Abstract
Centered in the Chinese hamster dihydrofolate reductase origin of replication is a prominent nuclear matrix attachment region (MAR). Indirect lines of evidence suggested that this MAR might be required for origin activation in early S phase. To test this possibility, we have deleted the MAR from a Chinese hamster ovary variant harboring a single copy of the dihydrofolate reductase locus. However, 2D gel replicon mapping shows that removal of the MAR has no significant effect either on the frequency or timing of initiation in this locus. Rather, fluorescence in situ hybridization studies on cells swollen under either neutral or alkaline conditions show that deletion of the MAR interferes with local separation of daughter chromatids. This surprising result provides direct genetic evidence that at least a subset of MARs performs an important biological function, possibly related to chromatid cohesion and separation.
In metaphase, chromatin loops are radially attached to a proteinaceous scaffolding running through the center of each chromosome arm (1). The sequences at the bases of each loop are AT rich, and their juxtaposition on the scaffold generates a so-called AT queue that can be visualized microscopically (2). Presumably, differential compaction of the scaffolding gives rise to characteristic chromosome bands (2). In the interphase nucleus, chromosomes remain constrained, occupying distinct chromosome territories (3) separated by interchromosome domains (reviewed in ref. 4). Many active genes are located at the surface of the chromosome territories (5), whereas other active domains protrude into the interchromosome domain as large loops (6). These observations are consistent with the view that, within each territory, the chromatin folding that characterizes metaphase chromosomes is maintained largely in interphase, with the ≈100-kb loop probably constituting the basic building block. Electron microscopic observation of a distinct fibrillo-granular network in chromatin-depleted interphase nuclei suggests the existence of an internal matrix (7), which would explain why replication and transcription foci remain in place after chromatin removal (8). As in metaphase, DNA appears to be attached to this nuclear scaffolding via AT-rich matrix attachment regions (MARs; ref. 9). However, direct evidence for a nuclear matrix in interphase cells in vivo is still lacking. Indeed, it has been argued that the insoluble structure remaining after extraction of nuclei with high salt or isotonic lithium 3,5-diiodosalicylate (LIS) is an artifact of aggregation of normally soluble proteins (10, 11). Yet, given the observed compartmentalization of the nucleus, even artifactual aggregations may reflect some underlying organizational principles within the living cell. Thus, it is yet to be determined whether the matrix (operationally defined here as the insoluble structure remaining after LIS extraction of nuclei) and the AT-rich MARs that associate with them, have any functional significance.
We and others have extensively characterized the Chinese hamster dihydrofolate reductase (DHFR) domain, which consists of two convergently transcribed genes (DHFR and 2BE2121; Fig. 1A) flanking a 55-kb intergenic spacer. Replication initiates at any of a large number of sites scattered throughout the intergenic spacer (12–15), with at least three sites within the spacer being preferred (ori-β, ori-β′, and ori-γ; refs. 15–21). We previously identified a MAR near the center of this complex origin and localized it to a 3.4-kb PvuII fragment (Fig. 1A; ref. 22). Additionally, we showed that two of the most efficient initiation sites in the spacer display an altered chromatin structure (ori-β and ori-γ; Fig. 1), but only in the subset of origins that partitions with the matrix when chromatin loops are excised with an endonuclease (23). Thus, the possibility arose that attachment to the intergenic MAR might be required for origin activity in early S phase. This model would be consistent with studies showing that DNA synthesized at the onset of S phase remains close to the matrix throughout S phase, but DNA synthesized at later times migrates into the DNA loops (24, 25).
Figure 1.
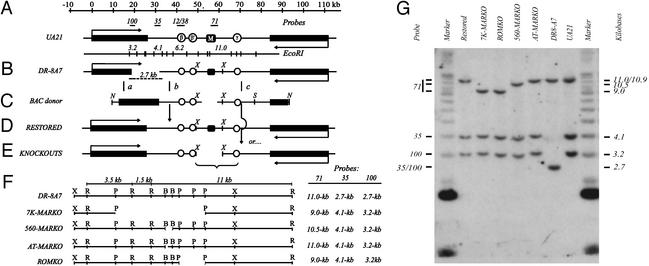
The DHFR locus and the ROKO approach. (A) The 120-kb region encompassing the DHFR and 2BE2121 (53, 54) genes in the WT hemizygote, UA21, showing the position of three preferred replication initiation sites (ori-β, ori-β′, and ori-γ; refs. 16, 17, and 21) and the intergenic MAR (black square; ref. 22). Positions of relevant hybridization probes and an EcoRI map of the region are shown above and below the map, respectively. (B) The DHFR-deficient variant, DR-8A7, with the diagnostic deletion junction EcoRI fragment shown below. (C) The BAC donor used to restore the DHFR gene and knock out the downstream sequence of interest, showing the XhoI fragment into which each deletion was engineered (in the example shown, a 7-kb region encompassing the MAR; see text). (D and E) Recombination near sites a and b (C) leads to a restored WT derivative (D), whereas recombination near sites a and c leads to restoration of the gene and simultaneous deletion of the downstream target (E). (F) Detailed maps showing positions of deletions (see text) and sizes of resulting diagnostic EcoRI fragments. X, XhoI; R, EcoRI; P, PstI; B, BstEII. (G) Southern analysis of EcoRI digests of each cell line, hybridized with a mixture of probes 100, 35, and 71 (see A and F). Size markers are a mixture of a 1-kb ladder and high molecular weight standard (Invitrogen).
In the present study, we have defined the intergenic MAR more precisely and have deleted it by using a reach-out-and-knock-out (ROKO) homologous recombination strategy (26). The unexpected phenotypic consequences of this deletion are described.
Materials and Methods
Cell Culture and Synchronization.
Chinese hamster ovary (CHO), CHOC400, DR-8A7, and DHFR+ derivatives of DR-8A7 were maintained and synchronized as described (26–29).
In Vivo and in Vitro MAR Assays.
MAR assays were performed as described (22) on matrix/halo structures prepared by extracting nuclei with lithium 3,5-diiodosalicylate (LIS) (9). For the in vivo assay (9, 22), ≈1 × 108 CHOC400 or 2–3 × 108 CHO cell equivalents were digested with a combination of PvuII and HinfI to yield matrix-attached and loop DNA fractions. For the in vitro assay (22, 30), 2–4 × 106 matrices from CHOC400 cells were digested with DNase I (GIBCO/BRL), and naked matrices were incubated with ≈10 ng of the appropriate end-labeled digest and increasing concentrations of nonspecific competitor DNA (pBR322 digested with HinfI, to yield fragments of similar average size as the labeled probes). See figure legends for details.
Construction of Donor Bacterial Artificial Chromosome (BACs) for Performing in Loco Mutagenesis.
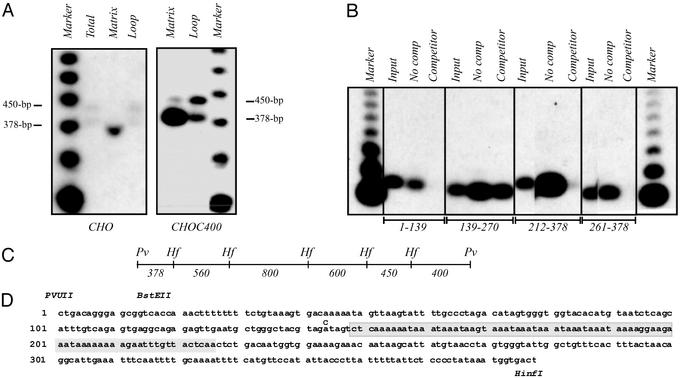
A recombinant BAC was constructed in the pBeloBAC-11 vector (31) that contains the entire intergenic region except for the 12-kb XhoI fragment that harbors the MAR (Fig. 1C; L.D.M., unpublished data). This BAC then served as the acceptor for the same 12-kb XhoI fragment from which regions of interest had been deleted (Fig. 1 C and F). The 78-bp AT-MARKO donor construct was generated by performing PCR on a closed-circular plasmid into which a MAR-containing 1.9-kb EcoR1/PstI fragment was cloned by using primers oriented away from the MAR (primers: positions 148–130 and 227–246; Fig. 2D). For diagnostic purposes (see text), a new SpeI site was engineered into this region by insertion of a C residue at position 144 (Fig. 2D). The product was circularized, and the insert was excised and exchanged with the corresponding WT fragment in the donor XhoI fragment. Maps, detailed protocols, and clones are available on request.
Figure 2.
Defining the minimal MAR-binding sequence. (A) Matrix-halo structures were prepared from CHO and CHOC400 cells as described in Materials and Methods, and loop DNA was removed with a combination of PvuII and HinfI. Equal weights of matrix and loop DNA fractions were separated on an agarose gel and blotted to a nylon membrane. The transfer was then hybridized with a combination of 450- and 379-bp PvuII/HinfI subfragments. The marker is an end-labeled 123-bp ladder (BRL). (B) Matrix/halo structures were prepared from CHOC400 cells and all attached DNA was completely removed with DNaseI. Isolated DNA-free matrices were then incubated with end-labeled fragments from the 3.4-kb PvuII MAR-containing fragment in the presence and absence of cold competitor DNA. The bound radioactive DNA was analyzed by Southern blotting. (C) HinfI subfragments of the 3.4-kb PvuII MAR-binding fragment (22), which were tested for association with the matrix in the in vivo assay illustrated in A (additional probings not shown). (D) Primary sequence of the 378-bp PvuII/HinfI MAR-binding fragment. Relevant restriction enzyme sites are shown above the sequence, and the 78-bp AT-rich deletion in the AT-MARKO cell line is shaded. At position 144, a C was inserted in the donor BAC that gave rise to the AT-MARKO cell line to create an SpeI site for diagnosis of recombinants. The sequence shown is from PCR products obtained from DR8A-7, as well as a subclone from a CHOC400 cosmid library. Sequence was also obtained from the AT-MARKO cell line to confirm deletion of the shaded region.
Transfection and Screening of Potential Recombinants.
Transfections were performed as described (26), after linearizing donor BACs with NotI and SalI. DR-8A7 cells were electroporated with 10 μg of DNA per 108 cells in a volume of 250 μl. DHFR+ cells were selected on minimal medium as described (26), and surviving clones were screened by Southern blotting.
2D Gel Replicon Mapping.
Cells were released from a G1/S mimosine block (27) and sampled 90, 180, 360, and 540 min later (2–3 × 108 cells per sample). Replication intermediates were prepared with EcoRI as described (29), separated on neutral/neutral 2D gels (29, 32), and transferred to Hybond N+ (Amersham Pharmacia). Probes were (i) a mixture of probe 12 (a 0.3-kb BamH1/PvuII fragment) and probe 38 (a 0.47-kb PvuII/XmnI fragment), and (ii) a 1.2-kb BamH1 fragment that recognizes a 6.5-kb fragment in the rhodopsin locus (Fig. 1A; ref. 33).
Fluorescence in Situ Hybridization (FISH) Analysis.
Determination of the kinetics of replication and segregation of the DHFR domain by FISH was performed as described (26), except that, in addition to swelling in 40 mM KCl (neutral FISH), a separate aliquot of cells was swollen in 40 mM KCl, 5 mM glycine, pH 9.5 (alkaline FISH) before fixation (34). Only those alkaline FISH samples that showed no appreciable loss in the number of cells compared with neutral FISH samples were analyzed.
Results
Characterization of the Intergenic MAR.
In a previous study, a 3.4-kb PvuII fragment near the center of the intergenic spacer was shown to be enriched in the matrix-attached DNA fraction of both CHOC400 and CHO cells (labeled M in Fig. 1A; ref. 22). Additional mapping was performed by an in vivo assay to further localize the matrix-binding activity. Matrix/halo structures were prepared from CHO and CHOC400 cells and digested to completion with a combination of PvuII and HinfI, which yields fragments in a narrow size range. The matrix and loop fractions were then analyzed by Southern blotting and hybridization with radioactive probes to identify those selectively retained by the matrix. For example, a combination of 450- and 378-bp PvuII/HinfI probes (Fig. 2C) demonstrates the preferential retention of the 378-bp fragment by the matrix in both cell lines (Fig. 2A; other probings not shown). This fragment has an AT-rich core, within which the sequence AAAT is tandemly repeated almost perfectly nine times (Fig. 2D; 155–190 nt). (Note that matrix preparations normally contain considerable amounts of RNA, which was not removed completely from the sample in Fig. 2A, resulting in faster migration of the DNA fragments. The 378-bp fragment migrates normally in the total and loop DNA samples in the same gel and in the CHOC400 samples.)
The matrix-binding element was further localized within the 378-bp PvuII/HinfI fragment by an in vitro assay in which various 32P-labeled subfragments were incubated with naked matrices in the presence or absence of unlabeled competitor. The bound DNA was isolated, purified, and separated on an agarose gel along with an aliquot of labeled input DNA. As shown in the resulting autoradiogram (Fig. 2B), all of the subfragments have some affinity for the matrix in the absence of nonspecific competitor DNA. However, only subfragment 139–270 remains attached to the matrix in the presence of 10 μg/ml competitor. Because the overlapping fragment 212–378 has little, if any, specific affinity for the matrix, binding can be attributed primarily to the region between residues 139 and 212, which encompasses the AT-rich core.
Deleting Sequences in and Around the MAR Region by the ROKO Approach.
Because we could not predict whether any biologically relevant sequence would be completely contained within the minimal matrix-binding element, a series of MAR deletions was performed, culminating in the removal of a 78-bp sequence encompassing the AT-rich core (Figs. 1F and 2D, shaded region). In the ROKO approach (26), the truncated DHFR gene in the DHFR-deficient variant, DR-8A7 (35), is restored to WT by recombination with a donor BAC that supplies a small region of homologous overlap with the truncated DHFR gene in DR-8A7, the missing 3′ end of the gene, and the downstream region from which the targeted sequence has been deleted (Fig. 1 B and C). DHFR+ cells that have undergone exchanges near regions a and either b or c are selected on minimal medium, which does not supply the thymidine, hypoxanthine, or glycine needed by cells lacking a functional DHFR gene (36). Those DHFR+ survivors that have recombined downstream from the target sequence in region c are then identified by Southern blotting (Fig. 1G).
The sequences deleted from the DHFR locus are as follows (Fig. 1F): (i) an ≈7-kb region corresponding to the 5.0-, 1.1-, and 0.9-kb PstI fragments in which the MAR is centered (7K-MARKO); (ii) a 560-bp BstEI fragment encompassing the 378-bp PvuII/HinfI fragment and ≈150-bp downstream (560-MARKO); (iii) a 78-bp sequence consisting of the AT-rich MAR core (AT-MARKO; shaded region in Fig. 2D); and (iv) a control 2-kb region lying to the right of the MAR that corresponds to 1- and 0.9-kb PstI fragments (ROMKO). Shown in Fig. 1G are the patterns obtained when a mixture of probes 100, 35, and 71 (Fig. 1A) were hybridized with EcoRI digests of DNA from the relevant cell lines (compare with expected diagnostic fragment sizes in Fig. 1F). An AT-MARKO cell line was identified first by Southern analysis of an SpeI digest, which yields a 12-kb fragment in the wild-type configuration, but a 6-kb variant fragment in the AT-MARKO resulting from the introduction of a C at position 144 (Fig. 2D; see Materials and Methods). Sequencing of a PCR product generated with primers flanking the shaded sequence in Fig. 2D confirmed the deletion. The WT controls are the restored derivative and the hemizygote, UA21 (Fig. 1 A and D; ref. 36).
FISH Analysis Suggests That Deletion of the Intergenic MAR Delays Replication of the DHFR Locus.
To determine whether deletion of the MAR has any effect on initiation in the DHFR origin, each of the KO cell lines was first analyzed by a FISH-based assay that has been used to determine replication timing (Fig. 3A; refs. 26 and 37). If early-firing origin activity is negatively affected by deletion of the MAR, then the locus should replicate later than the WT arrangement. Synchronized cells were sampled 90, 180, 360, and 540 min after entry into S-phase cells were swollen at neutral pH and spread on microscope slides. The spreads were hybridized with a combination of (i) digoxygenin-labeled cosmid KD504, which spans the central 45 kb of the intergenic region, and (ii) biotin-labeled cosmid C3B, which contains sequences from the control, early-replicating, rhodopsin origin (15, 33). The hybridization probes were detected with sheep antidigoxygenin and fluorescein-labeled donkey anti-sheep IgG or Texas red-labeled streptavidin, respectively. The numbers of fluorescent dots of each color were then counted microscopically to determine whether, in a given cell, the particular loci had doubled and separated (see Fig. 3A). A replication index was obtained by dividing the number of S-phase cells that display two fluorescent DHFR signals by the number that have doubled one or both early-replicating rhodopsin loci (box G). Representative FISH images and data tabulation can be found in Tables 1–3 and Fig. 5, which are published as supporting information on the PNAS web site, www.pnas.org.
Figure 3.
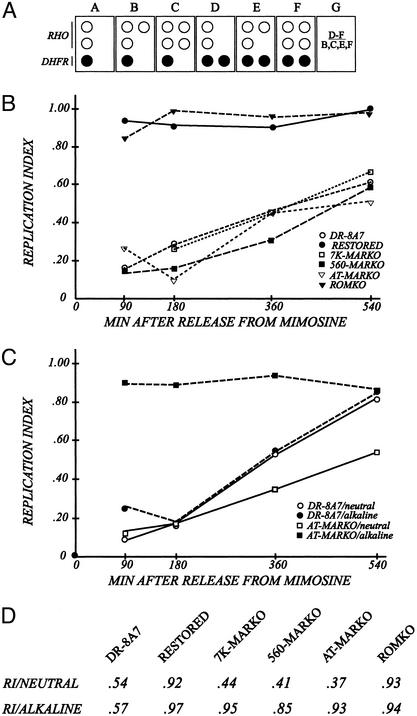
The MARKO deletion variants appear to be very late replicating by the criterion of the FISH-based replication timing assay. (A) Principle of the FISH-based replication timing assay (37). Cells are swollen spread on microscope slides (34, 40), and hybridized with a digoxygenin-labeled cosmid specific for the intergenic region (KD504) and with a biotin-labeled cosmid specific for the control, early-replicating, rhodopsin origin. The respective signals are detected with fluorescein-labeled antidigoxygenin or Texas red-labeled antiavidin, as outlined in Materials and Methods (40). The number of dots of each color is recorded for at least 100 interphase nuclei from each cell line. Boxes B–F represent cells in different stages of the S-period or G2. Box G defines a replication index, which is calculated by dividing the number of cells that have replicated the DHFR locus (as in boxes D–F) by the number that has doubled the rhodopsin control locus (as in boxes B, C, E, and F). (B) Cells were sampled from synchronized cultures at the indicated times and swollen under neutral conditions. Resulting replication indices for each cell line are plotted as a function of time after release from the G1/S mimosine block. (C) Synchronized AT-MARKO and DR8-A7 cell lines were harvested at the indicated times, divided into two, and prepared for the FISH assay under either neutral or alkaline conditions. After hybridization with the mixed DHFR/rhodopsin probe, replication indices were determined. (D) Unsynchronized cultures of each of the indicated cell lines were divided into two and prepared for FISH under either neutral or alkaline swelling conditions. Replication indices were determined as described above.
In Fig. 3B, the calculated replication indices of the various cell lines are plotted as a function of time in the S period. It is clear that the DHFR locus in most DR-8A7 cells (Fig. 3B, open circles) does not replicate until very late in S phase relative to the rhodopsin control, in agreement with earlier FISH and 2D gel studies (26). However, when the missing 3′ part of the DHFR gene is restored by homologous recombination at positions a and b (Fig. 1 B–D), the locus reverts to an early-replicating phenotype, with most cells doubling the locus at the same time as rhodopsin (i.e., with replication indices for the restored cell line close to unity at all time points; Fig. 3B, filled circles). The DHFR locus in the ROMKO control was similarly early replicating (Fig. 3B, filled triangles). In contrast, the DHFR locus in the 7K-MARKO, 560-MARKO, and AT-MARKO cell lines did not appear to double until very late in S phase (540 min). This finding initially suggested that deletion of the MAR might inhibit early-firing origin activity in the intergenic spacer.
2D Gel Analysis Suggests That DHFR Origin Activation Is Not Affected by the MAR Deletion.
To further test the possibility that the MAR deletion might affect initiation per se, samples from the synchronized cell preparations were also analyzed by a 2D gel replicon mapping technique (32). Replication intermediates were prepared at each time point by using EcoRI to digest the DNA, and after separation on a 2D gel, were analyzed with a radiolabeled probe specific for ori-β, one of the two most active initiation sites in the intergenic spacer (15–17, 19, 21). The principle of the 2D gel method is outlined in Fig. 4A Right.
Figure 4.
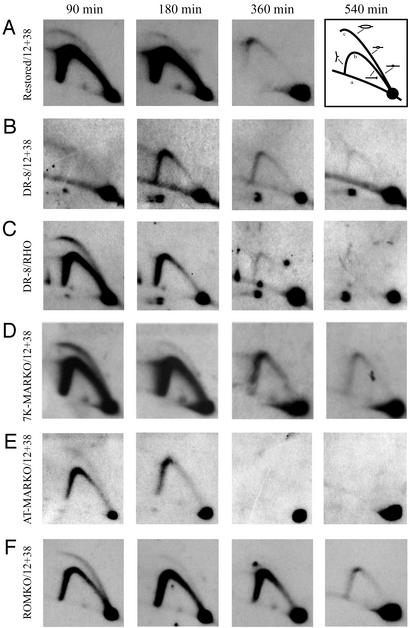
2D gel analysis of the MARKO variants suggests that deletion of the MAR has no effect on either the efficiency or timing of initiation in the DHFR locus. Replication intermediates were purified from aliquots of the same synchronized cell populations analyzed by the FISH-based assay in Fig. 3B. After digestion with EcoRI, intermediates were separated on a neutral/neutral 2D gel, transferred to Hybond N+, and hybridized with a combination of probes 12 and 38, which are specific for a fragment containing ori-β. Each cell line was additionally analyzed with a probe specific for the early-replicating rhodopsin standard (shown here for the DR8-A7 variant only). The principle of the method is outlined in A Right. Replication intermediates are separated in the first dimension according to molecular mass, which for any fragment will vary from 1n (unreplicated) to just less than 2n. The first dimension lane is excised, turned through 90°, and separated in the second dimension according to both mass and shape (32). Linear nonreplicating fragments trace a diagonal (curve a). If a fragment is replicated passively by a fork originating from a site outside of the fragment, it will display a single fork arc (curve b). However, if the fragment contains a centered initiation site, it will display an arching bubble arc that extends from the 1n to the 2n positions (curve c).
In the WT restored control (Fig. 1D), a composite pattern consisting of a bubble arc and a single fork arc is observed in both the 90- and 180-min time points (Fig. 4A). This pattern characterizes fragments located in an early-firing, broad, initiation zone, because any given fragment will sometimes be replicated from an internal initiation site (contributing to the bubble arc) but most often by forks emanating from sites in a neighboring fragment in the same zone (contributing to the single fork arc) (12). By 360 min, very few single forks are detected in the ori-β region, showing that most copies of the locus have finished replicating (13, 14, 27). In contrast, the DR-8A7 cell line (the late-replicating control) displays neither a bubble arc nor a single fork arc in the 90-min sample (Fig. 4B). A single fork arc is visible at 180 and 360 min, which diminishes by 540 min. When the same transfer was stripped and rehybridized with a probe for the early-firing rhodopsin control origin, an early-firing pattern was detected (Fig. 4C), showing that the cells were well synchronized in this experiment. Thus, the 2D gel data on the restored and DR8-A7 cell lines are in agreement with results from the FISH-based assay, indicating that the origin in DR8-A7 no longer fires in early S phase, but can be restored to an early-firing phenotype by restoration of the deletion (Fig. 1 B and D).
Surprisingly, when the three MARKO cell lines were analyzed at the same time points, all three were seen to initiate replication in early S phase and replicate the locus with similar kinetics as the early replicating restored control and the ROMKO cell line from which a sequence lying to the right of the MAR was deleted (Fig. 4 D–F; data for the 560-MARKO not shown). (Note that the films for the AT-MARKO cell line are somewhat underexposed relative to those for the other cell lines, but the overall pattern is very similar to that of the early-firing rhodopsin control.)
The Intergenic MAR Is Required for Local Chromatid Separation.
Thus, there is a clear dissociation between results of the FISH-based assay and 2D gels when applied to the MARKOs, but not to any other cell lines analyzed in this or other studies from our laboratory (e.g., ref. 26). The source of this discrepancy could be related to the fact that the FISH-based replication timing assay depends for its validity not only on duplication of a locus, but also on enough local separation of chromatids to resolve the two fluorescent dots by microscopy. In fact, previous studies have shown that structural features of some chromosomal loci can result in the inability to discern two dots in the FISH-based assay, even though independent replication assays show that the locus has doubled (38, 39). To test whether the MARKO cell lines might be impaired in their ability to separate daughter strands in the vicinity of the DHFR locus, the FISH-based assay was repeated on nuclei prepared for fixation and spreading either by the standard neutral swelling conditions (40) or an alkaline swelling regimen that is known to decompact chromatin and expand the nuclei (34, 41).
FISH data on synchronized cultures of the DR8-A7 and AT-MARKO cell lines are presented in Fig. 3C. The DHFR locus in DR-8A7 appears to replicate at the same time in mid- to late S phase regardless of whether nuclei are prepared under neutral or alkaline conditions (Fig. 3C, open and filled circles, respectively). However, the AT-MARKO cell line displays either a late- or an early-replicating phenotype, depending on whether nuclei were prepared under neutral or alkaline conditions, respectively (Fig. 3C, open and filled squares). The 7K-MARKO and 560-MARKO cell lines behaved identically to the AT-MARKO variant (data not shown). Furthermore, a comparison with unsynchronized cells swollen under the two conditions (Fig. 3D) recapitulates the data on synchronized cultures (Fig. 3C): the replication indices of the three MARKOs are similar to that of DR-8A7 (≈0.5) when log cells were swollen at neutral pH, but are similar to WT (i.e., are close to unity) when swollen in alkali.
Discussion
In the present study, we have used in vivo and in vitro assays to narrow the minimal matrix-binding element to a 131-bp fragment containing nine contiguous AAAT repeats (fragment 139–270; Fig. 2B). If a demonstrable phenotype were to result from the deletion of this element, a failure to attach to the matrix would likely be responsible. Although deletion of either the largest or smallest MAR-containing fragments had no demonstrable effect on initiation in the intergenic origin, a visible separation of daughter DNA strands was dramatically delayed until just before mitosis. Importantly, however, all of the MARKO cell lines display two fluorescent dots at this locus on mitotic chromosomes (L.D.M., unpublished observations).
We have demonstrated a clear phenotype associated with deletion of a matrix attachment element from its native chromosomal position, although effects of certain MARs on the activity of colinear genes in transfected DNA have been reported (e.g., refs. 42 and 43). An important additional conclusion is that the in vivo and in vitro assays used to define the binding element appear to measure a physiologically relevant interaction that can be shown to be so by in loco mutagenesis.
It is somewhat surprising that deletion of the intergenic MAR has no demonstrable effect on initiation of replication per se, in view of the proposed association of replication forks and active replication origins with the matrix (24, 44). Rather, the MAR seems to be involved in the structural organization of the replicating genome. Because topoisomerase II might be involved in anchoring DNA loops to the nuclear matrix (45, 46), and because some MARs contain topoisomerase II cleavage sites (47), it has been proposed that MARs could be involved in relieving torsional stress that accumulates during replication and/or in deconcatenating daughter strands when forks from adjacent origins meet (45–47). However, the AAAT repeat in the intergenic element does not conform to a proposed topoisomerase II consensus sequence (47), nor have we detected stalled termination structures in the region of the MAR deletion on 2D gels (P.A.D., unpublished observations).
We consider it more likely that the MAR is involved directly in interphase chromatid cohesion and/or separation (reviewed in ref. 48). Cohesion is mediated by cohesin (49), which is loaded onto chromosomes during replication, at least in yeast (48). Cohesin prevents extracentromeric chromatid separation until the majority of the cohesin is lost from chromosomes before metaphase (48). Perhaps in mammalian cells, the chromatids begin to separate locally soon after replication forks pass, but are held together periodically by cohesin until the early stages of mitosis. In this scenario, the matrix itself presumably would also be duplicating and separating concomitantly, and periodic attachment of DNA to the matrix at noncohesed sites would provide the mechanical leverage for local chromatid separation. A related possibility is that some MARs might correspond to periodic binding sites for a factor that can reverse local cohesin-mediated chromatid interactions.
Finally, MARs can affect chromatin compaction over relatively large distances (50), suggesting that the intergenic MAR may be involved indirectly in sister chromatid separation. In this regard, AT-hook proteins have been identified that bind with high affinity to some MARs in vitro (51) and, when expressed in Drosophila larvae, have been shown to affect chromatin architecture (52). Having refined the critical matrix-binding element in the DHFR domain to a 78-bp sequence in the present study will help to distinguish among the many mechanistic possibilities by helping to identify cognate binding partners.
Supplementary Material
Acknowledgments
We thank Lawrence Chasin and Adelaide Carothers (Columbia University, New York) for sharing the UA21 and DR8-A7 cell lines, respectively, with us. We also thank Carlton White and Kevin Cox for technical help with this project and Swati Saha for critical reading of the manuscript. This work was supported by National Institutes of Health Grants RO1 GM26108 (to J.L.H.) and RO1 GM55675 (to P.A.D.).
Abbreviations
- DHFR
dihydrofolate reductase
- MAR
matrix attachment region
- RO
reach out
- KO
knock out
- CHO
Chinese hamster ovary
- BAC
bacterial artificial chromosome
- FISH
fluorescence in situ hybridization
References
- 1.Marsden M P, Laemmli U K. Cell. 1979;17:849–858. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- 2.Saitoh Y, Laemmli U K. Cell. 1994;76:609–622. doi: 10.1016/0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 3.Boveri T. Arch Zellforschung. 1909;3:181–191. [Google Scholar]
- 4.Cremer T, Kreth G, Koester H, Fink R H, Heintzmann R, Cremer M, Solovei I, Zink D, Cremer C. Crit Rev Eukaryotic Gene Expression. 2000;10:179–212. [PubMed] [Google Scholar]
- 5.Mahy N L, Perry P E, Gilchrist S, Baldock R A, Bickmore W A. J Cell Biol. 2002;157:579–589. doi: 10.1083/jcb.200111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volpi E V, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell R D, Goldsworthy M, Powis S H, Ragoussis J, et al. J Cell Sci. 2000;113:1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- 7.Nickerson J A, Krockmalnic G, Wan K M, Penman S. Proc Natl Acad Sci USA. 1997;94:4446–4450. doi: 10.1073/pnas.94.9.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma H, Siegel A J, Berezney R. J Cell Biol. 1999;146:531–542. doi: 10.1083/jcb.146.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirkovitch J, Mirault M E, Laemmli U K. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 10.Tan J H, Wooley J C, LeStourgeon W M. Mol Biol Cell. 2000;11:1547–1554. doi: 10.1091/mbc.11.5.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pederson T. Mol Biol Cell. 2000;11:799–805. doi: 10.1091/mbc.11.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaughn J P, Dijkwel P A, Hamlin J L. Cell. 1990;61:1075–1087. doi: 10.1016/0092-8674(90)90071-l. [DOI] [PubMed] [Google Scholar]
- 13.Dijkwel P A, Vaughn J P, Hamlin J L. Mol Cell Biol. 1991;11:3850–3859. doi: 10.1128/mcb.11.8.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dijkwel P A, Vaughn J P, Hamlin J L. Nucleic Acids Res. 1994;22:4989–4996. doi: 10.1093/nar/22.23.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dijkwel P A, Wang S, Hamlin J L. Mol Cell Biol. 2002;22:3053–3065. doi: 10.1128/MCB.22.9.3053-3065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anachkova B, Hamlin J L. Mol Cell Biol. 1989;9:532–540. doi: 10.1128/mcb.9.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leu T H, Hamlin J L. Mol Cell Biol. 1989;9:523–531. doi: 10.1128/mcb.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handeli S, Klar A, Meuth M, Cedar H. Cell. 1989;57:909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- 19.Pelizon C, Diviacco S, Falaschi A, Giacca M. Mol Cell Biol. 1996;16:5358–5364. doi: 10.1128/mcb.16.10.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burhans W C, Vassilev L T, Caddle M S, Heintz N H, DePamphilis M L. Cell. 1990;62:955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi T, Rein T, DePamphilis M L. Mol Cell Biol. 1998;18:3266–3277. doi: 10.1128/mcb.18.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dijkwel P A, Hamlin J L. Mol Cell Biol. 1988;8:5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pemov A, Bavykin S, Hamlin J L. Proc Natl Acad Sci USA. 1998;95:14757–14762. doi: 10.1073/pnas.95.25.14757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dijkwel P A, Wenink P W, Poddighe J. Nucleic Acids Res. 1986;14:3241–3249. doi: 10.1093/nar/14.8.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carri M T, Micheli G, Graziano E, Pace T, Buongiorno-Nardelli M. Exp Cell Res. 1986;164:426–436. doi: 10.1016/0014-4827(86)90041-8. [DOI] [PubMed] [Google Scholar]
- 26.Kalejta R F, Li X, Mesner L D, Dijkwel P A, Lin H B, Hamlin J L. Mol Cell. 1998;2:797–806. doi: 10.1016/s1097-2765(00)80294-4. [DOI] [PubMed] [Google Scholar]
- 27.Dijkwel P A, Hamlin J L. Mol Cell Biol. 1992;12:3715–3722. doi: 10.1128/mcb.12.9.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dijkwel P A, Hamlin J L. Mol Cell Biol. 1995;15:3023–3031. doi: 10.1128/mcb.15.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosca P J, Dijkwel P A, Hamlin J L. Mol Cell Biol. 1992;12:4375–4383. doi: 10.1128/mcb.12.10.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cockerill P N, Garrard W T. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 31.Kim U-J, Birren S W, Slapak T, Mancino V, Boysen C, Kang H-L, Simon M I, Shizuya H. Genomics. 1996;34:213–216. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- 32.Brewer B J, Fangman W L. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 33.Dijkwel P A, Mesner L D, Levenson V V, d'Anna J, Hamlin J L. Exp Cell Res. 2000;256:150–157. doi: 10.1006/excr.2000.4809. [DOI] [PubMed] [Google Scholar]
- 34.Yokota H, van den, E. G, Mostert M, Trask B J. Genomics. 1995;25:485–491. doi: 10.1016/0888-7543(95)80049-r. [DOI] [PubMed] [Google Scholar]
- 35.Jin Y, Yie T A, Carothers A M. Carcinogenesis. 1995;16:1981–1991. doi: 10.1093/carcin/16.8.1981. [DOI] [PubMed] [Google Scholar]
- 36.Urlaub G, Kas E, Carothers A M, Chasin L A. Cell. 1983;33:405–412. doi: 10.1016/0092-8674(83)90422-1. [DOI] [PubMed] [Google Scholar]
- 37.Kitsberg D, Selig S, Keshet I, Cedar H. Nature. 1993;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- 38.Hansen R S, Canfield T K, Gartler S M. Hum Mol Genet. 1995;4:813–820. doi: 10.1093/hmg/4.5.813. [DOI] [PubMed] [Google Scholar]
- 39.Kawame H, Gartler S M, Hansen R S. Hum Mol Genet. 1995;4:2287–2293. doi: 10.1093/hmg/4.12.2287. [DOI] [PubMed] [Google Scholar]
- 40.Trask B J. In: Mapping Genomes, Genome Analysis: A Laboratory Manual Series. Birren B, Green E, Hietar P, Klaphole S, Myers R, Riethman H, Roskams J, editors. Vol. 4. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 303–413. [Google Scholar]
- 41.Heng H H, Squire J, Tsue L C. Proc Natl Acad Sci USA. 1992;89:9509–9513. doi: 10.1073/pnas.89.20.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klehr D, Maass K, Bode J. Biochemistry. 1991;30:1264–1270. doi: 10.1021/bi00219a015. [DOI] [PubMed] [Google Scholar]
- 43.Phi-van L, Stratling W H. Nucleic Acids Res. 1999;27:3009–3017. doi: 10.1093/nar/27.15.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaughn J P, Dijkwel P A, Mullenders L H, Hamlin J L. Nucleic Acids Res. 1990;18:1965–1969. doi: 10.1093/nar/18.8.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adachi Y, Kas E, Laemmli U K. EMBO J. 1989;8:3997–4006. doi: 10.1002/j.1460-2075.1989.tb08582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berrios M, Osherof N, Fisher P A. Proc Natl Acad Sci USA. 1985;82:4142–4146. doi: 10.1073/pnas.82.12.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miassod R, Razin S V, Hancock R. Nucleic Acids Res. 1997;25:2041–2046. doi: 10.1093/nar/25.11.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nasmyth K. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- 49.Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 51.Strick R, Laemmli U K. Cell. 1995;83:1137–1148. doi: 10.1016/0092-8674(95)90140-x. [DOI] [PubMed] [Google Scholar]
- 52.Girard F, Bello B, Laemmli U K, Gehring W J. EMBO J. 1998;17:2079–2085. doi: 10.1093/emboj/17.7.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foreman P K, Hamlin J L. Mol Cell Biol. 1989;9:1137–1147. doi: 10.1128/mcb.9.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crouse G F, Leys E J, McEwan R N, Frayne E G, Kellems R E. Mol Cell Biol. 1985;5:1847–1858. doi: 10.1128/mcb.5.8.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.