium for 1 h. (C) Role of Pitx2 in c-Myc expression. C2C12 cells were microinjected with αPitx2 IgG and a c-Myc promoter carrying LacZ as a reporter. αPitx2 IgG inhibits the serum-dependent stimulation of the LacZ expression by 70%.
Indeed, ChIP analysis with primers that span the E2F sites and the adjacent Pitx2 sites revealed that Pitx2 was bound to c-Myc regulatory sequences in C2C12 cells (Fig. 1B). As expected, some HDAC1 also was present on the promoter under these cultures (Fig. 1B). In addition, binding of LEF1, but not TCF1, TCF3, or TCF4, could be detected further 5′ in the c-Myc promoter (data not shown), consistent with previous suggestions of c-Myc as a Wnt-regulated gene (24). Therefore, Pitx2, as well as LEF1, binds to the c-Myc promoter in specific cell types and may participate in the recruitment of β-catenin and the dismissal of HDAC1 upon activation of the Wnt pathway. In addition, in transient cotransfection assays in αT3–1 pituitary cells or C2C12 myoblast cells, Pitx2 could stimulate the c-Myc promoter (−1,100/+500), ≈2- to 2.5-fold, comparable to the effects of addition of lithium, requiring the presence of the three defined Pitx2 sites (data not shown). When the single-cell nuclear microinjection assay with the c-Myc reporter was used, serum-dependent stimulation was abolished by αPitx2 IgG in C2C12 cells (Fig. 1C). These data led us to further investigate the potential interactions and roles of β-catenin in the action of Pitx2 and determine whether a more general antirepressor function of β-catenin might serve functions in growth stimulation by the Wnt pathway.
Linkage of β-Catenin to HDAC1 and Nuclear Receptor Corepressor (N-CoR) Repressive Actions.
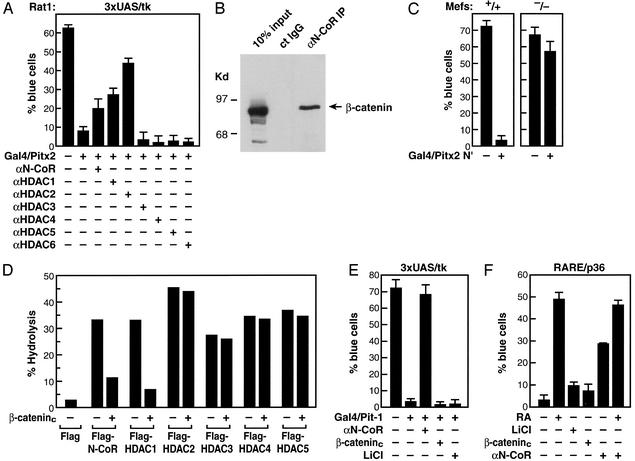
Many homeodomain activators have been shown to serve as repressors dependent on promoter context and culture conditions, based on their interactions with a specific corepressor (25). For example, Pit-1 can recruit, directly or indirectly, an N-CoR-containing corepressor that is required for cell type-specific gene restriction events (26, 27). Consistent with these findings, Pitx2 or a Gal4/Pitx2 fusion protein can function as a repressor on a Pitx2 RE/tk or a UAS/tk promoter (Fig. 2A and data not shown), even though it is clearly an activator on many promoters (28). Based on the ability of the Wnt pathway to reverse TCF/LEF-dependent repression, we evaluated whether activation of this pathway could reverse Pitx2-dependent repression.
Figure 2.
Dismissal of HDAC activity by β-catenin. (A) Coinjection of specific IgGs against HDAC1 or HDAC2, but not HDAC3, HDAC4, HDAC5, or HDAC6, and, to a lesser extent, αN-CoR IgGs relieves repression by Gal4/Pitx2 in a single-cell nuclear microinjection assay in Rat1 cells. (B) N-CoR and β-catenin interactions. Based on isolation of β-catenin in a yeast two-hybrid screen with RDIII of N-CoR, immunoprecipitation experiments were performed in HCT116 colorectal tumor cells, revealing robust interactions, by using specific αβ-catenin and αN-CoR (Upper). Coimmunoprecipitation from 293 cells transfected with HDAC1 and β-catenin and Western blot analysis were performed by using the indicated IgGs. (C) Expression of Gal4/Pitx2-N′ causes repression in MEFs from N-CoR−/− but not N-CoR+/+ littermates. (D) β-Catenin inhibits N-CoR and HDAC1 deacetylase activity. 293 cells were transfected with expression vectors for Flag-tagged N-CoR or Flag-tagged HDACs in the presence or absence of the β-cateninc expression vector. After immunoprecipitation with αFlag-IgG, HDAC activity was assessed as reported previously (17). N-CoR and HDAC levels were equivalent in all immunoprecipitates, as assessed by using αFlag IgG. (E and F) Failure of β-catenin to reverse β-RAR- or Pit-1-dependent repression. Gal4/Pit-1 repression or β-RAR repression on a DR5-dependent lacZ reporter could not be overcome by expression of β-cateninc or administration of lithium. αN-CoR reversed both β-RAR- and Pit-1-dependent repression. All specific cell nuclear microinjection experiments are mean ± SEM with >300 cells microinjected, and experiments were repeated independently a minimum of three times.
We found that either lithium or constitutively active β-catenin (β-cateninc) could, in fact, reverse Gal4/Pitx2-dependent repression (9). This Pitx2-dependent repression also can be overcome partially by injection of purified antibodies against N-CoR, HDAC1, or HDAC2 (Fig. 2A). We considered whether β-catenin could associate with N-CoR as well as specific HDACs and exert functional effects. This was considered, in part, because yeast two-hybrid screens performed with repressor domain III of N-CoR as bait yielded multiple, independent isolates encompassing the armadillo repeat region of β-catenin that interacted specifically with repressor domain III (data not shown). Coimmunoprecipitation assays performed in HCT116 cells derived from colorectal tumors, which express a constitutively active form of β-catenin (29), by using αN-CoR or αβ-catenin IgGs revealed a strong interaction between N-CoR and β-catenin (Fig. 2B), which is consistent with previous observations (30). Indeed, a Gal4/Pitx2 N-terminus fusion protein (Gal4/Pitx2-N′), a dominant-negative form of Pitx2 (9) that is capable of resulting repression by UAS/lacZ reporter in MEFs from N-CoR+/+ mice, was not an effective repressor in MEFs from N-CoR−/− mice (Fig. 2C).
We therefore evaluated the potential effects of β-catenin on the HDAC enzymatic activity of N-CoR or HDAC complexes by coimmunoprecipitation assay by expressing Flag-tagged HDACs in the presence or absence of β-cateninc. The HDAC activity associated with immunoprecipitation of Flag-tagged N-CoR also was inhibited 3- to 4-fold by expression of β-cateninc (Fig. 2D). The activity of HDAC1 complexes also was inhibited 4- to 5-fold by β-cateninc, but the activities of HDAC2, HDAC3, HDAC4, and HDAC5 were not affected (Fig. 2D). Interestingly, the ability of β-catenin to inhibit the associated deacetylase activity of HDAC1, as well as N-CoR, is consistent with the actions of αHDAC1 and αN-CoR to potentially reverse Pitx2-dependent repression.
We did not observe reversal of the repressive actions of Pit-1 or unliganded retinoic acid receptor (RAR) by LiCl or β-cateninc in similar nuclear microinjection assays (Fig. 2 E and F), whereas αN-CoR did reverse repression, revealing specificity of the antirepressor action of β-catenin, based on recruitment to Pitx2 but not to Pit-1 or RAR.
β-Catenin Causes a Dismissal of HDAC1 Associated with Pocket Proteins from the c-Myc Promoter.
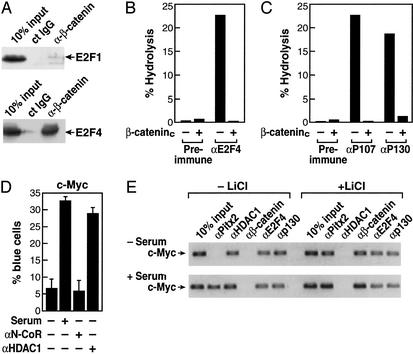
Based on the complex regulation between E2F factor exchanges and the known association of HDAC1 with p130 (31), we next evaluated the potential association of E2F1 or E2F4 with β-catenin. Interestingly, coimmunoprecipitation assays revealed a robust, selective interaction between β-catenin and E2F4, whereas little if any interaction was observed between β-catenin and E2F1 (Fig. 3A). Furthermore, the presence of β-catenin virtually eliminated the deacetylase activity normally associated with the immunoprecipitated E2F4/p130 complex, because coimmunoprecipitation of E2F4, p107, or p130 in the presence of β-cateninc caused a striking inhibition of associated enzymatic activity (Fig. 3 B and C). If HDAC1 enzymatic activity is a critical component of E2F4-dependent repression, then αHDAC1 IgG might be expected to increase expression of c-Myc promoter-driven reporters. Indeed, under serum-free conditions for 48 h in C2C12 cells, we found that nuclear microinjection of αHDAC1, but not αN-CoR, caused a stimulation of the c-Myc promoter-driven reporter similar to that obtained with serum (Fig. 3D).
Figure 3.
Role of β-catenin in derepression. (A) β-Catenin interacts with E2F4. HA-E2F1 and HA-E2F4 expression vectors were cotransfected with β-cateninc-expressing vectors, and immunoprecipitation was performed with control IgG of αβ-catenin IgG. β-Catenin was interacting strongly with E2F4. (B) β-Catenin dismisses HDAC activity associated with E2F4. 293 cells were transfected with β-cateninc expression vectors and immunoprecipitated with preimmune IgG or αE2F4 IgG, and HDAC activity was measured. (C) β-Cateninc also inhibits HDAC activity on p107- and p130-immunoprecipitated material from 293 cells. (D) Effects of αHDAC1 and αN-CoR IgGs on expression of the c-Myc promoter in Rat1 cells. After 48 h under serum-free conditions, cells were microinjected with either αN-CoR or αHDAC1 IgGs, and serum was added 8 h before assay. Each point is mean ± SEM of >300 microinjected cells; similar results were obtained in two additional experiments of similar design by using C2C12 or Rat1 cells. (E) Effects of lithium stimulation on binding of E2F4, p130, and HDAC1 to the c-Myc promoter in C2C12 cells. C2C12 cells were placed in a serum-free medium for 48 h to synchronize cells, and serum, lithium, or both were added for 1–16 h before harvest for ChIP assay. Consistent with serum-dependent regulation of Pitx2 gene expression in C2C12 cells, Pitx2 was not detected at 1 h on the c-Myc promoter in C2C12 cells under the serum-free conditions (Upper) but was detected in serum-treated cells (Lower). Under both conditions, E2F4, p130, and HDAC1, but not β-catenin, were detected. However, 1 h after lithium addition, HDAC1 was no longer detected and β-catenin was present on the promoter.
Therefore, we wanted to determine whether β-catenin caused a dismissal of HDAC1 or p130 from the c-Myc promoter in response to activation of the Wnt/β-catenin pathway. To evaluate this issue, the effects of activation of the β-catenin pathway were evaluated in C2C12 cells by using the ChIP assay. C2C12 cells were placed under serum-free conditions for 48 h to synchronize cells, and then either serum or lithium or both were added for 1–16 h. Under serum-free conditions, no Pitx2 was detected initially on the c-Myc promoter, consistent with its down-regulation in serum-starved C2C12 cells. However, E2F4, p130, and HDAC1 all were present on the c-Myc promoter. With lithium treatment for 1 h, Pitx2 and β-catenin now were present on the c-Myc promoter, but both p130 and E2F4 remained bound (Fig. 3E). However, HDAC1 was almost completely dismissed. This profile of factors bound to the c-Myc promoter remained similar at 16 h (data not shown). With addition of serum as well as lithium, more Pitx2 appeared bound at 1 h, and, again, no alterations were observed in p130 and E2F4; however, HDAC1 again was not detected (Fig. 3E). These data suggest that β-catenin does not interfere with binding of E2F4 and does not displace the pocket proteins from E2F4, but recruitment of β-catenin does result in dismissal of HDAC1 from E2F4 on the c-Myc promoter, analogous to its action in derepressing Pitx2.
Regulation of Cyclin D1 and Cyclin D2 by the Wnt Pathway in Human Colon Cancer Cells.
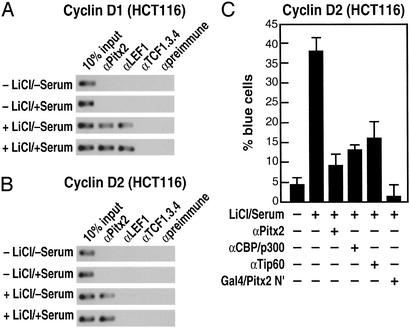
Regulation of cyclin D1 by the Wnt/β-catenin pathway has been reported in human colon cancer cells based on the presence of LEF/TCF sites in the promoter (32), raising the question of which cell-specific factor might operate in these cells. In response to lithium alone or lithium and serum, β-catenin is recruited to the cyclin D1 promoter (data not shown). However, we found that Pitx2 is expressed robustly in human colon cancer cells, consistent with its expression in developing colon and block of colon development in Pitx2−/− mice (data not shown). We find that Pitx2 is recruited to both the cyclin D2 and cyclin D1 promoters in colon cancer cells (HCT116) but that, in these cells, in contrast to C2C12 cells, the cyclin D1 promoter binds LEF1 (Fig. 4 A and B), consistent with previous reports (18). Antibodies against Pitx2, coactivators of Pitx2 including CBP/p300 and Tip60 (9), or Gal4/Pitx2-N′ block the cyclin D2 activation in these cells from the single-cell nuclear microinjection assay, indicating that Pitx2 again seems to serve as a cell-specific factor involved in G1 progression (Fig. 4C), as is the case for pituitary and muscle we reported recently (9).
Figure 4.
Regulation of cyclin D1 and cyclin D2 by the Wnt pathway in human colon cancer cells. (A and B) ChIP analysis of Cyclin D1 and Cyclin D2 promoters in HCT116 cells, treated as indicated. On the Cyclin D2 promoter, Pitx2 is detected but LEF1 is not detected. On the Cyclin D1 promoter, LEF1 is detected, as is Pitx2. (C) A block of Cyclin D2 expression in HCT116 cells in single-cell nuclear microinjection by αPitx2 IgG or by expression of a dominant-negative form of Pitx2 (Pitx2 N terminus).
Discussion
In this article, we have probed further the molecular mechanism by which a cell-restricted DNA-binding transcription factor, Pitx2, serves as an important modulator of growth control genes, indicating that c-Myc and cyclin D1 as well as cyclin D2 can serve as target genes. Using well studied C2C12 muscle cell and αT3–1 pituitary cell lines, we suggested that inhibitory E2F4, Pitx2, and LEF1 might all serve as synergistic regulated repressors that, upon binding β-catenin, have dismissal of corepressors including HDAC1.
Investigation of Pitx2-dependent gene regulation has revealed an additional level of control of Pitx2 by the Wnt pathway that may represent its effects on cellular proliferation events. As is the case for many homeodomain factors, Pitx2 can act alternatively as a repressor and/or activator depending on promoter context or cell type. The Wnt pathway not only induces Pitx2 gene expression via β-catenin pathway as we reported (9) but also reverses its ability to function as a repressor. This effect is conferred, in part, by actions of HDAC1 and, possibly to some extent, by N-CoR. These data are consistent with the hypothesis that β-catenin can modulate Pitx2 repressor function based on direct interactions, causing both dismissal and inhibition of associated HDAC1. Thus, our data suggest that β-catenin interacts with several additional transcription factors in addition to LEF1 (18) and Pitx2 (9), in each case dismissing HDAC1 or N-CoR-dependent complexes on specific promoters. These actions are specific, because other N-CoR/HDAC1-dependent repressors, such as nuclear receptors or Pit-1, are not affected by β-catenin.
Our data suggest that the role of β-catenin may provide an additional molecular mechanism for growth regulation based on interactions with E2F repressor complex. A critical aspect of growth-promoting genes is the switch between the activator E2Fs (E2F1–3) and repressor E2Fs (E2F4–6), which are associated with the pocket proteins, particularly p130, which, in turn, recruit HDAC1-containing complexes (33). Thus, a component of the β-catenin-dependent effects on proliferation possibly could involve its ability to reverse E2F4/p130/HDAC1-dependent repression on some promoters as well as the ability of Pitx2 to directly act on growth regulatory genes, an example of which is provided by c-Myc and cyclin D1. In this regard, our data suggest that β-catenin might serve to both inhibit HDAC1 activity and to dismiss HDAC1 from the E2F4/p130 complex while not affecting binding of E2F4 and the associated pocket proteins. These data are consistent with the well established requirement of hyperphosphorylation for their dismissal.
c-Myc has been reported to be regulated by the Wnt pathway via β-catenin (24) and is regulated in a multifactorial and tissue-specific fashion. Pitx2, which binds to regulatory regions in the c-Myc promoter, is suggested to function as a component of Wnt control in specific cell types. Indeed, E2F-dependent regulation of c-Myc is modulated by a series of tissue-specific repressors, including the inducer of differentiation Blimp-1 in B cells (34) and the ets repressor METS in macrophages (23) that also requires the presence of E2F-associated pocket proteins to mediate repression. It is tempting to speculate that Pitx2 and E2F4, perhaps in concert with LEF1, act to stabilize association of the E2F4/pocket protein/HDAC1 corepressor complex and, conversely, that their interactions with β-catenin then may participate in the regulation of the exchange of E2F1–3 to E2F4, 5.
Our observation that β-catenin can selectively associate strongly with E2F4, but only weakly, if at all, with E2F1, and can inhibit the HDAC activity recruited by the E2F4-associated pocket proteins p107/p130 would suggest a possible regulatory program that also may be influenced by the activation of the Wnt pathway, perhaps serving as an aspect of its proliferative effects in many organ systems. This observation is consistent with previously reported interactions between p130 and HDAC1 (31). In cases such as c-Myc regulation, one then might suggest that Pitx2, with the more distally bound LEF1, via binding of β-catenin could cooperate in the exchange of E2F4-associated corepressor complexes containing HDAC1, providing an additional “anticorepressor mechanism” for c-Myc gene activation.
Interestingly, cyclin D1 may use distinct regulatory machinery in response to activation of this Wnt pathway depending on cell type. In C2C12 cells, Pitx2 but not LEF1 is detected, but in colon cancer cell lines, both Pitx2 and LEF1 are detected on the cyclin D1 promoter. These data raise the question of how these factors might operate differently between C2C12 myoblast cells and HCT116 colon cancer cells.
Acknowledgments
We particularly thank Amy Gonzalez (Santa Cruz Biotechnology) for providing the information to select crucial specific antibodies, C. Nelson for technical assistance, and P. Myer and M. Fisher for figure and manuscript preparation. S.H.B. is supported by a California Breast Cancer Research Program fellowship; M.G.R. is an Investigator with the Howard Hughes Medical Institute; and A.W.-B. is supported by an institutional grant from the Howard Hughes Medical Institute. This work was supported by grants from the National Institutes of Health (to A.W.-B., D.W.R., C.K.G., and M.G.R.).
Abbreviations
- LEF
lymphoid enhancer factor
- TCF
T cell factor
- ChIP
chromatin immunoprecipitation
- HDAC
histone deacetylase
- N-cor
nuclear receptor corepressor
- β-cateninc
constitutively active β-catenin
- RAR
retinoic acid receptor
References
- 1.Cadigan K M, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 2.Ikeya M, Lee S M, Johnson J E, McMahon A P, Takada S. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 3.Eastman Q, Grosschedl R. Curr Opin Cell Biol. 1999;11:233–240. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 4.Hsu S C, Galceran J, Grosschedl R. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutros M, Mlodzik M. Mech Dev. 1999;83:27–37. doi: 10.1016/s0925-4773(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 6.Chan S K, Struhl G. Cell. 2002;111:265–280. doi: 10.1016/s0092-8674(02)01037-1. [DOI] [PubMed] [Google Scholar]
- 7.Cavallo R A, Cox R T, Moline M M, Roose J, Polevoy G A, Clevers H, Peifer M, Bejsovec A. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 8.Fisher A L, Caudy M. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 9.Kioussi C, Briata P, Baek S H, Rose D W, Hamblet N S, Herman T, Ohgi K, Lin C, Gleiberman A, Wang J, et al. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- 10.Jurata L W, Pfaff S L, Gill G N. J Biol Chem. 1998;273:3152–3157. doi: 10.1074/jbc.273.6.3152. [DOI] [PubMed] [Google Scholar]
- 11.Bach I, Carriere C, Ostendorff H P, Andersen B, Rosenfeld M G. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- 12.Baek S H, Ohgi K A, Rose D W, Koo E H, Glass C K, Rosenfeld M G. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 13.Ikura T, Ogryzko V V, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 14.Chen S, Wang Q L, Nie Z, Sun H, Lennon G, Copeland N G, Gilbert D J, Jenkins N A, Zack D J. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 15.Lin C R, Kioussi C, O'Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte J C, Rosenfeld M G. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 16.Semina E V, Reiter R, Leysens N J, Alward W L, Small K W, Datson N A, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel B U, et al. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann K, Brosch G, Loidl P, Jung M. Nucleic Acids Res. 1999;27:2057–2058. doi: 10.1093/nar/27.9.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Driever W, Thoma G, Nusslein-Volhard C. Nature. 1989;340:363–367. doi: 10.1038/340363a0. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi Y, Rayman J B, Dynlacht B D. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H, Moulton K, Horvai A, Parik S, Glass C K. Mol Cell Biol. 1994;14:2129–2139. doi: 10.1128/mcb.14.3.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaubatz S, Lindeman G J, Ishida S, Jakoi L, Nevins J R, Livingston D M, Rempel R E. Mol Cell. 2000;6:729–735. doi: 10.1016/s1097-2765(00)00071-x. [DOI] [PubMed] [Google Scholar]
- 23.Klappacher G W, Lunyak V V, Sykes D B, Sawka-Verhelle D, Sage J, Brard G, Ngo S D, Gangadharan D, Jacks T, Kamps M P, et al. Cell. 2002;109:169–180. doi: 10.1016/s0092-8674(02)00714-6. [DOI] [PubMed] [Google Scholar]
- 24.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 25.Asahara H, Dutta S, Kao H Y, Evans R M, Montminy M. Mol Cell Biol. 1999;19:8219–8225. doi: 10.1128/mcb.19.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu L, Lavinsky R M, Dasen J S, Flynn S E, McInerney E M, Mullen T M, Heinzel T, Szeto D, Korzus E, Kurokawa R, et al. Nature. 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- 27.Scully K M, Jacobson E M, Jepsen K, Lunyak V, Viadiu H, Carriere C, Rose D W, Hooshmand F, Aggarwal A K, Rosenfeld M G. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- 28.Amendt B A, Sutherland L B, Semina E V, Russo A F. J Biol Chem. 1998;273:20066–20072. doi: 10.1074/jbc.273.32.20066. [DOI] [PubMed] [Google Scholar]
- 29.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 30.Billin A N, Thirlwell H, Ayer D E. Mol Cell Biol. 2000;20:6882–6890. doi: 10.1128/mcb.20.18.6882-6890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. Proc Natl Acad Sci USA. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tetsu O, McCormick F. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 33.Lai A, Kennedy B K, Barbie D A, Bertos N R, Yang X J, Theberge M-C, Tsai S C, Seto E, Zhang Y, Kuzmichev A, et al. Mol Cell Biol. 2001;21:2918–2932. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y, Wong K, Calame K. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]