Abstract
Olfactory receptor (OR) genes constitute the basis for the sense of smell and are encoded by the largest mammalian gene superfamily of >1,000 genes. In humans, >60% of these are pseudogenes. In contrast, the mouse OR repertoire, although of roughly equal size, contains only ≈20% pseudogenes. We asked whether the high fraction of nonfunctional OR genes is specific to humans or is a common feature of all primates. To this end, we have compared the sequences of 50 human OR coding regions, regardless of their functional annotations, to those of their putative orthologs in chimpanzees, gorillas, orangutans, and rhesus macaques. We found that humans have accumulated mutations that disrupt OR coding regions roughly 4-fold faster than any other species sampled. As a consequence, the fraction of OR pseudogenes in humans is almost twice as high as in the non-human primates, suggesting a human-specific process of OR gene disruption, likely due to a reduced chemosensory dependence relative to apes.
Olfactory receptors (ORs), the largest gene family in the human genome, underlie an exquisite capacity for odor perception (1–3). One of the most surprising features of the human olfactory gene repertoire is that >60% of human OR genes bear one or more sequence disruption, likely resulting in the functional inactivation of the encoded protein (4, 5). By contrast, the mouse, which has a similar number of OR genes, has only ≈20% pseudogenes (6–8).
The increased rate of OR gene disruption in humans relative to the mouse may be a general feature of the primate lineage. Alternatively, different primates may accumulate OR coding region disruptions at different rates. In particular, we are interested in the comparison of the olfactory repertoire of humans and that of the great apes and other non-human primates.
To date, there is only suggestive evidence of heterogeneity among primates in the size of the olfactory gene repertoire. A previous study examined partial coding sequences of 14 OR loci from one gene cluster on human chromosome 17 in humans and apes (9). Although three to five of these 14 OR loci were found to carry coding region disruptions in one or more ape species, all 14 OR genes were inferred to be intact in the common ancestor of all apes (9). A different study (10) used OR degenerate primers to examine the OR gene repertoire in different mammals. On the basis of a small number of genes in non-human primates (18–23), Rouquier et al. (10) concluded that humans and apes have significantly more pseudogenes than Old World monkeys. A specific comparison of human and the great ape species was not possible in their study due to the small number of genes in non-human species. Additionally, it was not possible to estimate species-specific rates of gene disruption due to the use of degenerate primers. Furthermore, even the apes and the Old World monkey comparisons may have not been accurate, because the use of degenerate primers may have biased the results. Indeed, Rouquier et al. (10) report zero pseudogenes in mouse and 70% pseudogenes in humans (where they looked at 99 ORs). These values are significantly different from the true values obtained once the entire OR gene repertoire was reported for these species (20% and 58%, respectively).
To determine whether the high fraction of nonfunctional OR genes is specific to humans or is also present among non-human apes, we compared 50 OR loci in humans, three great apes and one Old World monkey. The results point to a more rapid accumulation of OR coding region disruptions in the human lineage than in any other primate lineage.
Methods
OR Genes.
OR genes were obtained from the Human Olfactory Receptor Data Exploratorium (HORDE) database (http://bioinformatics.weizmann.ac.il/HORDE/), which contains the inferred protein sequence for every OR gene and pseudogene as mined from the public database (4). ORs were selected at random (by using a random number generator), with a sole constraint that the coding region length is >870 bp, ignoring functional annotation. OR genes of the 7E subfamily were excluded from the sample. This OR gene subfamily consists of 127 known loci in human; all but one are pseudogenes. It has been suggested that this gene subfamily has expanded in primates (4, 11). Excluding these genes from our sample is conservative regarding our conclusions of a higher fraction of OR pseudogenes in humans.
PCR and DNA Sequencing.
Primers for PCR amplification and sequencing were designed as the first and last 22 base pairs of each OR coding region to amplify the entire ORF. The same primers were used for all species. In all cases, the amplified PCR product was specific (no more than three polymorphic sites were found in any of the genes, and none of the polymorphic sites caused a coding region disruption). In 10 cases, we failed to amplify a product (Table 1). PCR was performed in a total volume of 25 μl, containing 0.2 μM of each deoxynucleotide (Promega); 50 pmol of each primer; 1.5 mM MgCl2; 50 mM KCl; 10 mM Tris, pH 8.3; two units of Taq DNA polymerase; and 50 ng of genomic DNA. PCR conditions were as follows: 35 cycles of denaturation at 94°C, annealing at 53, 55, or 57°C, depending on the primers, and extension at 72°C, each step for 1 min. The first step of denaturation and the last step of extension were 3 and 10 min, respectively. PCR products were separated and visualized in a 1% agarose gel and purified by using the High Pure PCR Product Purification Kit (Boehringer Mannheim). Sequencing reactions were performed on PCR products in both directions with a dye-terminator cycle sequencing kit (Applied Biosystems) on an ABI 3700 automated sequencer (Applied Biosystems).
Table 1.
OR genes analyzed
| Locus | Human | Chimp | Gorilla | Orang | Rhesus |
|---|---|---|---|---|---|
| 10A3 | 824-stop | ||||
| 10A5 | 396-stop | ||||
| 10AA1p | 302-del | 302-de | 302-del | ||
| 10J5 | 238-stop | ||||
| 10T1p | 391-stop | ||||
| 11H7p | 691-stop | ||||
| 11i1p | 274-stop | 274-stop | 274-stop | ND | |
| 11K1p | 176-stop | 176-stop | 176-stop | 176-stop | |
| 12D1p | 557-del | 235-ins | |||
| 13C3 | 180-stop | ||||
| 13C6p | 235-del | 744-del | |||
| 13D1 | 105-stop | ||||
| 13E1p | 182-del | 182-del | |||
| 13H1 | 300-del | 404-del | |||
| 1C1 | |||||
| 1J2 | 317-stop | ||||
| 1L3 | ND | ||||
| 1S1 | 96-del | ||||
| 2Ai1p | 528-del | 528-del | 203-del | ||
| 2J3 | 530-ins | ||||
| 2L8 | 444-stop | 245-ins | |||
| 2Q1p | 505-del | 507-del | 518-del | ||
| 2T7p | 77-del | ||||
| 4A13p | 174-stop | 406-stop | |||
| 4A4 | 309-del | ||||
| 4E1p | |||||
| 4F15 | ND | 328-del | |||
| 4G3p | 178-del | 178-del | 178-del | 178-del | 178-del |
| 4K15 | |||||
| 4L2p | 249-ins | ||||
| 51A5p | 706-stop | ||||
| 51A6p | 373-del | 373-del | 373-del | 84-stop | 414-del |
| 51G2 | |||||
| 51J1p | 302-ins | ||||
| 51R1p | 679-stop | 679-stop | 176-stop | ||
| 52B1p | 723-del | 723-del | 723-del | 723-del | 723-del |
| 52H2p | 270-del | 270-del | 270-del | 270-del | 540-stop |
| 52L1 | 847-del | ||||
| 5AK4p | 236-stop | 236-stop | 236-stop | 236-stop | 236-stop |
| 5AL2p | 346-del | ND | |||
| 5E1p | 335-stop | 384-del | |||
| 5H5p | 212-del | 212-del | 212-del | ||
| 5H8p | 678-ins | ND | |||
| 5K1 | |||||
| 5M13p | 471-ins | 252-del | 238-stop | ||
| 5P4p | 355-del | 143-stop | 203-del | ||
| 5U1 | |||||
| 6F1 | |||||
| 6K2 | ND | ||||
| 6M1 | 61-ins | 294-del | |||
| 6Q1 | |||||
| 6Y1 | |||||
| 7A8p | 430-del | 430-del | ND | ||
| 7D4p | 43-ins | ||||
| 8B5p | 71-stop | 71-stop | 71-stop | ND | 71-stop |
| 8D1 | 261-del | ||||
| 8F1p | 605-del | 605-del | 605-del | 605-del | 605-del |
| 8J2p | 208-stop | 718-stop | |||
| 9A2 | ND | 255-ins | |||
| 9i2p | 257-del | 257-del | ND |
We list the names of all OR loci in our sample. The nomenclature follows the suggestion of ref. 26. OR names consist of the subfamily to which the OR belongs and a serial number of the OR within the subfamily. The positions and nature of the disruptions are given for each OR gene analyzed. Empty spaces indicate intact coding regions. For the last 10 genes, DNA sequences could not be determined for the species indicated. ND, not determined.
Sequence Analysis.
After base calling with Applied Biosystems analysis software (Ver. 3.0), the data were edited and assembled by using the sequencher program, Ver. 4.0 (Gene Codes, Ann Arbor, MI). At both ends of each coding sequence, ≈40 base pairs including the PCR primers were excluded from the analysis. Because OR genes share high degrees of similarity, we compared the consensus sequence of each gene from the two individuals sequenced for each species against the HORDE database. In all cases but two, the best hit was the desired gene. In two cases, OR12D1P and OR4L2P, the best hits for all non-human primates were OR12D2 and OR4L1, respectively. However, assembling both reference sequences with the actual data revealed that we indeed amplified OR12D1P and OR4L2P in all species. OR12D2 and OR4L1 were the first hit in the blast due to length. These are probably gene duplications that predate the human–rhesus separation, and where both copies remained functional in most non-human primates.
Coding region disruptions were identified separately for each species. When more than one coding region disruption was identified in the same species, we inferred which occurred first by identifying disruptions shared between species. We considered only one disruption per gene to determine the gene silencing rate in each lineage.
Mouse gene annotation was done by comparing inferred human OR protein sequences to the nonredundant (nr) division of GenBank (www.ncbi.nih.gov/GenBank) by using the tBLASTN algorithm (12). The protein sequence of the highest hit, corresponding to a mouse sequence and spanning at least 290 residues, was used as a query in a blast search against the HORDE database. Orthology was deduced for 33 locus pairs (66%) when the best hit in the second search was the human OR gene, which served as the query sequence for the first search. For the other 17 loci, we chose the best mouse hit for the human query as the “ortholog.” If only the first set of orthologous pairs is considered, the fraction of OR pseudogenes in the mouse would be 12% (compared with 16% in the entire sample).
Recent common ancestor sequences for every node were inferred by maximum likelihood by using the paml software package (13). Divergence was estimated by using either the Jukes–Cantor model or Kimura's two-parameter model in DNAsp (14) by using the paml output. The choice of mutation model did not affect the qualitative conclusions. Divergence values (Fig. 2) are presented for the Jukes–Cantor model.
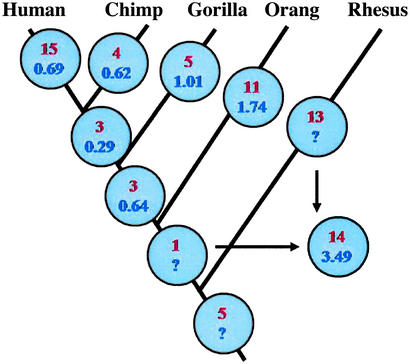
Figure 2.
OR pseudogene accumulation and DNA sequence divergence in primates. The number of OR pseudogenes inferred to have arisen on each branch is given in red. The inferred divergence (in percentage) for each branch is indicated in blue. Because we used the rhesus as an outgroup, we could not infer the divergence specific to the rhesus lineage. A combined divergence value for the outgroup is indicated by the arrows. We inferred gene silencing events on the rhesus lineage by assuming that the mutations have always occurred to disrupt an intact coding region rather than to reverse a disruption.
Results and Discussion
We selected 60 full-length OR genes at random from the human genome sequence repositories (Table 1), irrespective of whether their coding regions are annotated as disrupted or intact. PCR amplification was performed with primers positioned at the extreme ends of the OR coding regions based on the database human sequences. The relatively low paralog conservation in these sequence regions ensures a high probability of ortholog specific PCR amplification (20). This was performed in two humans, two chimpanzees (Pan troglodytes), two gorillas (Gorilla gorilla), two orangutans (Pongo pygmaeus), and two rhesus macaques (Macaca mulatta). Fifty of the ORs were successfully amplified from all five primate species and were subjected to DNA sequence analysis. The 50 ORs are located on 14 different chromosomes and belong to 13 different OR gene families (Fig. 1), thus providing an adequate coverage of the OR repertoire. The DNA sequences of these genes were determined from all individuals. Consistent with previous results (4), 54% of the ORs sequenced in humans contained at least one stop codon in the reading frame and are thus pseudogenes (Tables 1 and 2). The coding region disruptions identified were identical to those reported in the HORDE database except in one case (OR4E1P), where we found the OR gene to be intact. This may be a sequencing error incorporated in the database or may represent a human polymorphism. We found no polymorphism that created an OR-coding region disruption in any of the studied species.
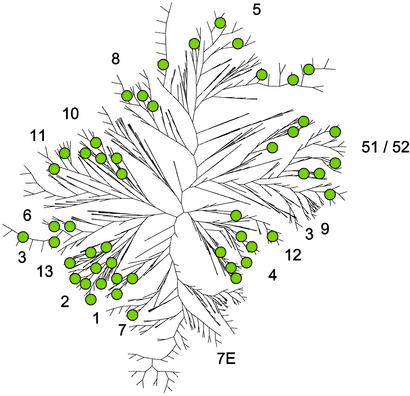
Figure 1.
A neighbor-joining tree of all human OR loci in the HORDE database. The names of the OR gene families are indicated next to the branches, which represent the majority of members from each subfamily. Few OR members of any subfamily may be dispersed elsewhere in the tree (not specified). The 50 ORs chosen at random for this study are indicated as circles.
Table 2.
Relative rates of OR gene silencing
| Human | Chimp | Gorilla | Orang | Rhesus | |
|---|---|---|---|---|---|
| Fraction of OR pseudogenes, % | 54 | 32 | 28 | 32 | 36 |
| Gene silencing rate relative to the mean* | 3.28 | 0.92 | 0.72 | 0.89 | 0.66 |
| FET† | 0.00003 | 1 | 0.675 | 0.871 | 0.213 |
| Gene silencing rate relative to mean, human excluded‡ | 4.29 | 1.20 | 0.94 | 1.17 | 0.87 |
| FET | 0.00001 | 0.771 | 1 | 0.715 | 0.757 |
Gene silencing rate on a specific lineage relative to the mean rate of the entire phylogeny.
P values for Fisher's exact tests (FET) for the difference between the mean rate of OR pseudogene accumulation and the lineage-specific rates.
All specific lineages rates are relative to a mean rate, which is calculated excluding the human lineage.
The fraction of pseudogenes in the apes and the rhesus macaque was 28–36% (Table 2). This is significantly more than in the mouse (P < 0.04 for all comparisons) but significantly fewer than in the humans (P < 0.03 for all comparisons). Even if we include the 10 genes that we could not amplify, and conservatively assume that they are all pseudogenes in the non-human primates for which the amplification failed (Table 1), the observation that humans have more pseudogenes than apes remains statistically significant (P < 0.05). We inferred on which lineage each gene silencing event occurred by estimating the ancestral sequences of each node in a tree representing the phylogenetic relationships of the species (Fig. 2). Nine OR genes were intact in all primate species examined, whereas six were pseudogenes in all species. Of these, five OR loci were inferred to have been pseudogenes in the common ancestor of all five species (Fig. 2). In one case (OR11K1P), an ancestral stop codon seems to have been lost on the orangutan lineage.
The estimated interspecies DNA sequence divergence values (Fig. 2) are consistent with other reports (15–17) and do not reject the hypothesis of equal evolutionary rates on all lineages. In contrast, the hypothesis of equal OR coding region disruption rates among lineages is rejected for the human lineage (P = 0.00003) but not for the other lineages (Table 2). If the mean rate of accumulating OR pseudogenes in the entire phylogeny is calculated as the ratio of the number of gene disruption events to the number of nucleotide substitutions, humans have a rate of OR gene disruption 3.3 times higher then the mean rate throughout the phylogeny (Table 2). If the human lineage is excluded, and mean rate is estimated for the non-human primate phylogeny only, the rate of OR gene disruption for all non-human primates is practically identical, whereas the human rate is 4.3 times greater than the mean (Table 2). Thus, although monkeys and apes have about twice as many pseudogenes in their OR gene repertoire as the mouse, humans have accumulated OR pseudogenes significantly faster than other apes, such that they currently have >50% more pseudogenes. Assuming a human–mouse separation of ≈110 million years (18) and conservatively assuming that 20% of the mouse OR genes accumulated coding region disruptions since the human–mouse divergence, the rate of OR gene silencing in mice would be half as low as in the non-human apes and approximately nine times lower than in humans.
Nine OR genes were intact in all primate species examined (Table 1). Along the human evolutionary lineage, only one amino acid change has occurred in the putative OR-binding sites [a total of 261 amino acids (19); O.M., Y.G. and D.L., unpublished results] of these nine OR genes. This compares with 11 changes in the 14 OR genes (406 amino acids) intact in humans but not in one or more of the other primates examined (P = 0.034). This observation may suggest that evolutionary constraints differ among human OR genes. We propose that OR genes in the human genome belong to three functional groups: (i) OR genes that are essential to all primates and therefore are under selective pressure to remain intact in humans as well; (ii) OR genes that are not important for humans but are essential for other primates; (iii) OR pseudogenes that have lost their function in humans. The two latter categories of genes would accumulate coding region disruptions at a neutral rate in humans, thus explaining the high rate of OR gene silencing observed in the human evolutionary lineage.
In conclusion, our results show that a much faster functional deterioration of the largest mammalian gene superfamily occurred in the human lineage. This process is probably still ongoing in humans, as indicated by the presence of many OR genes carrying a polymorphism for an intact/disrupted coding region (20). It cannot be excluded that a reduction in the efficiency of purifying selection as a result of the smaller effective population size in humans relative to the other primates (15, 16, 21) has contributed to the high rate of OR gene disruption in humans. However, previous reports indicate that the difference in population size between humans and other apes is 2- or 3-fold (15, 16, 21). For this difference to explain our observation, the selection coefficients associated with OR gene silencing must be within a narrow range in all non-human primates across a large fraction of the OR gene repertoire (1 < N⋅s < 3, where N is the effective population size and s the selection coefficient), which seems unlikely. Instead, we suggest that humans do not rely on their sense of smell as much as apes. For example, certain aspects of monkey social behavior and mating choice have been suggested to be influenced by the olfactory system (22–25). Although it has not been established that the OR genes are responsible for these functions, it is tempting to speculate that a lesser need for the sense of smell in humans may be manifested in relaxed evolutionary constraints, resulting in a higher rate of OR coding region disruption in humans. Further work into the functional properties of OR genes as well as into the genome-wide patterns of gene silencing in humans and apes is necessary to clarify whether this is the case.
Acknowledgments
We thank C. Allen and T. Insel of the Yerkes Primate Center, Atlanta, for primate tissue samples; Birute Galdikas (Simon Fraser University, Burnaby, BC, Canada) for orangutan blood samples; and M. Przeworski for helpful discussions and comments on the manuscript. The experimental work was financed by the Bundesministerium für Bildung und Forschung (01KW9959-4) and by the Max Planck Gesellschaft. D.L. holds the Ralph and Lois Silver Chair in Human Genomics supported by the Crown Human Genome Center at the Weizmann Institute of Science.
Abbreviation
- OR
olfactory receptor
Footnotes
References
- 1.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 2.Lancet D, Ben-Arie N. Curr Biol. 1993;3:668–674. doi: 10.1016/0960-9822(93)90064-u. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Arie N, Lancet D, Taylor C, Khen M, Walker N, Ledbetter D H, Carrozzo R, Patel K, Sheer D, Lehrach H, North M A. Hum Mol Genet. 1994;3:229–235. doi: 10.1093/hmg/3.2.229. [DOI] [PubMed] [Google Scholar]
- 4.Glusman G, Yanai I, Rubin I, Lancet D. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- 5.Zozulya S, Echeverri F, Nguyen T. Genome Biol. 2001;2:RESEARCH0018. doi: 10.1186/gb-2001-2-6-research0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young J M, Friedman C, Williams E M, Ross J A, Tonnes-Priddy L, Trask B J. Hum Mol Genet. 2002;11:535–546. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Firestein S. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 8.Young J M, Trask B J. Hum Mol Genet. 2002;11:1153–1160. doi: 10.1093/hmg/11.10.1153. [DOI] [PubMed] [Google Scholar]
- 9.Sharon D, Glusman G, Pilpel Y, Khen M, Gruetzner F, Haaf T, Lancet D. Genomics. 1999;61:24–36. doi: 10.1006/geno.1999.5900. [DOI] [PubMed] [Google Scholar]
- 10.Rouquier S, Blancher A, Giorgi D. Proc Natl Acad Sci USA. 2000;97:2870–2874. doi: 10.1073/pnas.040580197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rouquier S, Friedman C, Delettre C, van den Engh G, Blancher A, Crouau-Roy B, Trask B J, Giorgi D. Hum Mol Genet. 1998;7:1337–1345. doi: 10.1093/hmg/7.9.1337. [DOI] [PubMed] [Google Scholar]
- 12.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 14.Rozas J, Rozas R. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 15.Jensen-Seaman M I, Deinard A S, Kidd K K. J Hered. 2001;92:475–480. doi: 10.1093/jhered/92.6.475. [DOI] [PubMed] [Google Scholar]
- 16.Kaessmann H, Wiebe V, Weiss G, Paabo S. Nat Genet. 2001;27:155–156. doi: 10.1038/84773. [DOI] [PubMed] [Google Scholar]
- 17.Ebersberger I, Metzler D, Schwarz C, Paabo S. Am J Hum Genet. 2002;70:1490–1497. doi: 10.1086/340787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S, Hedges S B. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 19.Pilpel Y, Lancet D. Protein Sci. 1999;8:969–977. doi: 10.1110/ps.8.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gilad, Y. & Lancet, D. (2003) Mol. Biol. Evol., in press. [DOI] [PubMed]
- 21.Hacia J G. Trends Genet. 2001;17:637–645. doi: 10.1016/s0168-9525(01)02494-5. [DOI] [PubMed] [Google Scholar]
- 22.Michael R P, Zumpe D, Bonsall R W. J Comp Physiol Psychol. 1982;96:875–885. [PubMed] [Google Scholar]
- 23.Michael R P, Zumpe D. J Endocrinol. 1982;95:189–205. doi: 10.1677/joe.0.0950189. [DOI] [PubMed] [Google Scholar]
- 24.Savic I, Berglund H, Gulyas B, Roland P. Neuron. 2001;31:661–668. doi: 10.1016/s0896-6273(01)00390-7. [DOI] [PubMed] [Google Scholar]
- 25.Ferris C F, Snowdon C T, King J A, Duong T Q, Ziegler T E, Ugurbil K, Ludwig R, Schultz-Darken N J, Wu Z, Olson D P, et al. NeuroReport. 2001;12:2231–2236. doi: 10.1097/00001756-200107200-00037. [DOI] [PubMed] [Google Scholar]
- 26.Glusman G, Bahar A, Sharon D, Pilpel Y, White J, Lancet D. Mamm Genome. 2000;11:1016–1023. doi: 10.1007/s003350010196. [DOI] [PubMed] [Google Scholar]