Abstract
Single human bone marrow-derived mesodermal progenitor cells (MPCs) differentiate into osteoblasts, chondrocytes, adipocytes, myocytes, and endothelial cells. To identify genes involved in the commitment of MPCs to osteoblasts we examined the expressed gene profile of undifferentiated MPCs and MPCs induced to the osteoblast lineage for 1–7 days by cDNA microarray analysis. As expected, growth factor, hormone, and signaling pathway genes known to be involved in osteogenesis were activated during differentiation. In addition, 41 transcription factors (TFs) were differentially expressed over time, including TFs with known roles in osteoblast differentiation and TFs not known to be involved in osteoblast differentiation. As the latter group of TFs coclustered with osteogenesis-specific TFs, they may play a role in osteoblast differentiation. When we compared the gene expression profile of MPCs induced to differentiate to chondroblasts and osteoblasts, significant differences in the nature and/or timing of gene activation were seen. These studies indicate that in vitro differentiation cultures in which MPCs are induced to one of multiple cell fates should be very useful for defining signals important for lineage-specific differentiation.
Osteogenesis is a strictly controlled developmental process in which numerous extrinsic factors, including hormones and growth factors, activate osteoblast-specific signaling proteins and transcription factors (TFs) required for osteoblast differentiation (1, 2). The most extensively studied TF important in osteogenesis is the master TF, core binding factor alpha 1 (Cbfa1), also called runt-related gene 2 (Runx2). Cbfa1 activates osteocalcin, an osteoblast-specific gene expressed by fully differentiated osteoblasts (3). Cbfa1-deficient mice lack bone formation because of a maturation arrest of osteoblasts (4, 5), demonstrating that Cbfa1 is necessary for osteogenesis. The signals that regulate Cbfa1 expression are not fully defined. Members of the transforming growth factor (TGF)-β family including bone morphogenetic protein (BMP)-2, BMP-4, and BMP-7, which induce osteoblast differentiation, up-regulate Cbfa1 expression (1). However, Cbfa1 is not sufficient for osteogenesis as differentiation of C2C12 cells to mature osteoblasts cannot be induced by activation of Cbfa1 alone (6). Thus, aside from Cbfa1, other TFs are needed to activate the genetic pathways controlling osteoblast differentiation.
We have recently identified a rare bone marrow cell that differentiates at the single-cell level not only into mesenchymal cell types such as osteoblasts, chondroblasts, and adipocytes, but also into cells of visceral mesodermal origin (7). We have termed this cell a mesodermal progenitor cell (MPC). Because MPCs can be expanded without obvious senescence, they may be an ideal source of cells to generate osteoblasts for treating common bone diseases such as osteoporosis or nonhealing fractures or inherited bone disorders such as osteogenesis imperfecta (8). To optimally use MPCs, a better understanding of processes that govern initial commitment and further differentiation from a mesodermal progenitor to an osteoblast fate will be needed. In vitro differentiation of MPCs to either osteoblasts, chondroblasts, myoblasts, or endothelial cells appears to be specific, as most cells recovered after 14–21 days of differentiation express lineage-specific markers (7). Specifically for osteoblast differentiation, we showed that 90% of cells obtained after 14 days from dexamethasone-, ascorbic acid-, and β-glycerophosphate-treated cultures expressed osteocalcin. In contrast, when MPCs were treated with TGF-β to induce a chondroblast phenotype, >90% of cells recovered after 14 days had a chondroblast phenotype. This finding led us to speculate that the in vitro differentiation model may allow identification of TFs, aside from Cbfa1, that are important in osteogenesis. To assess the genetic pathways involved in osteoblast differentiation, we used cDNA microarray analysis (9).
Materials and Methods
MPC Culture and Osteoblast or Chondrocyte Differentiation.
Bone marrow was obtained from three healthy donors after informed consent according to guidelines from the University of Minnesota Committee on the Use of Human Subjects in Research. Isolation and culture of human bone marrow-derived MPCs and differentiation of MPCs to the osteoblast and chondroblast lineage were done as described (7). Immunohistochemical staining for osteocalcin, collagen type I, and collagen type II was done as described (7). For Alzarin red staining, cells were fixed with 4% paraformaldehyde at room temperature for 15 min, rinsed in PBS, and then stained in Alzarin red for 15 min. After rinsing in deioined H2O, slides were dehydrated and mounted.
cDNA Microarray Analysis.
Total RNA was isolated from MPCs or MPCs induced to the osteoblast or chondroblast lineage for 1, 2, and 7 days by using a RNeasy mini-kit (Qiagen, Chatsworth, CA) per the manufacturer's protocol. Total RNA was digested with DNase I (Promega) at 37°C for 1 h followed by repurification with Qiagen RNeasy mini-columns to eliminate genomic DNA. α-33P-dCTP-labeled cDNA probes were generated and hybridized to a Human GeneFilters microarray (Research Genetics, Huntsville, AL, catalogue no. GF211) at 42°C for 18–20 h as recommended in the protocol from the manufacturer. Blots were washed twice in 2× SSC, 1% SDS at 55°C for 20 min and once in 0.5× SSC, 1% SDS at 55°C for 15 min. Arrays were read by a PhosphorImager screen scanner (Molecular Dynamics, Storm 860). Software PATHWAY 3.0 (Research Genetics) and SPOTFIRE 7.0 (Spotfire, Cambridge, MA) were used to analyze the data. In brief, the raw data from each array were normalized according to the manufacturer's guidelines by using PATHWAY 3.0. Paired t test on normalized intensity with P value ≤0.05 followed by ratio change (ratio of normalized intensity ≥2.0 or ≤0.5) was used to generate the list of genes with significant change in expression profile during osteoblast differentiation. For hierarchical and K-means cluster analysis, data with normalized intensity from excel spreadsheets were imported into SPOTFIRE 7.0. Hierarchical clustering was done by using the unweighted average method. The dendrogram was calculated based on the similarity between genes by using Euclidean distance as a similarity measurement. K-means clustering was done by using an evenly spaced profile and correlation as a similarity measure.
Real-Time RT-PCR.
Total RNA from MPCs and MPCs differentiated to the osteoblast lineage for 1, 2, 4, 7, and 14 days was reverse-transcribed by using Superscript II (GIBCO). Quantitative RT-PCR (ABI PRISM 7700, Perkin–Elmer/Applied Biosystems) was done as follows: 40 cycles of PCR (95°C for 15 s, 60°C for 1 min) after initial denaturation for 10 min at 95°C by using 12.5 μl of 2× Syber green universal mix (Perkin–Elmer/Applied Biosystems), 100 nmol primers, 1 μl cDNA, and H2O to 25 μl. mRNA levels were normalized by using β-actin as a housekeeping gene. We compared levels in undifferentiated MPCs with levels in MPCs induced to the osteoblast lineage by using ABI PRISM 7700 software DETECTOR 1.6. Primers used for RT-PCR are as follows: muscle segment homolog Drosophilia homeobox 2 (MSX2), 5′-ctacccgttccatagacctgtgctt-3′, 5′-gagagggagaggaaaccctttgaa-3′; PTHr1, 5′-catcttttggtccatctgtccatc-3′, 5′-ctggagaccctcgagaccaca-3′; POU2F1, 5′-agtttgcggctggaggtgcctta-3′, 5′-agtcacgatgttgggggctcctc-3′; KIAA0407, 5′-catgggaagcttgagtatttcac-3′, 5′-ttaatccctcgaaagagcatgta-3′; KIAA0220, 5′-aaggccatggtaacaaccaa-3′, 5′-accctcatgatggagagtcg-3′; CBFA1, 5′-agaggtaccagatgggactgtggtt-3′, 5′-ggtagctacttggggaggatttgtg-3′; KLF1, 5′-gcgcgtgacacaatgctg-3′, 5′-ttgggaacgcgagtcca-3′; GATA1, 5′-ttcttgcccgccatgc-3′, 5′-cgaggaatcatccctggct-3′; BMP8, 5′-cagtccaacagggagtctgacttgt-3′, 5′-cccgtcctcagtctccacataga-3′; and β-actin, 5′-tacctcatgaagatcctca-3′, 5′-ttcgtggatgccacaggac-3′.
Results and Discussion
MPCs Differentiate to the Osteoblast Lineage.
We have recently shown that rare cells copurifying with mesenchymal stem cells from human bone marrow can be extensively expanded and induced to differentiate to most mesodermal cell types in vitro (7). The in vitro differentiation process appears highly specific as >90% of cells after 14–21 days express lineage-specific marker. We therefore hypothesized that this in vitro differentiation system may be useful for identifying genetic mechanisms underlying lineage commitment and differentiation. To test this hypothesis we compared changes in expressed gene profile in MPCs induced to differentiate into two closely related cell lineages, osteoblasts and chondroblasts.
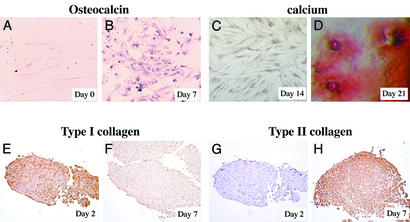
We isolated MPCs from the bone marrow of three healthy donors (two male, one female, ages 25, 31, and 22) by depleting CD45+ cells and GlyA+ cells and culture under MPC-specific conditions. The phenotype of MPCs from the three donors was CD45, MHC class I and class II, and β2-microglobulin negative, similar to what we have described (7). MPCs were expanded by subculturing every 2–3 days at a 1:2/1:4 dilution until 28 population doublings were done. Cells were then induced to differentiate to chondroblasts or osteoblasts as described (7). Immunohistochemistry showed that the osteoblast-specific marker, osteocalcin, was not present in undifferentiated MPCs (Fig. 1A), but could be detected from day 7 on in cultures of MPCs treated with dexamethasone, ascorbic acid, and β-glycerophosphate (Fig. 1B). Calcium deposition was seen after 21 days, as shown by Alzarin red staining (Fig. 1 C and D). Osteoblast-differentiated MPCs stained positive for collagen type I but not collagen type II (not shown). In contrast, MPCs in chondroblast differentiation cultures stained positive for collagen type II (Fig. 1 E–H) and aggrecan (not shown) but did not express osteocalcin or collagen type I and did not contain calcium deposits (not shown). These studies confirm that MPCs could differentiate specifically into osteoblasts when treated with dexamethasone, ascorbic acid, and β-glycerophosphate, and chondroblasts when treated with TGF-β I.
Figure 1.
Immunohistochemistry of undifferentiated MPCs (A) or MPCs treated with ascorbic acid, β-glycerophosphate, and dexameasone for 7–21 days (B–D), and TGF-β I for 2 days (E and G) and 7 days (F and H). Cells were stained with antibodies against osteochalcin (A and B), type I (E and F), or type II collagen (G and H) or stained with Alzarin red dye (C and D).
To identify genes involved in the commitment of MPCs to the osteoblast lineage, we isolated RNA from undifferentiated MPCs and MPCs induced to the osteoblast lineage or to the chondroblast lineage for 1, 2, and 7 days from the three donors. cDNAs were hybridized to a microarray that contains 4,000 human genes, mostly with known functions. We compared the expressed gene profile over time and between the three donors. Genes known to be expressed in osteoblasts were consistently up-regulated in bone differentiation cultures compared with undifferentiated MPCs. For instance, alkaline phosphatase (AP) transcripts were elevated >2-fold on days 2 and 7, and osteomodulin and cadherin-11 transcripts were increased 2-fold on day 7. Likewise, genes involved in chondroblast commitment and differentiation, such as cartilage paired-class homeoprotein 1 (CART-1) and cartilage oligometrix protein (COMP) were consistently up-regulated in TGF-β I-treated cultures. These results confirmed that MPCs underwent differentiation to the osteoblast or chondroblast lineage.
Data Generated from Microarrays Are Repeatable.
Because of the genetic variability among normal individuals, many investigators have chosen to pool RNA samples from different donors before use in cDNA array analysis (10). Other studies have evaluated RNA samples separately and used either average changes in gene expression or changes in gene expression found in the majority of their samples (11). We elected to test the RNA samples from the different donors separately. We found that the expressed gene profile of MPCs or MPCs induced to differentiate to the osteoblast lineage varied when comparing the three donors. For instance, for undifferentiated MPCs, the correlation coefficient of intensity was 0.92 when we compared donors 1 and 3, but only 0.5 and 0.6 when donor 2 was compared with either donor 1 or donor 3. When evaluating MPCs induced to the osteoblast lineage for 1 or 7 days, the correlation coefficient of intensity was 0.9 between donors 2 and 3, but only 0.5 between donor 1 and either donor 2 or donor 3.
We do not believe that differences between the three donors are caused by technical shortcomings of the assay leading to nonreproducible results. We compared results from studies in which the same cDNA was hybridized two or three times, either to the same membrane that had been stripped between hybridizations or to new membranes. Correlation coefficients of intensity ranged between 0.92 and 0.97. In addition, 77 cDNAs were spotted in duplicate on the arrays. The correlation coefficient of intensity of these repeated genes within an array was >0.9 for all three donors at all three time points. These studies indicate that the hybridization studies are repeatable. Another possibility is technical differences in the differentiation cultures, despite our best efforts at maintaining rigorous standards during differentiation. This idea would be consistent with our finding that transcripts for some TFs, such as short stature homeobox gene (SHOX-2), SRY-box 22 (Sox-22), and MSX2, known to play a role in osteogenesis, were not elevated in one of the three donors. The fact that on day 2 expression of Sox-22 and SHOX-2 was not significantly increased in donor 2, whereas on day 7, transcripts for MSX2 were not elevated in donor 1, suggests that differences in culture technique may not underlie differences seen in gene expression. A third possibility is differences in gender, as donor 2 was female and donors 1 and 3 were male. However, the observation that for some TFs differences were noted between donor 1 and donor 2 or 3 suggests that this might not be the case. In any case, much larger numbers of donors would need to be evaluated to conclude that gender may be a factor in genetic mechanisms governing osteoblast differentiation. A final possibility is inherent genetic differences between individuals, independent of gender, leading to alterations in gene expression profile in response to the osteoblast-inducing factors.
Because of this variability, we used paired t test on normalized intensity changes between MPCs and MPCs induced to osteoblast differentiation at various time points to analyze the data. Using this selection criterion alone, 219 of the 4,000 genes showed significant changes (P ≤ 0.05) in expression levels in day 1-differentiated osteoblasts compared with undifferentiated MPCs. A total of 377 genes and 595 genes showed significant changes in expression levels on day 2 and day 7 differentiated osteoblasts, respectively. Because of the small sample size (n = 3) we did not do any formal adjustment for multiple comparison, which may be too conservative. Rather, we filtered this list by using a simple data filter that selects genes for which the average change in intensity among all three donors was >2-fold relative to the data for MPCs. Combining filter 1 and filter 2, we found that a total of 535 genes was differentially expressed at any time point. Among them, 24 genes were up-regulated and 52 down-regulated on day 1, 43 genes were up-regulated and 130 down-regulated on day 2, and 274 were up-regulated and 122 down-regulated on day 7 compared undifferentiated MPCs (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org).
One potential drawback in using such a highly stringent set of filters is that this may lead to underestimation of genes that are differentially expressed on osteoblast differentiation. For instance, the TF MSX2, an upstream regulator of Cbfa1 (12), was up-regulated in two donors, (2-fold increase for donor 1, 3-fold for donor 2) but showed only an 1.45-fold increase in donor 3 on day 7. Thus, although the average change of intensity between undifferentiated MPCs and day 7 osteoblasts was >2-fold, the P value for MSX2 from paired t test was only 0.09. Larger numbers of donor sample would be necessary to decrease the variability.
Genes Differentially Regulated During Osteoblast Differentiation.
We divided differentially expressed genes in 12 categories based on their function: genes involved in apoptosis, cell cycle control, DNA repair and RNA splicing, extracellular matrix components, protein synthesis and degradation, cell metabolism, nuclear proteins, receptor/membrane proteins, secreted molecules, secretion pathway, signal transduction molecules, and TFs.
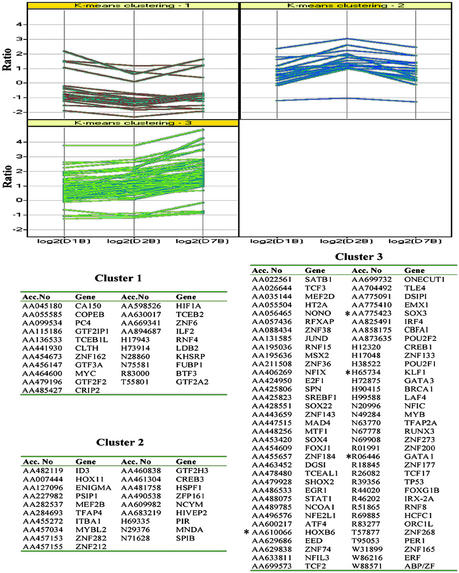
We used K-means clustering to determine the expression pattern of TFs. Different TFs were up- or down-regulated immediately after differentiation was induced (day 1 and/or 2), only late after differentiation induction (day 7) or throughout differentiation (days 1–7) (Fig. 2). Several differentially expressed TFs have known function in osteoblast differentiation. These included Sox-4 (13, 14) (P = 0.017, ratio = 2.35), Sox-22 (13) (P = 0.007, ratio = 2.39), SHOX-2 (15) (P = 0.008, ratio = 3.01), and MSX2 (12) (P = 0.098, ratio = 3). As expected, we also found that the basal transcription machinery was up-regulated early during differentiation. For instance expression of general TF II H (GTF2H3) (P = 0.035, ratio = 2.36), CCAAT-binding TF, nuclear factor I/X (P = 0.1, ratio = 2.01), and nuclear factor I/C (P = 0.0566, ratio = 3.4) were up-regulated. Transcriptional repressor such as zinc finger protein 133 (ZNF 133) (P = 0.004, ratio 3.07) (16) was up-regulated at day 7. In addition, several growth repressors, including Ets-2-repressor factor (ERF) (P = 0.005, ratio = 2.01) (17) and delta sleep-inducing peptide immunoreactor (DSIP) (P ≤ 0.05, ratio >6.05 on days 1 and 2), were up-regulated during differentiation. A number of TFs required for cell proliferation were suppressed as cells differentiated. A good example is Myc mRNA, which was decreased on days 1, 2, and 7 (P < 0.05, ratio >2) (see Table 2, which is published as supporting information on the PNAS web site).
Figure 2.
TFs differentially expressed during osteoblast differentiation clustered with K-means clustering. The similarity was measured by correlation. Three clusters showed TFs that are up- or down-regulated immediately after differentiation was induced (day 1 and/or 2), only late after differentiation induction (day 7), or throughout differentiation (days 1–7). *, TFs known to be important for other lineage differentiation other than osteoblast differentiation.
However, we also found increased expression of TF previously not known to be involved in osteogenesis. These included homeobox (Hox)-B6 (P = 0.02, ratio = 2.11), GATA-1 (P = 0.009, ratio = 2.63 on day 2 and P = 0.001, ratio = 3.40 on day 7), and Kruppel-like factor (KLF)-1 (P = 0.046, ratio = 2.58). GATA-1 (18, 19), and erythroid KLF-1 (20) are involved in eryhtropoiesis. Increased expression of TFs important in lineages other than the osteoblast lineage may signify that the initial step of differentiation causes promiscuous activation of multiple signal transduction pathways. Specific osteoblast differentiation conditions would then re-enforce signal pathways required for osteoblast differentiation, leading to a >90% specific osteoblast phenotype at later time points. This hypothesis is consistent with what we saw during chondroblast differentiation. Early during TGF-β I-induced differentiation we also saw up-regulation of the hematopoietic TF GATA-1 and KLF-1. However, if some of the MPCs started differentiation to a nonosteoblast lineage, one might expect significant cell death during the initial phase of differentiation, which was not seen in either osteoblast or chondroblast differentiation. Consistent with this notion is the finding that a number of genes, including death-associated protein 3 (DAP3) (P = 0.02, ratio = 0.40) (21) and caspase 4 (CASP 4) (P = 0.025, ratio = 0.46) (22), known to be involved in apoptosis induction were down-regulated during the differentiation (see Table 2). An alternative explanation is that MPCs are not homogeneous, but consist of multiple lineage-specific stem cells, in which cell cycle arrest as a result of the high density in differentiation cultures causes initial differentiation. However, we have shown that osteoblasts, chondroblasts, myoblasts, and endothelial cells are derived from single progenitors in MPC cultures (23), suggesting that this hypothesis is not correct. TFs involved in differentiation to lineages other than osteoblast differentiation may also play a role in osteoblast differentiation if one evaluates this, using progenitors that precede the osteoblast-committed progenitor cell. Finally, the finding that GATA-1 and KLF-1 are up-regulated may signify that, like during development, the “chondroblast/osteoblast” microenvironment supports initial commitment and differentiation of primitive progenitors to the hematopoietic lineage.
We also found up- or down-regulation of many zinc finger proteins whose function is not yet known. Because these zinc finger proteins have a similar expression profile as TFs important for osteogenesis, such as MSX2 and Cbfa1 (Fig. 3), they may play a role in osteoblast differentiation. Future studies aimed at defining their function during osteoblast differentiation may lead to the identification of additional TFs that are important in osteogenesis.
Figure 3.
Several zinc finger proteins with not well known function are expressed in a pattern similar to MSX2 and cbfa1, two TFs known to be important for osteoblast differentiation.
Aside from TFs, expression of several other classes of genes was modulated during differentiation. Genes important for modulating chromatin structure such as histone deacetylase 2 (HDAC2) (24) were down-regulated on day 7 (P = 0.02, ratio = 0.41). Zinc finger protein (CMPX1) 6 (25, 26) was down-regulated on days 1, 2, and 7 (P = 0.05 ratio = 0.43 on day 1, 0.28 on day 2). TATA box binding protein (TBP)-associated factor, RNA polymerase IIN (TAF2N), and TAF3C were up-regulated at days 1, 2, and 7 (P ≤ 0.05, ratio >2, see Table 2). Finally, expression of a number of cell cycle inhibitors was up-regulated during differentiation, even though this was not true for all cell cycle inhibitors. For instance, p27Kip mRNA levels were 7- to 15-fold increased over MPCs on days 1, 2, and 7, and BTG2, recently implicated in the inhibition of G1 to S progression of the cell cycle by down-regulating on cyclin D1 transcription (27), was increased 2.9-fold on days 2 and 7 (P < 0.05). In contrast, mRNA levels of BTG1 (antiproliferative B-cell translocation gene 1) were increased on days 1, 2, and 7 (P < 0.05 and ratio >2), and inhibitor of growth family member 1 (ING1) increased 2.19-fold on day 7 (P = 0.0009). Moreover, cell cycle stimulators such as cell division cycle 2 (CDC2) and mitotic arrest deficient like 1 (MAD2L1) were significantly decreased (P ≤ 0.05, ratio ≤0.5, see Table 2). These data are in agreement with the notion that differentiation is associated with arrest of cell cycle progression.
mRNA levels for other regulators of osteoblast cell proliferation and differentiation were also increased, including several TGF-β signaling pathway members [TGF-β receptor III increased on day 7 (P = 0.07, ratio = 2.14) and BMP-8 on days 1, 2, and 7 (P = 0.1, ratio = 24.84)]. The parathyroid hormome receptor (PthR)1 (P = 0.05, ratio = 7.82), the estrogen-related receptor alpha (ESRRA) (P = 0.19, ratio = 2.18), the retinoic acid (RA) receptor responder 2 (RARRES2) (P = 0.02, ratio = 2.91), cellular RA-binding protein 1 (CRABP1) (P = 0.027, ratio 4.29), and the vitamin D3 receptor (P = 0.18, ratio = 10.88) were up-regulated during the differentiation. Although some of these genes, whose intensity changed >2-fold over undifferentiated MPCs even though the P values are in some instances >0.05 by paired t test did not attain significance levels. However, we included them in the analysis, as several of these genes were confirmed to be overexpressed when tested by quantitative RT-PCR, or because the level of overexpression, although variable between the three donors, was significantly >3-fold.
Functional Pathways Involved in Osteoblast Differentiation.
We next tested whether the array analysis could delineate functional pathways involved in osteoblast differentiation. We examined the data set to identify upstream regulators of the master TF, Cbfa1. Shown in Fig. 4, MSX2, an up-stream regulator of Cbfa1, was up-regulated on day 7. Activation of TGF-β/BMP-Smad signaling promotes Cbfa1 expression and osteoblast differentiation. Transcripts of several components of the TGF-β/BMP-Smad pathway were up-regulated during differentiation: BMP-8, Smad-3, and the TGF-β receptor III were up-regulated on day 7. In addition, we found changes in expression profile of a number of genes in signaling pathways known to affect the TGF-β/BMP-Smad pathway. Smad 7, an inhibitory Smad, can be activated by the activated nuclear factor, NF-κB, in response to cytokine stimulation through tumor necrosis factor (TNF)-α and IL-1α (29, 30), and by the Janus kinase (JAK1)/signal transducer and activator of transcription 1 (Stat-1) pathway after stimulation with IFN-γ (31). We found that transcripts of STAT-induced STAT inhibitor-1 (SSI-1), an inhibitor for Jak-1/Stat-1 signaling (32), were elevated on days 2 and 7 (P = 0.0016, ratio = 2.91 on day 7), whereas levels of Stat-1 transcripts were down-regulated on days 1 and 2 (P = 0.026, ratio = 0.415 on day 1). Levels of IκB, an inhibitor of NF-κB, were increased on day 7 (P = 0.08, ratio = 3.03). Among the IL genes, IL-1RL1 and IL-II RA were up-regulated on days 2 and 7 (P < 0.05, ratio >2), and TNF superfamily 12 (P = 0.0001, ratio = 3.54), TNF receptor superfamily (RSF)-8, -11B (osteoprotegerin), and -12 all were up-regulated during differentiation (P < 0.05, ratio >2), see Table 2). Smad7 can also be activated by Smad3, downstream from TGF-β, establishing a negative feedback loop (32).
Figure 4.
Pathways involved in osteoblast differentiation. ↑ indicates a gene that is up-regulated and ↓ indicates a gene that is down-regulated on array. *, confirmed by quantitative RT-PCR; †, interact with each other.
We next evaluated the signal pathways involved in bone protein expression such as AP and osteopontin. Expression of the TF v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) (Ets1), important in osteogenesis (33, 34), can be induced via retinoic acid, estrogen, and vitamin D3 signaling. Ets1 is responsible for activating expression of AP and osteopontin. Ets1 can also up-regulate expression of PthR1, which is important for controlling the calcium levels in osteoblasts (35). We found that the receptors for retinoic acid, estrogen, VD3, PTHr1, and AP all were up-regulated during differentiation. Finally, Ets1 can interact with Cbfa1, and together they regulate the expression of downstream pathways for both Ets1 and Cbfa1 (37).
Thus, using cDNA array it is possible to identify changes in gene expression pattern in a number of functional pathways needed for commitment and differentiation of MPCs to the osteoblast lineage.
Independent Confirmation by Quantitative RT-PCR.
To confirm the data generated from microarray studies we performed quantitative RT-PCR by using the Syber green method on total RNA derived from MPCs and MPCs induced to the osteoblast lineage for 1, 2, 7, and 14 days from the same three donors. Genes chosen were BMP8, MSX2, Cbfa1, PTHr1, POU2F1, KIAA0407, KIAA0220, KLF-1, and GATA-1. Results of the average of the three donors are shown in Table 3, which is published as supporting information on the PNAS web site, compared with the average change in transcript levels for the three donors detected by cDNA array analysis. The genes found to be differentially expressed in the microarray analysis were confirmed to be differentially expressed by quantitative RT-PCR. However, the degree of increased or decreased expression differed for some genes, likely because of differences in the sensitivity of the two assays.
Changes in Gene Expression Differ Significantly Between Osteoblast and Chondrocyte Differentiation from MPCs.
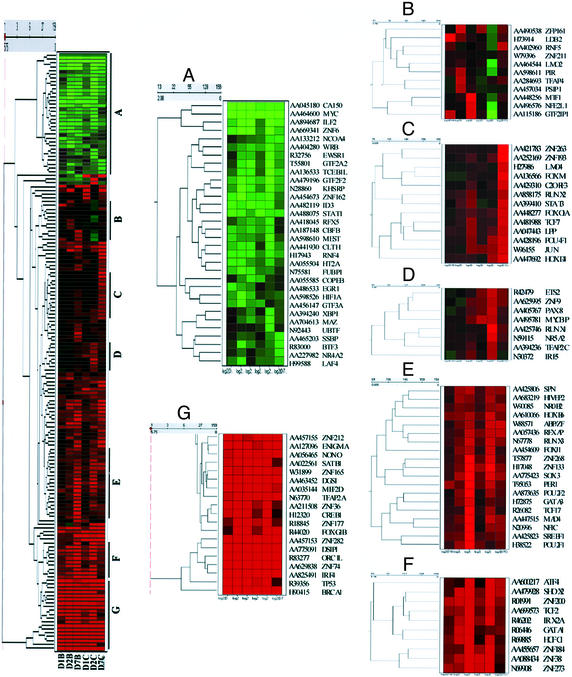
Finally, we compared the pattern of TF expression during osteoblast and chondroblast differentiation, using hierarchical clustering and tree view analysis (Fig. 5). For these studies we used the average change in gene expression in all three donors. As can be seen in Fig. 5, the 159 TFs whose expression pattern changed during differentiation were grouped according to their expression similarity measured by Euclidean distance over the time course. Seven groups of TFs appeared to be differentially expressed during osteoblast vs. chondroblast differentiation (Fig. 5). Cluster A represents TF down-regulated >2-fold during the differentiation in chondroblast and/or osteoblasts, whereas cluster G represents TFs up-regulated during differentiation to osteoblasts and chondroblasts. Clusters B, C, and D contained TFs either up-regulated during osteoblast differentiation, whereas they remained constant or were suppressed in chondroblast differentiation cultures, and TFs were up-regulated during chondroblast differentiation on days 2 or 7 but unchanged or down-regulated in osteoblast cultures. These included TF Lim binding domain 2 (LDB2), which was up-regulated 2.77-fold (P = 0.04) in osteoblast culture but was never up-regulated during chondroblast differentiation. Likewise, some TFs up-regulated during chondroblast differentiation on day 7, such as PAX-8 and HOXD4, were not up-regulated at any point during osteoblast differentiation. Groups E and F contained TFs whose expression pattern differed between osteoblast and chondroblast culture because of differences in timing of activation. For instance, HoxB6, MAD4, POU2F1 (POU domain class 2 TF 1), ZNF 133, GATA-1, and SHOX2 all belong to these two groups (P < 0.05, ratio >2; see Table 2 and Fig. 5). MSX-2 transcripts were up-regulated >2-fold from day 2 on in chondroblast cultures, but not until day 7 in osteoblasts. This result is consistent with the finding that Cbfa1, whose expression is regulated by MSX2, was up-regulated >2-fold on day 7 in chondroblast cultures, whereas >2-fold up-regulation in osteoblast cultures did not occur until day 14 (see Fig. 5).
Figure 5.
Hierachical clustering of TFs reveals significant differences between osteoblast and chondroblast differentiation from MPCs. A total of 159 TFs whose expression changed ≥2-fold during differentiation were clustered according to their similarity measured by Euclidean distance. Color change scale is continuous from minimum of −3-fold decreased (green) to maximum of 3-fold increased (red) and represents down- and up-regulation, respectively. Seven clusters were selected, and each cluster is enlarged to show representative genes.
Conclusion
We used gene array analysis of MPCs induced to differentiate to osteoblasts and chondroblasts to identify TFs and signaling pathways important in the differentiation of mesodermal progenitors to the osteoblast lineage. We confirmed changes in expression of key regulators of osteogenesis, including Cbfa1, and could demonstrate that upstream regulators of this osteogenesis master gene are also activated. In addition, although we used arrays that contained mostly genes with known function, we identified a series of zinc-finger TFs with not yet defined functions that were coregulated with MSX2 and Cbfa1. Studies in which the function of these TFs or others that might be similarly regulated are tested should help to determine a possible role in osteogenesis. We also made the somewhat surprising observation that a series of TFs important for hematopoietic commitment and differentiation were up-regulated, suggesting that the developing “bone” system may be conducive to hematopoietic differentiation. Finally, we demonstrate that the pattern of gene expression in two closely related lineages, osteoblasts and chondroblasts, can be easily discriminated by using this in vitro model of bone and cartilage development. This finding suggests strongly that MPCs, that not only differentiate into osteoblasts and chondroblasts, but also myoblasts, endothelium, and possibly neuroectodermal and endodermal lineages, may be an important tool for starting to decipher signal pathways involved in lineage-specific differentiation. This work ultimately may lead to the development of protein or small molecule “drugs” that enhance these differentiation paths in vitro or even in vivo.
Supplementary Material
Acknowledgments
We thank Dr. Vivek Kapur for critically reading the manuscript and Dr. Kapur and Michael Paustian for help with gene clustering analysis. This work was supported by National Institutes of Health Grant RO1-DK 58295, the McKnight Foundation, and the Tulloch Family Foundation.
Abbreviations
- MPC
mesodermal progenitor cell
- TF
transcription factor
- Cbfa1
core binding factor alpha 1
- MSX2
muscle segment homolog Drosophilia homeobox 2
- TGF
transforming growth factor
- BMP
bone morphogenetic protein
- KLF
Kruppel-like factor
- TNF
tumor necrosis factor
- AP
alkaline phosphatase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Yamaguchi A, Komori T, Suda T. Endocr Rev. 2000;21:393–411. doi: 10.1210/edrv.21.4.0403. [DOI] [PubMed] [Google Scholar]
- 2.Ducy P, Schinke T, Karsenty G. Science. 2000;289:1501–1504. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- 3.Ducy P. Dev Dyn. 2000;219:461–471. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1074>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 4.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson R T, Gao Y H, Inada M, et al. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 5.Otto F, Thornell A P, Crompton T, Denzel A, Gilmour K C, Rosewell I R, Stamp G W, Beddington R S, Mundlos S, Olsen B R, et al. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 6.Lee M H, Javed A, Kim H J, Shin H I, Gutierrez S, Choi J Y, Rosen V, Stein J L, van Wijnen A J, Stein G S, et al. J Cell Biochem. 1999;73:114–125. [PubMed] [Google Scholar]
- 7.Reyes M, Lund T, Lenvik T, Aguiar D, Koodie L, Verfaillie C M. Blood. 2001;98:2615–2625. doi: 10.1182/blood.v98.9.2615. [DOI] [PubMed] [Google Scholar]
- 8.Horwitz E M, Prockop D J, Fitzpatrick L A, Koo W W, Gordon P L, Neel M, Sussman M, Orchard P, Marx J C, Pyeritz R E, Brenner M K. Nat Med. 1999;5:309–313. doi: 10.1038/6529. [DOI] [PubMed] [Google Scholar]
- 9.Schulze A, Downward J. Nat Cell Biol. 2001;3:E190–E195. doi: 10.1038/35087138. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y W, Zhao P, Borup R, Hoffman E P. J Cell Biol. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka T S, Jaradat S A, Lim M K, Kargul G J, Wang X, Grahovac M J, Pantano S, Sano Y, Piao Y, Nagaraja R, et al. Proc Natl Acad Sci USA. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, et al. Nat Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- 13.Wegner M. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reppe S, Rian E, Jemtland R, Olstad O K, Gautvik V T, Gautvik K M. J Bone Miner Res. 2000;15:2402–2412. doi: 10.1359/jbmr.2000.15.12.2402. [DOI] [PubMed] [Google Scholar]
- 15.Clement-Jones M, Schiller S, Rao E, Blaschke R J, Zuniga A, Zeller R, Robson S C, Binder G, Glass I, Strachan T, et al. Hum Mol Genet. 2000;9:695–702. doi: 10.1093/hmg/9.5.695. [DOI] [PubMed] [Google Scholar]
- 16.Vissing H, Meyer W K, Aagaard L, Tommerup N, Thiesen H J. FEBS Lett. 1995;369:153–157. doi: 10.1016/0014-5793(95)00728-r. [DOI] [PubMed] [Google Scholar]
- 17.Sgouras D N, Athanasiou M A, Beal G J, Jr, Fisher R J, Blair D G, Mavrothalassitis G J. EMBO J. 1995;14:4781–4793. doi: 10.1002/j.1460-2075.1995.tb00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto M, Takahashi S, Onodera K, Muraosa Y, Engel J D. Genes Cells. 1997;2:107–115. doi: 10.1046/j.1365-2443.1997.1080305.x. [DOI] [PubMed] [Google Scholar]
- 19.Orkin S H, Shivdasani R A, Fujiwara Y, McDevitt M A. Stem Cells. 1998;16:79–83. doi: 10.1002/stem.5530160710. [DOI] [PubMed] [Google Scholar]
- 20.Coghill E, Eccleston S, Fox V, Cerruti L, Brown C, Cunningham J, Jane S, Perkins A. Blood. 2001;97:1861–1868. doi: 10.1182/blood.v97.6.1861. [DOI] [PubMed] [Google Scholar]
- 21.Kissil J L, Deiss L P, Bayewitch M, Raveh T, Khaspekov G, Kimchi A. J Biol Chem. 1995;270:27932–27936. doi: 10.1074/jbc.270.46.27932. [DOI] [PubMed] [Google Scholar]
- 22.Kamada S, Washida M, Hasegawa J, Kusano H, Funahashi Y, Tsujimoto Y. Oncogene. 1997;15:285–290. doi: 10.1038/sj.onc.1201192. [DOI] [PubMed] [Google Scholar]
- 23.Reyes M, Dudek A, Jahagirdar B, Koodie L, Marker P H, Verfaillie C M. J Clin Invest. 2002;109:337–346. doi: 10.1172/JCI14327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai S C, Valkov N, Yang W M, Gump J, Sullivan D, Seto E. Nat Genet. 2000;26:349–353. doi: 10.1038/81671. [DOI] [PubMed] [Google Scholar]
- 25.Lloyd S L, Sargent C A, Chalmers J, Lim E, Habeebu S S, Affara N A. Nucleic Acids Res. 1991;19:4835–4841. doi: 10.1093/nar/19.18.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valleley E M, Muller U, Ferguson M W, Sharpe P T. Gene. 1992;119:221–228. doi: 10.1016/0378-1119(92)90275-t. [DOI] [PubMed] [Google Scholar]
- 27.Guardavaccaro D, Corrente G, Covone F, Micheli L, D'Agnano I, Starace G, Caruso M, Tirone F. Mol Cell Biol. 2000;20:1797–1815. doi: 10.1128/mcb.20.5.1797-1815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bitzer M, von Gersdorff G, Liang D, Dominguez-Rosales A, Beg A A, Rojkind M, Bottinger E P. Genes Dev. 2000;14:187–197. [PMC free article] [PubMed] [Google Scholar]
- 29.Itoh S, Itoh F, Goumans M J, Ten Dijke P. Eur J Biochem. 2000;267:6954–6967. doi: 10.1046/j.1432-1327.2000.01828.x. [DOI] [PubMed] [Google Scholar]
- 30.Ulloa L, Doody J, Massague J. Nature. 1999;397:710–713. doi: 10.1038/17826. [DOI] [PubMed] [Google Scholar]
- 31.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, et al. Nature. 1997;387:924–929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 32.von Gersdorff G, Susztak K, Rezvani F, Bitzer M, Liang D, Bottinger E P. J Biol Chem. 2000;275:11320–11326. doi: 10.1074/jbc.275.15.11320. [DOI] [PubMed] [Google Scholar]
- 33.Raouf A, Seth A. Oncogene. 2000;19:6455–6463. doi: 10.1038/sj.onc.1204037. [DOI] [PubMed] [Google Scholar]
- 34.Tolon R M, Castillo A I, Jimenez-Lara A M, Aranda A. Mol Cell Biol. 2000;20:8793–8802. doi: 10.1128/mcb.20.23.8793-8802.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goltzman D, White J H. Crit Rev Eukaryotic Gene Expression. 2000;10:135–149. [PubMed] [Google Scholar]
- 36.Sato Y. Hum Cell. 1998;11:207–214. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.