Abstract
Wnt/β-catenin signaling plays key roles in several developmental and pathological processes. Domains of Wnt expression have been extensively investigated in the mouse, but the tissues receiving the signal remain largely unidentified. To define which cells respond to activated β-catenin during mammalian development, we generated the β-catenin-activated transgene driving expression of nuclear β-galactosidase reporter (BAT-gal) transgenic mice, expressing the lacZ gene under the control of β-catenin/T cell factor responsive elements. Reporter gene activity is found in known organizing centers, such as the midhindbrain border and the limb apical ectodermal ridge. Moreover, BAT-gal expression identifies novel sites of Wnt signaling, like notochord, endothelia, and areas of the adult brain, revealing an unsuspected dynamic pattern of β-catenin transcriptional activity. Expression of the transgene was analyzed in mutant backgrounds. In lipoprotein receptor-related protein 6-null homozygous mice, which lack a Wnt coreceptor, BAT-gal staining is absent in mutant tissues, indicating that BAT-gal mice are bona fide in vivo indicators of Wnt/β-catenin signaling. Analyses of BAT-gal expression in the adenomatous polyposis coli (multiple intestinal neoplasia/+) background revealed βcatenin transcriptional activity in intestinal adenomas but surprisingly not in normal crypt cells. In summary, BAT-gal mice unveil the entire complexity of Wnt/β-catenin signaling in mammals and have broad application potentials for the identification of Wnt-responsive cell populations in development and disease.
Keywords: transgenic mice‖lipoprotein receptor-related protein 6‖adenomatous polyposis coli‖colon cancer
Proteins of the Wnt family are secreted factors regulating cell proliferation, fate, and behavior in contexts ranging from early embryonic development to stem cell homeostasis (1–3). Wnts exert many of their effects by activating the expression of target genes through a pathway controlled by β-catenin, a protein originally identified as a component of the cadherin cell adhesion complex and later shown to be a key mediator for Wnt signaling. Wnt ligands bind a receptor complex composed of Frizzled and lipoprotein receptor-related protein (LRP) family members (4). This complex leads to the activation of downstream effectors ultimately promoting β-catenin accumulation in the cytoplasm (3). In the absence of Wnt stimulation, free β-catenin levels are kept low by a β-catenin degrading complex that includes the adenomatous polyposis coli (APC) and Axin tumor suppressors (1, 5). APC/axin activities are inhibited in the presence of Wnt signaling, leading to β-catenin accumulation. Stabilized β-catenin translocates into the nucleus, where it activates its target genes by binding to sequence-specific transcription factors of the lymphoid enhancer factor/T cell factor (LEF/TCF) family (6).
Wnt proteins are involved in cell-to-cell signaling in almost every process of mammalian development. To fully understand how this family of growth factors can elicit such diverse cellular responses, methods are needed to identify cells in which the Wnt pathway is active at a given time during development and in adult tissues. Although contributing important information, genetic analyses do not provide an unequivocal solution to this problem for several reasons.
First, the Wnt pathway entails a complex set of molecules: in humans, for example, at least 19 Wnt factors and 10 Frizzled receptors have been identified. Ligands and receptors are distributed in complex, often overlapping expression patterns (for details, see the Wnt web site by the Nusse laboratory, www.stanford.edu/∼rnusse). Second, not all Wnt–Frizzled combinations end up with the activation of the canonical transduction cascade, because two β-catenin independent pathways have been identified (4). Third, Wnt signaling is modulated extracellularly by a variety of secreted antagonists that bind either to the ligands or the LRP coreceptors inhibiting their activity (4). Finally, it must be stressed that the identification of cells with nuclear-localized β-catenin does not imply functional signaling. Indeed, the activities of β-catenin are constantly held in check even within the nucleus; at least in some cellular contexts, LEF/TCF members recruit transcriptional inhibitors and maintain the promoter in an OFF state even in the presence of activated β-catenin (7, 8).
Here we report the generation of mouse lines [β-catenin-activated transgene driving expression of nuclear β-galactosidase reporter (BAT-gal)] that can be used as a general read-out of β-catenin activity. In several tissues, the expression of this transgene mirrors known domains of Wnt signaling. Inhibition of the canonical Wnt pathway in LRP6 mutant mice results in the concomitant lack of BAT-gal expression, indicating that these transgenic mice are reliable reporters of Wnt/β-catenin activity. We then explored BAT-gal potential in revealing the status of Wnt signaling in cancer cells. In humans, mutations of the APC tumor suppressor are associated with the formation of intestinal tumors (1, 9). To gain insight into the role of β-catenin signaling in normal and neoplastic colon, we assayed BAT-gal expression in the APC [multiple intestinal neoplasia (Min)/+] mouse (10), a murine model of the human familial adenomatous syndrome (OMIM 175100). In APC(Min/+)/BAT-gal crosses, we detected high levels of β-catenin-induced transcription in the epithelium of adenomas. Normal intestinal epithelium, however, did not stain for lacZ, indicating that β-catenin activity is tightly controlled during normal crypt homeostasis.
Materials and Methods
Generation and Analyses of Transgenic Mice.
BAT-gal was constructed by fusing seven TCF/LEF-binding sites upstream of a 0.13-kb fragment containing the minimal promoter–TATA box of the gene siamois (11). Details of the synthesis of this construct and the initial controls for β-catenin-specific activation of BAT-gal can be found in Supporting Text and Fig. 6, which are published as supporting information on the PNAS web site, www.pnas.org. Transgenic BAT-gal mouse lines and embryos were produced from B6D2F1 females mated with B6D2F1 males (Charles River Breeding Laboratories) by using standard procedures (12). DNA was microinjected into the pronuclei of one-cell embryos, and the surviving embryos were implanted into CD1 pseudopregnant foster mothers. Transgenic mice were identified by analysis of genomic DNA from tail biopsies by PCR and Southern blot to detect lacZ. lacZ primers: forward, 5′-CGGTGATGGTGCTGCGTTGGA-3′; reverse, 5′-ACCACCGCACGATAGAGATTC-3′. Other experimental procedures are published as supporting information on the PNAS web site.
Results and Discussion
Generation of Wnt/β-Catenin Reporter Mice.
To identify Wnt/β-catenin responding cells during mammalian development, we generated a new transgenic mouse strain, here referred to as BAT-gal, which drives expression of nuclear β-galactosidase under the control of multimerized LEF/TCF-binding sites. In vitro, this construct leads to a specific and robust reporter gene activation on Wnt/β-catenin stimulation (see Fig. 6). To assay the efficacy and specificity of BAT-gal expression in vivo, we first performed transient analyses of mouse embryos injected with BAT-gal DNA at the one-cell stage and collected at embryonic day (E)8.5 and E10.5 for 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) staining. At E8.5, transgenic embryos displayed positivity at high frequency in the midbrain, a hallmark of Wnt-1 activity (13) and, at E10.5, showed strong staining in the central nervous system, correlating with the expression pattern of several Wnts (data not shown, see Figs. 1E and 2A) (14). To check for β-catenin-independent reporter activity, we injected a construct with mutated LEF/TCF-binding sites, which resulted in no expression in any embryo (n = 77, data not shown). From this control, we concluded that BAT-gal transcription reflects endogenous Wnt/β-catenin activity, because it relies exclusively on functional LEF/TCF enhancer elements, with the minimal promoter and flanking sequences having no tissue-specific effect on transcription. Next, we generated three BAT-gal mouse lines that were bred to F2 and analyzed for whole-mount X-Gal staining at several stages of development. The three lines displayed qualitatively identical expression patterns, although quantitative differences could be observed, likely reflecting variations in the transgene copy number or chromosomal context.
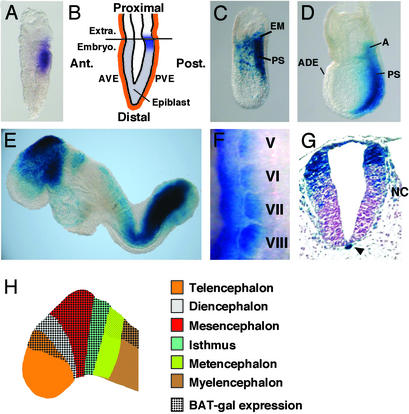
Figure 1.
BAT-gal expression during early development. (A) First expression in the posterior epiblast of prestreak stage embryo detected by in situ hybridization with antisense β-galactosidase probe. (B) Diagram indicating the location of lacZ-positive cells (in blue) at the prestreak stage. Ant., anterior; Post., posterior; Extra., extraembryonic region; Embryo., embryonic region; AVE, anterior visceral endoderm; PVE, posterior visceral endoderm. (C and D) Whole-mount X-Gal staining of embryos at early- and late-streak stage, respectively. A, allantois; ADE, anterior definitive endoderm; PS, primitive streak. (E) E8.5 embryo shows staining in the fore-, mid-, and hindbrain, tailbud, newly formed somites, and cardiac region. (F) Lateral view of lacZ in situ hybridization of 9.5-days postcoitum somites V–VIII (anterior is up and dorsal is left). Note the progressive restriction of BAT-gal staining in the dorsal medial portion of the dermomyotome. (G) Cross section through the caudal spinal cord. Arrowhead points to the notochord. Note the migrating neural crest cells (NC). (H) Diagram indicating the expression of BAT-gal in different regions of the brain at E8.5.
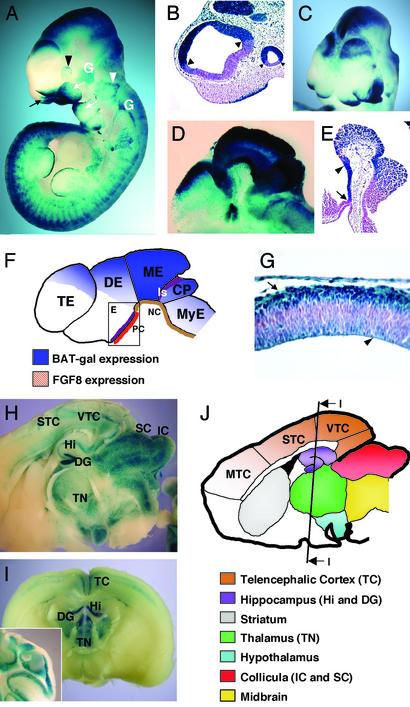
Figure 2.
Expression of BAT-gal in the developing brain and neural tissue. (A) Lateral view of whole-mount stained E10.5 embryo. Indicated are nasal folds (black arrow), optic vesicle (black arrowhead), otic vesicle (white arrowhead), and first and second branchial arches (white arrows). Note positive staining of cranial nerve ganglia (white, G). (B) Histological section through telencephalic and optic vesicles. Arrowheads point to the sharp borders of BAT-gal expression. (C) Anterior–top view of X-Gal-stained embryo (E9.5). Note the enhanced expression of BAT-gal in the dorsal side of the telencephalic hemispheres. (D) Sagittal section of head from a whole-mount-stained E10.5 embryo. Note expression in fore-, mid-, and hindbrain. (E) Section through ventral diencephalon showing lacZ expression in the region comprised between the infundibulum (arrow) and the mammilary body (arrowhead). (F) Schematic diagram of the β-galactosidase-positive areas of the developing brain at E10.5. PC, prechordal plate; NC, notochord; Is, isthmus; TE, telencephalon; DE, diencephalon; ME, mesencephalon; CP, cerebellar plate; MyE, myelencephalon. The square in the diencephalons is a diagram for E. (G) Section of telencephalic vesicle at E11.5. Staining is restricted in the cortical (arrow) and ventricular (arrowhead) layers of the tissue. (H and I) Expression of the BAT-gal transgene in the adult encephalon. Vibratome sagittal (H) and coronal (I) sections of the organ are shown. Staining is detected in distinct areas of the adult brain (J). Note the unprecedented detection of Wnt signaling in the sensory (STC) and visual (VTC) telencephalic cortex, but not motor cortex (MTC). (I Inset) A section of the adult cerebellum showing staining in the granule cells.
Expression of BAT-gal During Early Development.
The onset of BAT-gal transcription is detected in pregastrulating embryos (Fig. 1A, E6.0) in the posterior side of the proximal epiblast, at the junction between the embryonic and extraembryonic ectoderm (Fig. 1B). The anterior and posterior visceral endoderm appear negative. At the beginning of gastrulation, lacZ staining is found in extraembryonic mesoderm and in the primitive streak, where it remains until the end of gastrulation (Fig. 1C). Therefore, BAT-gal expression is one of the earliest markers of regionalization in the mouse epiblast. The identification of functional Wnt signaling at the border between visceral endoderm and embryonic/extraembryonic ectoderm is intriguing, given the inductive interactions occurring between these germ layers of the egg cylinder (15, 16). The Wnt ligand responsible for such early β-catenin stabilization is unknown. In vertebrates, β-catenin signaling is required for the establishment of the organizer tissue (17). It is noteworthy that the BAT-gal expression domain corresponds to cells endowed with powerful signaling activities, given that, in transplantation experiments, this region of the embryo displays cell fate and tissue-inducing activities characteristic of vertebrate organizers (18). It is therefore tempting to speculate a conservation of β-catenin signaling in mammalian axis specification. As gastrulation proceeds, Wnt/β-catenin signaling remains localized at the posterior side of the embryo, in the primitive streak and node, but not in the prechordal plate and anterior definitive endoderm (Fig. 1D).
At E8.5, expression is detected in paraxial mesoderm (Fig. 1E). As somite differentiation proceeds, lacZ expression gradually increases in the dorsal domain of the dermomyotome and then fades in more anterior somites (Fig. 1 E and F). Previous work has demonstrated that Wnts diffusing from the neural tube are instrumental for the establishment of the myogenic lineage (19, 20). Our data indicate that Wnt ligands act directly on the somite and signal through the β-catenin pathway.
Histological analyses of E9.5 embryos reveal striking lacZ expression in the notochord, but not in the floor plate, suggesting that a still unknown Wnt molecule might signal to this organizer tissue derivative (Fig. 1G).
Expression in the Developing and Adult Neural Tissue.
Many Wnt genes, Wnt extracellular antagonists, and β-catenin modulators are expressed in the developing central nervous system, often in overlapping patterns (14, 21). Such redundancy complicates the genetic analyses, because the contribution of each Wnt ligand to β-catenin stabilization is difficult to sort out. Wnt-responsive cell populations are first detected in the dorsal midbrain at the one-somite stage (data not shown), and then lacZ staining spreads anteriorly, in the presumptive diencephalon and telencephalon and posteriorly into the prospective isthmus (Fig. 1E and the corresponding diagram in Fig. 1H). Strong staining is observed in an extended domain along the dorsal, lateral, and ventral midbrain. Expression in metencephalon appears less intense than in the myelencephalon and spinal cord (Fig. 1E). Perhaps this reflects quantitative differences in Wnt signaling within the hindbrain, due to the gaps of Wnt-1 and -3a expression in this region (22).
The final regional distribution of β-catenin signaling can be well appreciated by E10.5 (Fig. 2 A–F and data not shown). As shown in Fig. 2B, BAT-gal expression in telencephalon displays sharp anterior and dorsoventral boundaries; a considerable enhancement is found in the dorsal telencephalon (Fig. 2C), in a region including the prospective hippocampus (23). In diencephalon, expression is strong in the dorsal side (Fig. 2D), whereas in the ventral side, staining appears in a discrete domain extending from the infundibulum to the mammilary body (Fig. 2E). In this location, β-catenin activity overlaps with an FGF8 expression profile, abutting the rostral extension of the prechordal plate (Fig. 2F) (24). This association of Wnt signaling and FGF8 expression, already noted in the isthmus, may define a new inducing center within the diencephalon. lacZ expression persists in mesencephalon and is turned on in the cerebellar plate, but it is much lower in myelencephalon (Fig. 2 A and D). In the spinal cord, lacZ expression is localized in the dorsal neural tube (Fig. 1G) and progressively decreases after E10.5, becoming undetectable by E12.5, except in the more caudal regions (Fig. 2A).
Histological analysis shows that during neural differentiation of telencephalon, staining decreases in cells of the ventricular layer, while increasing in postmitotic cells of the cortical layer (Fig. 2G). This implies a potential role of Wnt/β-catenin activity in the differentiation of cortical neurons (23). In the adult, the encephalon is the most active tissue for Wnt signaling. The most evident Wnt-responsive tissues include dentate gyrus, the hippocampus, the sensory telencephalic cortex, several thalamic nuclei, collicula, and the cerebellar cortex (Fig. 2 H–J). In general, specific groups of cells are labeled within these sites. In the telencephalic cortex, for example, only layers 2, 3, and 6 are positive (data not shown). Thus, the pattern of Wnt/β-catenin activity in the adult brain is unexpectedly complex and likely mirrors the several functions that have been suggested for Wnt signaling in late neuronal development and maintenance, including synapse formation and plasticity, neurotrophin transcription, and cell proliferation (25).
X-Gal staining is found in the migratory neural crest cells and in their derivatives, such as cranial nerves and the dorsal root ganglia (Figs. 1G and 2A). Neural crest cells devoid of Wnt signaling undergo massive apoptosis (26). Thus, our data support the view of Wnt/β-catenin signaling as a positive modulator of neural crest cell survival (27).
Organogenesis.
Reporter gene activity is apparent in the cardiac region (Fig. 1E), where it persists up to E10.5. This domain of expression includes cells of the septum transversum mesenchyme forming short villous structures contacting the myocardium (Fig. 3A). Curiously, it has been proposed that similar cells give rise to part of the epicardium (28).
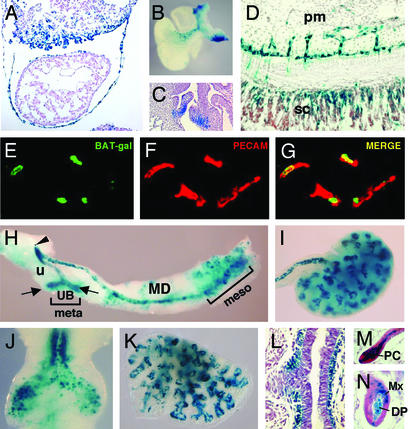
Figure 3.
Expression of the BAT-gal transgene during organogenesis. (A) Section of heart and septum transversum of E9.5 embryo. (B) Whole-mount staining of heart and large vessels. Ventral side is to the left. (C) Section of E16.5 embryo showing staining of aortic semilunar valves. (D) lacZ-positive endothelial cells of vessels invading the spinal cord (sc) from the perineural mesenchyme (pm) at E12.5. (E–G) Costaining of blood vessels of the mesenchyme surrounding the spinal cord for the endothelial marker platelet endothelial cell adhesion molecule (PECAM) and β-galactosidase. (E) Expression of the BAT-gal transgene driving nuclear β-galactosidase. (F) PECAM localization identifies endothelial cells. (G) Merge of the previous pictures. (H) Genitourinary organs. Epithelia of mesonephros (meso), mesonephric duct (MD), ureter (u), ureteral buds (UB, arrows), and urogenital sinus (arrowhead) express lacZ, whereas the mesenchyme of metanephros (meta) does not. (I) E13.5, kidney. (J) E11.5, trachea and main bronchi. (K) E13.5, lung. (L) E13.5, section of bronchus showing positive X-Gal reaction in the mesenchyme. (M and N) Histological sections of the skin. Postnatal day 4, hairs in active anagen phase show staining in dermal papilla (DP), matrix (Mx), and precortex (PC).
A novel domain of lacZ expression starts in mesenchymal cells at E10.5 in the ventral side of the outflow-tract and aortopulmonary bifurcation and persists up to E12.5 (Fig. 3B). At E10.5, scattered X-Gal positive cells are found in the cardiac cushions. This expression domain is maintained during cardiac valve morphogenesis and persists up to late gestation (Fig. 3C).
In blood vessels, expression is strong only at distinct sites of neoangiogenesis, such as the endothelial tubes invading the central nervous system from the perineural mesenchyme (starting at E10.5; Fig. 3D). This notion is supported by the colocalization of β-galactosidase and platelet endothelial cell adhesion molecule, an endothelium-specific marker, revealed by double immunohistochemistry labeling and confocal microscopy (Fig. 3 E–G), providing evidence that endothelial cells of some tissues respond positively to Wnt-signaling. A link between Wnt signaling and blood vessel morphogenesis has also been suggested from studies on the angiogenesis inhibitor endostatin (29), raising the intriguing possibility that Wnt signaling might be associated with vascular development and remodeling.
Epithelial–Mesenchymal Interactions.
The development of vertebrate organs requires reciprocal interactions between epithelial and mesenchymal cells that need to exchange inductive and permissive signals to coordinate the genetic programs of cell proliferation, survival, migration, and differentiation. Wnt signals are often key mediators of these interactions. Reproducible expression of the BAT-gal reporter was detected in all structures outgrowing from the main body axis, like ear pinnae, frontonasal process, branchial arches, and genital eminence (Fig. 2 A and C and data not shown).
Whole-mount and histological analyses of transgenic limb buds reveal weak lacZ expression in the lateral plate mesoderm at the site of bud formation starting at E8.5 (data not shown) and by E9.5, staining also appears in the apical ectodermal ridge (see Figs. 2A and 4 C and E).
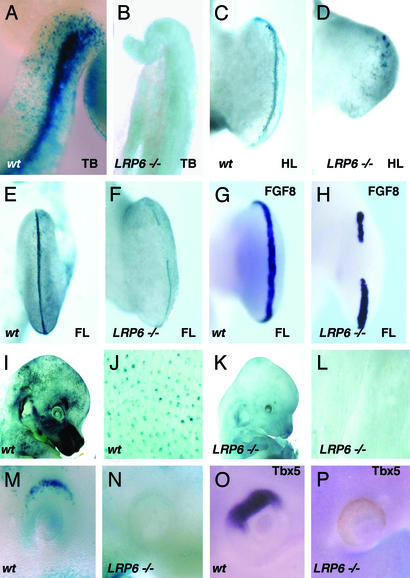
Figure 4.
Alterations of BAT-gal expression in LRP6 mutant mice. (A and B) Tailbuds (TB) of wild-type and mutant embryos at E10.5. (C–F) X-Gal staining of hindlimb (HL) or forelimb (FL) from wild-type (wt) and mutant (LRP6−/−) E11.5 embryos. (G and H) In situ hybridization with FGF8 antisense probe of normal (G) and mutant (H) forelimb buds. (I–L) Comparison of lacZ expression in the skin of wild-type and LRP6 mutant embryos at E14.5. (M) X-Gal staining of wild-type E9.5 embryos showing lacZ expression in the dorsal eye vesicle, which lacks in LRP6 mutants (N). (O and P) Corresponding in situ hybridization with a Tbx5 antisense probe of E10.5 embryos.
In the genitourinary system (Fig. 3H), early expression is noted in the mesonephric tubules, the mesonephric and paramesonephric ducts in the urogenital sinus, the ureter, and the ureteric buds. Several findings have implicated Wnt ligands as mediators of two key nephrogenic signals, one promoting ureteric bud outgrowth and a second stimulating tubulogenesis in the metanephric mesenchyme (30, 31). X-Gal-positive cells are found in the epithelium of the ureter and collecting ducts with a strong staining in the ureteric tips (Fig. 3I). In contrast, no β-catenin signal can be detected at various stages of differentiation of the metanephric mesenchyme, suggesting that nephron morphogenesis relies on a β-catenin-independent pathway.
The lung is another organ whose development is established by branching morphogenesis, although the role of Wnt signaling in this process has not been investigated yet. lacZ activity is first apparent in the lung bud (Fig. 3J) and continues in the epithelium during further development (Fig. 3K). In the proximal part of the bronchial tree, which has stopped undergoing branching morphogenesis, lacZ staining ceases in the epithelium and is turned on in the surrounding mesenchyme (Fig. 3L).
BAT-gal activity was detected in the developing skin. Wnt-responsive cells initially appear in the hair placodes and in the primordium of the dermal papilla (data not shown) (32). During hair growth, lacZ expression is detected in the precortex as well as in dermal papilla and matrix cells (Fig. 3 M and N). However, individual hairs display differences, because some are stained only in dermal papilla with weak or no labeling in matrix and vice versa. Such dynamic patterns of expression testify to the coordinated reciprocal signaling between the dermal papilla and the epidermis (33, 34).
BAT-gal Expression in LRP6 Mutant Mice.
If BAT-gal reports the endogenous status of β-catenin-mediated transcriptional activity, mice with defective Wnt signaling should reveal changes in the expression pattern of the BAT-gal transgene. LRP6 is a Wnt coreceptor essential for the activity of specific Wnts in some tissues (35). Embryos homozygous null for LRP6 exhibit developmental defects that are remarkably similar to those caused by mutations in some, but not all, Wnt genes (35). For example, LRP6 mutants develop axial truncation in the tail region due to depletion of mesodermal precursors in the tail bud. Thus, we examined β-catenin activity in the tail of wild-type and LRP6 mutant embryos carrying the BAT-gal transgene. Although very strong labeling was observed in wild-type animals, X-Gal staining was absent from the tail of LRP6 mutant embryos by E10.5 (Fig. 4 A and B).
Limb development is severely affected in LRP6 mutant embryos and, as revealed by BAT-gal staining, Wnt activity is defective in the apical ectodermal ridge (Fig. 4 C–F). Indeed, X-Gal staining in the normal apical ectodermal ridge typically entails only a few rows of cells at the border between dorsal and ventral limb ectoderm (Fig. 4 C and E). In LRP6 mutant limbs, X-Gal-positive cells are either absent (Fig. 4D, hindlimb) or discontinuous (Fig. 4F, forelimb). By in situ hybridization, we could detect gaps in the FGF8 expression pattern (compare Fig. 4 G and H), supporting the notion that Wnt/LRP6/β-catenin signaling is required for FGF8 expression.
Genetic deficiencies in β-catenin or LEF-1 are associated with defective skin appendage development (2). In LRP6 mutants, the epithelium appears devoid of hair follicles (data not shown). This defect has an early inception during the induction of hair germs, because we could detect placode staining in LRP6 mutant mice neither at E14.5 (compare Fig. 4 I and J with K and L) nor at earlier stages (data not shown).
Analyses of BAT-gal expression in LRP6 mutant embryos can help identify the location of new signaling centers. For instance, LRP6 knock-out mice are microphthalmic (35) and display severe retinal defects (V.B., unpublished work). We noted that BAT-gal is indeed expressed in the dorsal side of the optic vesicle in wild-type (Fig. 4M) but not in LRP6 mutant embryos (Fig. 4N). Tbx5 is an early inducer of dorsal retinal precursors (36), and in situ hybridization for this marker reveals that Wnt signaling is required for dorsoventral patterning of the eye (Fig. 4 O and P).
Activation of β-Catenin Signaling in Colon Cancer.
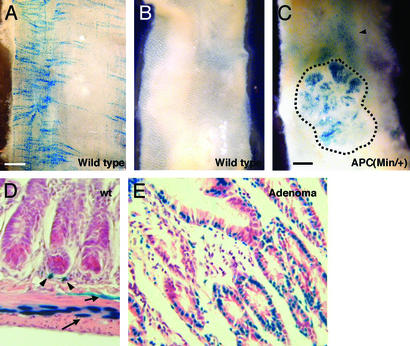
Although the homozygous APC(Min) mutation is early embryonic lethal, heterozygous mice develop intestinal adenomas at high frequency, due to the loss of APC heterozygosity (10). To establish whether the BAT-gal transgene can be activated during pathological activations of the Wnt cascade, we monitored BAT-gal expression in APC(Min/+) adenomas. When stained with X-Gal, intestine from control mice shows labeled cells in the abluminal side (Fig. 5A), whereas the epithelium of the luminal side is negative (Fig. 5B). In contrast, the adenomatous tissue of APC(Min/+) mice exhibited clear activation of the BAT-gal transgene (Fig. 5C).
Figure 5.
Activation of β-catenin/Wnt pathway in adenomas developing in APC(Min/+) mutant mice. (A) X-Gal staining of the intestine observed from the abluminal side. Note the network of perpendicular labeling characteristic of the mesenteric plexus. (B and C) View of the luminal side of the X-Gal-stained intestine from wild-type (B) and APC(Min/+) (C) mice. Adenomas (dashed line in C) express the lacZ reporter gene. (D) Transverse section of wild-type (wt) intestine. Arrowheads indicate scattered positive stromal cells surrounding the crypts. Arrows indicate positive cells arranged in longitudinal and circular networks (see A), probably belonging to the enteric plexus. (E) In APC(Min/+) adenomas, BAT-gal expression is greatly up-regulated in the epithelial neoplastic cells.
To identify the cells responding positively to Wnt signaling, we sectioned the intestine from wild-type mice and the neoplastic lesions of APC(Min/+) mutants (Fig. 5 D and E). Intriguingly, high transgene activation is found in scattered mesenchymal cells surrounding the crypts (Fig. 5D). The positive cells that appear distributed in longitudinal and circular threads in the whole-mount staining (Fig. 5A) are located within the smooth muscle layer and in the submucosa (Fig. 5D) and probably correspond to the enteric nervous system (37). Although in the normal intestine the crypt epithelium appeared devoid of BAT-gal activity (Fig. 5D), strong staining was observed in the epithelium of intestinal adenomas (Fig. 5E). Some stromal cells were also positive. To assay for quantitative differences in BAT-gal expression within the neoplastic tissue, we explanted epithelial cells from adenoma and phenotypically normal intestine from APC(Min/+) mice. Live cell suspensions were incubated with a fluorescent lacZ substrate. Importantly, only a proportion of adenoma cells incorporate high amounts of label (see Fig. 7, which is published as supporting information on the PNAS web site), suggesting that intestinal tumor cells might be heterogeneous in the levels of active β-catenin, in agreement with previous reports (38).
Final Comments
In conclusion, the BAT-gal mouse unveils the dynamic pattern of Wnt/β-catenin activation in mammals. Here we present a systematic tridimensional atlas of the spatial and temporal distribution of the endogenous status of β-catenin activation in several tissues and in intestinal tumors. These sites of activity could not be fully appreciated from previous studies on Wnt expression or gene knock-outs, given the complexity of the Wnt signal transduction mechanisms (4) and the early lethality of null β-catenin mice (39). The proof that BAT-gal mice are bona fide sensors of Wnt activity is 2-fold: first, the striking overlapping between lacZ reporter activation and expression of known Wnt molecules, and second, the correlation of BAT-gal transgene expression with loss and gain of Wnt signaling in defined mutant backgrounds.
The use of reporter constructs containing multiple LEF/TCF-binding elements joined to minimal promoter sequences was first introduced by Clevers and coworkers (40) to reveal transcriptional activation by β-catenin in mammalian cells. These constructs have been recently used in vivo to monitor Wnt signaling in transgenic zebrafish embryos (41) and in a transgenic mouse strain, Tcf optimal promoter (TOP)-galactosidase (TOP-gal) (33). In this latter study, the pattern of β-catenin activation was investigated only during skin development, whereas the present study investigates Wnt activity in multiple cellular contexts. Comparison of BAT- and TOP-gal during skin development suggests that BAT-gal activity in this specific tissue appears stronger than TOP-gal. For example, TOP-gal failed to reveal β-catenin activity in the dermal papilla, which instead was clearly labeled in BAT-gal mice. This finding is in agreement with the recent proposal that Wnt signaling is required to maintain such dermal organizing center in an active state (34).
BAT-gal expression visualizes the general status of Wnt activation within a given cell, which is the net effect of a variety of positive and negative inputs acting on β-catenin stability. This does not necessarily relate to the transcriptional status of individual target promoters, which might be sensitive to distinct thresholds of nuclear β-catenin or require specific cofactors to be activated (8).
The pattern of BAT-gal activation should allow monitoring of how the cellular context can affect β-catenin signaling. Extracellular signals other than Wnts can indeed converge on β-catenin stabilization. For example, mitogens and integrin-linked kinase can partially activate β-catenin-responsive genes (5). Moreover, it has been recently shown that transforming growth factor type β can also play a direct role in Wnt-mediated transcriptional responses (42).
In addition to confirming BAT-gal as a reliable Wnt signaling indicator, the data obtained in the APC(Min/+) strain contribute significant information on the role of APC and β-catenin in the formation of intestinal adenomas. In these mice, APC loss of heterozygosity in crypt stem cells has been shown to be the initial event in adenoma development (1, 10). However, the mechanism by which this initial alteration brings about tumor development is the focus of discussion (9). On the one hand, it has been proposed that APC loss induces cell fate changes due to transcriptional activity of β-catenin; alternatively, adenomas can be considered the consequence of a defect in cell migration and/or adhesion of APC mutant cells that is independent from nuclear β-catenin (1). The finding of lacZ-positive cells in adenomas certainly adds support to the first hypothesis. However, it seems reasonable to assume that local conditions within the adenoma tissue (such as tumor architecture, altered cell–cell or cell–matrix adhesion, and stromal influences) may play a key role in promoting β-catenin signaling or regulating its magnitude. The BAT-gal mouse may be instrumental in analyzing these local factors. No transgene expression has been observed in normal intestinal crypt cells, which is surprising in view of the observation that the homeostasis of the stem cell compartment of the crypts is critically dependent on signaling mediated by Tcf4 (43). Possible explanations for this unexpected finding are that β-catenin activation in these cells is below the level of detection by the BAT-gal transgene, or that Tcf4 may actually repress β-catenin signaling or act in a parallel pathway. Crossing BAT-gal into the Tcf4−/− background would certainly help in discriminating among these possibilities.
Finally, useful applications of BAT-gal mice can be envisaged in genetic or drug screens for Wnt signaling modulators, which may reveal further genes or therapeutic tools able to regulate the multiple roles of β-catenin in mammalian tissues.
Supplementary Material
Acknowledgments
We are grateful to W. Skarnes and K. Pinson (Department of Molecular and Cell Biology, University of California, Berkeley) for providing LRP6 mutant mice; to A. Vecchi (Istituto Ricerche Farmacologiche “Mario Negri,” Milan) for antibodies; to E. Colombo for support and animal crossings; and to S. Martinez, J. Rubenstein, O. Wessely, and K. McCullagh for comments. This work was supported by grants from Telethon, Associazione Italiana per la Ricerca sul Cancro, Agenzia Spaziale Italiana, Consiglio Nazionale delle Ricerche, and Ministero d'Istruzione, dell'Università, e della Ricerca.
Abbreviations
- APC
adenomatous polyposis coli
- BAT-gal
β-catenin-activated transgene driving expression of nuclear β-galactosidase reporter
- En
embryonic day n
- LEF/TCF
lymphoid enhancer factor/T cell factor
- LRP
lipoprotein receptor-related protein
- Min
multiple intestinal neoplasia
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bienz M, Clevers H. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 2.Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 3.Wodarz A, Nusse R. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 4.Niehrs C. Nature. 2001;413:787–788. doi: 10.1038/35101682. [DOI] [PubMed] [Google Scholar]
- 5.Peifer M, Polakis P. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 6.Hecht A, Kemler R. EMBO Rep. 2000;1:24–28. doi: 10.1093/embo-reports/kvd012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 8.Prieve M G, Waterman M L. Mol Cell Biol. 1999;19:4503–4515. doi: 10.1128/mcb.19.6.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson C B, Neufeld K L, White R L. Proc Natl Acad Sci USA. 2002;99:8683–8688. doi: 10.1073/pnas.122235399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su L K, Kinzler K W, Vogelstein B, Preisinger A C, Moser A R, Luongo C, Gould K A, Dove W F. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 11.Brannon M, Gomperts M, Sumoy L, Moon R T, Kimelman D. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogan B. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 13.Gavin B J, McMahon J A, McMahon A P. Genes Dev. 1990;4:2319–2332. doi: 10.1101/gad.4.12b.2319. [DOI] [PubMed] [Google Scholar]
- 14.Parr B A, Shea M J, Vassileva G, McMahon A P. Development (Cambridge, UK) 1993;119:247–261. doi: 10.1242/dev.119.1.247. [DOI] [PubMed] [Google Scholar]
- 15.Beddington R S, Robertson E J. Cell. 1999;96:195–209. doi: 10.1016/s0092-8674(00)80560-7. [DOI] [PubMed] [Google Scholar]
- 16.Lawson K A, Dunn N R, Roelen B A, Zeinstra L M, Davis A M, Wright C V, Korving J P, Hogan B L. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Robertis E M, Larrain J, Oelgeschlager M, Wessely O. Nat Rev Genet. 2000;1:171–181. doi: 10.1038/35042039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinder S J, Tsang T E, Wakamiya M, Sasaki H, Behringer R R, Nagy A, Tam P P. Development (Cambridge, UK) 2001;128:3623–3634. doi: 10.1242/dev.128.18.3623. [DOI] [PubMed] [Google Scholar]
- 19.Ikeya M, Takada S. Development (Cambridge, UK) 1998;125:4969–4976. doi: 10.1242/dev.125.24.4969. [DOI] [PubMed] [Google Scholar]
- 20.Rawls A, Wilson-Rawls J, Olson E N. Curr Top Dev Biol. 2000;47:131–154. doi: 10.1016/s0070-2153(08)60724-3. [DOI] [PubMed] [Google Scholar]
- 21.Oosterwegel M, van de Wetering M, Timmerman J, Kruisbeek A, Destree O, Meijlink F, Clevers H. Development (Cambridge, UK) 1993;118:439–448. doi: 10.1242/dev.118.2.439. [DOI] [PubMed] [Google Scholar]
- 22.Echelard Y, Vassileva G, McMahon A P. Development (Cambridge, UK) 1994;120:2213–2224. doi: 10.1242/dev.120.8.2213. [DOI] [PubMed] [Google Scholar]
- 23.Lee S M, Tole S, Grove E, McMahon A P. Development (Cambridge, UK) 2000;127:457–467. doi: 10.1242/dev.127.3.457. [DOI] [PubMed] [Google Scholar]
- 24.Crossley P H, Martin G R. Development (Cambridge, UK) 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- 25.Patapoutian A, Reichardt L F. Curr Opin Neurobiol. 2000;10:392–399. doi: 10.1016/s0959-4388(00)00100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikeya M, Lee S M, Johnson J E, McMahon A P, Takada S. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 27.Dorsky R I, Moon R T, Raible D W. Nature. 1998;396:370–373. doi: 10.1038/24620. [DOI] [PubMed] [Google Scholar]
- 28.Ho E, Shimada Y. Dev Biol. 1978;66:579–585. doi: 10.1016/0012-1606(78)90263-4. [DOI] [PubMed] [Google Scholar]
- 29.Hanai J, Dhanabal M, Karumanchi S A, Albanese C, Waterman M, Chan B, Ramchandran R, Pestell R, Sukhatme V P. J Biol Chem. 2002;277:16464–16469. doi: 10.1074/jbc.M112274200. [DOI] [PubMed] [Google Scholar]
- 30.Kispert A, Vainio S, McMahon A P. Development (Cambridge, UK) 1998;125:4225–4234. doi: 10.1242/dev.125.21.4225. [DOI] [PubMed] [Google Scholar]
- 31.Lin Y, Liu A, Zhang S, Ruusunen T, Kreidberg J A, Peltoketo H, Drummond I, Vainio S. Dev Dyn. 2001;222:26–39. doi: 10.1002/dvdy.1164. [DOI] [PubMed] [Google Scholar]
- 32.Fuchs E, Merrill B J, Jamora C, DasGupta R. Dev Cell. 2001;1:13–25. doi: 10.1016/s1534-5807(01)00022-3. [DOI] [PubMed] [Google Scholar]
- 33.DasGupta R, Fuchs E. Development (Cambridge, UK) 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 34.Kishimoto J, Burgeson R E, Morgan B A. Genes Dev. 2000;14:1181–1185. [PMC free article] [PubMed] [Google Scholar]
- 35.Pinson K I, Brennan J, Monkley S, Avery B J, Skarnes W C. Nature. 2000;407:535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- 36.Koshiba-Takeuchi K, Takeuchi J K, Matsumoto K, Momose T, Uno K, Hoepker V, Ogura K, Takahashi N, Nakamura H, Yasuda K, Ogura T. Science. 2000;287:134–137. doi: 10.1126/science.287.5450.134. [DOI] [PubMed] [Google Scholar]
- 37.Burns A J, Delalande J M, Le Douarin N M. Development (Cambridge, UK) 2002;129:2785–2796. doi: 10.1242/dev.129.12.2785. [DOI] [PubMed] [Google Scholar]
- 38.Chung G G, Provost E, Kielhorn E P, Charette L A, Smith B L, Rimm D L. Clin Cancer Res. 2001;7:4013–4020. [PubMed] [Google Scholar]
- 39.Huelsken J, Vogel R, Brinkmann V, Erdmann B, Birchmeier C, Birchmeier W. J Cell Biol. 2000;148:567–578. doi: 10.1083/jcb.148.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 41.Dorsky R I, Sheldahl L C, Moon R T. Dev Biol. 2002;241:229–237. doi: 10.1006/dbio.2001.0515. [DOI] [PubMed] [Google Scholar]
- 42.Nishita M, Hashimoto M K, Ogata S, Laurent M N, Ueno N, Shibuya H, Cho K W. Nature. 2000;403:781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- 43.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters P J, Clevers H. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.