Abstract
The vomeronasal organ (VNO) detects pheromones in many vertebrate species but is likely to be vestigial in humans. TRPC2(TRP2), a gene that is essential for VNO function in the mouse, is a pseudogene in humans. Because TRPC2 is expressed only in the VNO, the loss of selective pressure on this gene can serve as a molecular marker for the time at which the VNO became vestigial. By analyzing sequence data from the TRPC2 gene of 15 extant primate species, we provide evidence that the VNO was most likely functional in the common ancestor of New World monkeys and Old World monkeys and apes, but then became vestigial in the common ancestor of Old World monkeys and apes. We propose that, at this point in evolution, other modalities, notably the development of color vision, may have largely replaced signaling by pheromones.
The vomeronasal organ (VNO), a sensory organ that detects nonvolatile pheromones, is found in most terrestrial vertebrates but may be absent in humans (1, 2). In most species, the paired cigar shaped organ is present at the rostral end of the nasal cavity and sensory neurons within the VNO send projections to the accessory olfactory bulb. In humans a small pit is retained in the nasal septum, but cells within the presumptive VNO do not express many of the markers of mature sensory neurons (1, 2). The accessory olfactory bulb is not evident in humans or in any other Old World (OW) monkey or ape (3). Nonetheless, it has been claimed that in humans some pheromones are transduced by the VNO (4). Indeed, it has been difficult to determine unequivocally whether the VNO is indeed functional or nonfunctional in humans and higher primates (1).
Candidate signaling components for VNO sensory transduction that are specifically expressed in the VNO have been recently identified, including several large families of receptors (V1R/V3Rs and V2Rs) (5–7) and an ion channel TRPC2 (8). TRPC2 is localized to VNO sensory microvilli, suggesting a role in sensory transduction (8). Knockout studies show that TRPC2 is essential for a fully functioning VNO; in the absence of the TRPC2 gene, male mice initiate sexual rather than aggressive behavior toward other male mice, indicating an inability to detect some pheromones (9, 10). Similarly, deletion of V1R genes in the mouse leads to a loss of VNO sensitivity to a subset of signaling chemicals and to a loss of some pheromone-sensitive behaviors (11). The TRPC2 gene is a pseudogene in humans, as are the vast majority of V1R/V3R genes that have been identified in humans (5, 8, 12–18). The observed loss of molecular components of VNO signaling is perhaps the best evidence that the human VNO is vestigial (1). Because TRPC2 is expressed specifically in the VNO (8) and is essential for VNO function (9, 10), an examination of the gene across primate phylogeny can provide direct evidence concerning the functionality of the VNO in various species and through evolution.
Methods
Sequence of Human TRPC2 (hTRPC2).
Coding sequence for hTRPC2 was identified on 11p15.5 (GenBank accession no. AC060812) by comparison with mouse TRPC2 (mTRCP2) or homology to the partial cDNA for hTRPC2 (12), and the position of intron/exon boundaries was assigned based on similarity to consensus motifs. DNA sequences were assembled by using vector nti suite 6 (InforMax, Bethesda). A reconstructed full-length hTRPC2 cDNA consisting of 2,667 nt was obtained by conceptually splicing exons 1, 5, and 8 identified from the genomic sequence, and the partial cDNA sequence of hTRPC2, which contains the remaining exons.
PCR Amplification and Sequencing of Primate Genomic DNA.
Genomic DNA from the following primate species was obtained from Therion (Troy, NY): squirrel monkey (Saimiri sciureus), common marmoset (Callithrix jacchus), owl monkey (Aotus azarai), and black and white ruffed lemur (Varecia variegata). Genomic DNA from the following species was obtained from O. Ryder (San Diego Zoo, San Diego): sumatran orangutan (Pongo pygmaeus), bonobo (Pan paniscus), common chimpanzee (Pan troglodytes), western lowland gorilla (Gorilla gorilla), gibbon (Hylobates syndactylus), drill (Mandrillus leucophaeus), titi monkey (Callicebus moloch), spider monkey (Ateles geoffroyi), howler monkey (Alouatta seniculus), and ring-tailed lemur (Lemur catta). Genomic DNA from rhesus macaque (Macaca mulatta) and human was from CLONTECH. Note that by using chimp or bonobo DNA as template we were able to obtain amplification in only two of the five reactions; therefore, we do not include these data in our analyses.
PCR primers and conditions for this and other experiments in which primate DNA was amplified were as follows. PCR primer pairs: (i) AGCCTAACTGGACTGAGATCGTGAAC vs. GCTGCACTCGAGGCAGGCACAGG; (ii) GTAGCACACCTCATCTGCCAGCAAG vs. CCAGTAGCCAAGGCAGAGGAAGGG; (iii) either GCTGGGCTTGCCCACATGCACTGCC vs. GGCCAGGCGGGTGAAGCTGAGCATGC (most primates) or GCCACTGCGGCCCTCCTCCTGGCTGGGCTT vs. GAAGAGCACCTTCAGCCAAGAACTTGGG (human, macaque, langur); (iv) GGATGATGCTGACGTGGAGTGGA vs. GGCAAAAGTAGGGATCGGGGGATAGTC; and (v) GACCATATCTCGACTGCAAAGCGAGG vs. GCCAGACTCTCCGGAAGCCAGAGTCC. Conditions were as follows: 50 mM KCl, 10 mM Tris⋅HCl, 1.5 mM MgCl2, 200 mM dNTPs, 4 mM each primer, 0.05 units/μl Taq polymerase (Fisher), and 4 ng/μl template. PCR products were purified (QIAquick, Qiagen, Valencia, CA) and directly sequenced off of both strands by using the original PCR primers (Laragen, Los Angeles). Note that direct sequencing of the PCR products is less prone to artifacts than sequencing subcloned PCR products. Only sequence that was confirmed on both strands was used in the analysis. For 14 species, we were able to obtain 745–770 bp of sequence. Because of failure of one or more PCRs, common marmoset, squirrel monkey, and orangutan gave 605, 645, and 597 bp of sequence, respectively.
Analysis of Sequence Data.
Sequences were aligned and the ratio of nonsynonymous substitutions per nonsynonymous site vs. synonymous substitutions per synonymous sites, Ka/Ks, was determined by oden (19). The numbers of nonsynonymous and synonymous nucleotide differences (da and ds) and the numbers of nonsynonymous and synonymous sites (la and ls) were obtained for all possible pairs of sequences. Ka and Ks were estimated from da, ds, la, and ls with Jukes and Cantor's correction (20). To test the significance of the deviation of Ka/Ks from one, a two-tailed Fisher's exact test was conducted with the null hypothesis da:ds = la:ls. To examine the compatibility of two values of Ka/Ks, we conducted 2 × 2 Fisher's exact test with the null hypothesis that da/ds is constant. To test the heterogeneity in the rate of molecular evolution (measured by d = da + ds), a relative rate test was carried out (21).
Results
Identification of the Full-Length hTRPC2 Gene.
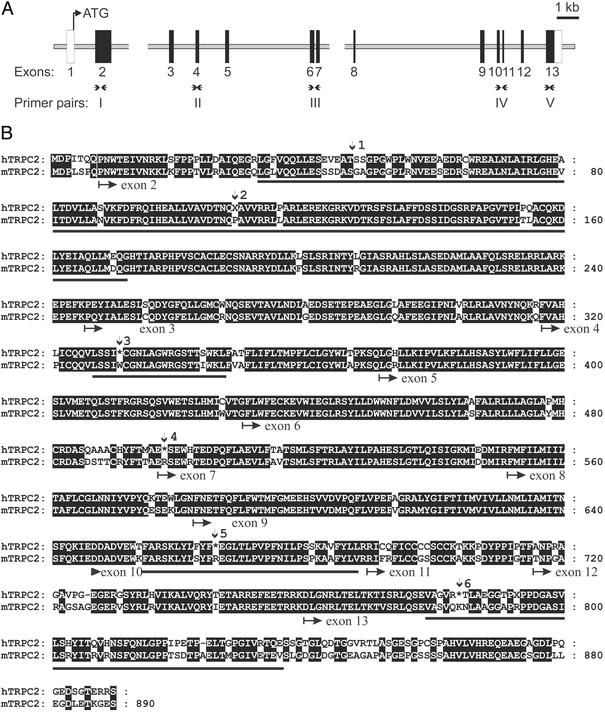
Based on sequence obtained from a partial cDNA clone, it was previously noted that hTRPC2 was a pseudogene (12). To obtain the full-length coding sequence of hTRPC2, we queried the human genome database with mouse and rat TRPC2 cDNA sequences. Only one region of the genome (11p15.5) showed a high degree of homology to rodent TRPC2 cDNAs (85% nucleotide identity). The coding region of hTRPC2 is contained on 13 exons (Fig. 1A). Within the deduced cDNA for hTRPC2, we have identified six deleterious mutations that generate stop codons (Fig. 1B); two of these are indels (insertion/deletions) and four are missense mutations.
Figure 1.
The genomic organization and deduced sequence of hTRPC2. (A) Structure of the TRPC2 gene. Thirteen exons were identified on chromosome 11p15.5 in a draft sequence of the human genome. Black boxes represent exons containing coding sequence. White boxes indicate exons that contain noncoding sequence; the size of these exons was not determined. Breaks in the sequence represent regions of the human genome for which the data are still ambiguous. The positions of primer pairs I–V are indicated. Note that two primer pairs (III and IV) span introns. (B) The deduced sequence of hTRPC2 aligned with the sequence of mouse TRPC2 (mTRPC2). Identical residues are boxed in black. The position of intron/exon boundaries is indicated below the sequence. Vertical arrows indicate the positions of deleterious mutations. Site 1 is a 4-bp insertion and site 2 is a 1-bp deletion. These nucleotides were removed to produce the coding sequence shown. Stop codons are indicated by an asterisk. Bars underlining sequences indicate the portion of the sequence used for determining Ka/Ks.
Deleterious Mutations in the TRPC2 Gene of Primates.
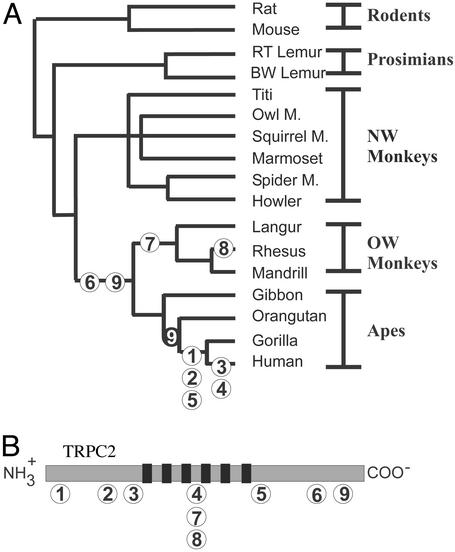
To ascertain when in primate evolution TRPC2 became a pseudogene, we determined when each of the deleterious mutations found in the human sequence occurred. Genomic DNA from 15 extant primate species was used in PCRs to obtain sequence of the TRPC2 gene. In the sequences of nonhuman primates, we identified three additional frameshift or non-sense mutations that were not found in the human sequence. Of the nine mutations, the earliest two mutations arose in the common ancestor of OW monkeys and apes (Fig. 2). These non-sense mutations both lead to a truncation of the cytoplasmic C terminus, a region that is likely to be important but may not be essential for the function of the protein (22). Occurring more recently were several mutations that would clearly generate a nonfunctional protein. (i) In the common ancestor of gorillas and humans, two frameshift mutations arose at the N terminus of the protein and a non-sense mutation arose after the sixth transmembrane domain. (ii) In the common ancestor of extant OW monkeys, there was a 1-bp insertion that is predicted to generate a stop codon between the third and fourth transmembrane domains (mutation 7 in Fig. 2). We did not detect any frameshift or non-sense mutations in sequences of TRPC2 from prosimians or New World (NW) monkeys. Thus, it appears that deleterious mutations in the TRPC2 gene began to accumulate in the common ancestor of OW monkeys and apes.
Figure 2.
Inferred time of occurrence of deleterious mutations in TRPC2 during primate phylogeny. (A) Mutations are numbered 1–9. Mutations 1–6 are found in human at the positions indicated in Fig. 1. Mutation 6 was found in all OW monkeys and hominoids but not in NW monkeys or prosimians; therefore, we infer that it occurred in the common ancestor of OW monkeys and apes. Mutation 9 is a missense mutation that was found in all OW monkeys and in gibbon but not in other hominoids, suggesting either that a reversion event occurred more recently in evolution (indicated by a black circle with a white 9 on the tree) or that the mutation arose independently more than once. Two frameshift mutations (7 and 8) were found only in OW monkeys. Mutation 7 is a 1-bp insertion at a position equivalent to mouse D483 and mutation 8 is a 13-bp deletion spanning the same position. The deletion event therefore restores the reading frame. (B) A schematic representation of the TRPC2 ion channel indicating the position of each mutation. Black bars represent the transmembrane domains.
Selective Pressure on the TRPC2 Gene During Primate Phylogeny.
To confirm the preceding analysis, and to gain more insight into the selective pressure on the TRPC2 gene across primate phylogeny, we measured rates of synonomous and nonsynonomous substitutions among TRPC2 sequences from a range of primate species. Synonomous substitutions are nucleotide substitutions that do not alter the amino acid sequence. In the absence of selective pressure and under neutral evolution, synonomous substitutions per synonomous site (denoted Ks) occur at a rate equal to nonsynonomous substitutions per nonsynonomous site (denoted Ka); i.e., Ka/Ks = 1. Any significant deviation from this prediction can be interpreted as indicating that the gene is under selective pressure and thus is likely to be functional. In the case of receptor proteins that interact with ligands in the environment, selection may favor adaptive changes in amino acid sequences, leading to an elevated value of Ka/Ks over part of the amino acid sequence. This is the case for mammalian odorant and vomeronasal receptors (23, 24). On the other hand, in the case of molecules that interact with intracellular messengers, such as TRPC2 and related ion channels, it might be expected that selection would be purifying, corresponding to a low value of Ka/Ks.
Because of the presence of a large number of introns, we generated sequence for analysis (745–770 bp for most species) by concatenating coding sequence from four exons. We eliminated the possibility that this analysis introduced a bias in two ways: (i) to eliminate ascertainment bias, before the analysis we removed codons for which the human had a non-sense mutation and we corrected frameshift mutations; and (ii) we confirmed that for the mouse and rat genes, comparison of the partial sequence gave a similar result, as a comparison of the full sequence (in both cases Ka/Ks = 0.08).
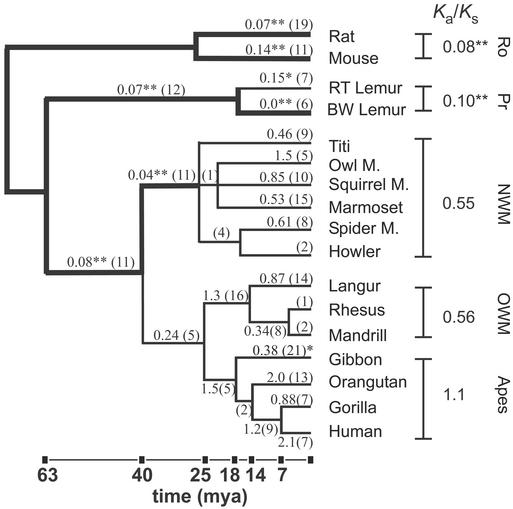
Ka/Ks values were then obtained for all pairwise comparisons of TRPC2 sequence data from 15 primate species and two rodent species (Fig. 3). In addition, we reconstructed the most likely ancestral sequence at each node of the phylogenetic tree by using maximum parsimony phylogeny analysis (phylip; ref. 25) and used these sequences to calculate Ka/Ks on each branch of the tree (Fig. 3; ref. 26). Parsimony analysis yielded three trees that differed only in branches leading from the common ancestor of NW monkeys to extant NW monkeys, and there was no significant difference in the evaluation of Ka/Ks by using these trees.
Figure 3.
Rates of synonymous and nonsynonomous substitutions in the TRPC2 gene across primate phylogeny. Pairwise comparisons of sequence from all species were performed and Ka/Ks values were computed. Within-group values show the average Ka/Ks ratio for all pairwise comparisons in a group. A double asterisk indicates that Ka/Ks is not compatible with neutral evolution (P ≤ 0.002). The Ka/Ks values computed by using predicted ancestral sequences are shown above the corresponding branch of the tree, and the total number of substitutions is shown in parentheses. Branches on which selective pressure was strongly evident (P ≤ 0.002) are shown with a thick line, and Ka/Ks values are marked with a double asterisk. This P value was derived by using the Bonferroni correction to ensure an overall type I error rate (false rejection of the null hypothesis) of 5% for each of the 22 tests conducted. On two branches, Ka/Ks values were significantly different from unity at P < 0.05. These branches are marked with a single asterisk. Lengths of branches are proportional to divergence dates for lineages of NW monkeys and OW monkeys and apes. Dates are based on ref. 34. Ro, rodent; Pr, prosimian; mya, million years ago.
The Ka/Ks value when comparing the two rodent sequences was significantly different from the neutral expectation (Ka/Ks = 0.08 P < 0.0001 Fisher's exact test) and was somewhat lower than the average value of 0.14 obtained when a large number of orthologous mouse and rat genes were examined (27). Thus, the TRPC2 gene exhibits a high degree of purifying selection in the rodent lineage, consistent with its known functional significance (9, 10).
The prosimians are considered the most primitive primates and are thought to have retained many of the characteristics of the common ancestor of modern primates. We find a high degree of purifying selection on the TRPC2 gene (Ka/Ks = 0.10; P < 0.0001) in comparing sequence from two strepsirrhine prosimians (black and white ruffed lemur and ring-tailed lemur; Fig. 3). In addition, we find a high degree of purifying selection on the branch of the phylogenetic tree leading to the common ancestor of these two species (Ka/Ks = 0.07, P < 0.0001). These data imply that the TRPC2 gene has been retained in prosimians.
NW monkeys comprise a diverse group of species, most of which have retained a VNO and an anatomically distinct accessory olfactory bulb (3). In pairwise comparisons of sequences of TRPC2 from NW monkeys, only one comparison yielded a Ka/Ks value that was significantly different from unity (owl monkey vs. titi: Ka/Ks = 0.33, P < 0.02). We found a significant relaxation of purifying selection on the TRPC2 gene in NW monkeys as evidenced by the significantly higher Ka/Ks values obtained from pairwise comparisons of NW monkey sequences (range, 0.33–0.94) as compared to the Ka/Ks value obtained in a comparison of rat and mouse TRPC2 [P < 0.03 for all (15/15) comparisons; 2 × 2 Fisher's exact test]. An examination of Ka/Ks values along the branches leading to the NW lineage shows a high degree of purifying selection up to a common ancestor of all NW monkeys (Fig. 3). A relaxation of selective pressure occurred more recently in evolution, suggesting that in some species of the NW monkeys the VNO may be vestigial or redundant. The recent relaxation of selective pressure may not have allowed sufficient time for deleterious mutations to accumulate in the TRPC2 gene.
Pairwise comparisons of sequence data among OW monkeys and apes shows no evidence for selective pressure on the TRPC2 gene (mean Ka/Ks = 0.86, range = 0.55–1.97; P > 0.05 for 20/21 comparisons). In the lineage leading to humans, we find that selective pressure on the TRPC2 gene was maintained in the anthropoid ancestor (the common ancestor of NW monkeys, OW monkeys, and apes) and was thereafter relaxed in a common ancestor of OW monkeys and apes (≈25–40 million years ago; Fig. 3). Following this time point, Ka/Ks values along branches leading to OW primates and apes are not significantly different from unity. Note that in one branch (leading from the common ancestor of apes to the gibbon), the Ka/Ks differs from unity at P < 0.05. However, the Bonferroni correction, which adjusts significance criteria in cases in which multiple significance tests are performed, requires a value of P < 0.002 (assuming 22 comparisons). The finding that selective pressure was relaxed on the TRPC2 gene in the common ancestor of OW monkeys and apes is consistent with our preceding analysis, which showed that at this same point in evolution deleterious mutations began to accumulate in the TRPC2 gene.
Genes that are under relaxed selective pressure will accumulate base substitutions at a higher rate than genes that are under purifying selection (28). To determine whether the rates of nucleotide substitutions within the TRPC2 gene varied among branches of the phylogenetic tree, we performed the relative rates test (21) on all pairwise comparisons of primates species, by using both rat and mouse as outgroups. The relative rates test confirms that mutations have accumulated in the OW monkey and ape lineage at a rate significantly higher than in the prosimians, consistent with the contention that in the OW monkey and ape lineage the TRPC2 gene is not under selective pressure (23/24 comparisons were significant at P < 0.05; one tailed relative rates test; 24 comparisons were from two prosimians compared to six OW monkeys and apes with two outgroups). Note that the test is very conservative, because the generation time for prosimians is shorter than for OW monkeys and apes, and therefore it would be expected that neutral substitutions would occur at a higher rate in prosimians.
Discussion
Our data show that among present-day primates there has been strong selective pressure on the TRPC2 gene only in a strepsirrhine prosimian, the lemur. These data are consistent with literature indicating that lemurs have retained a functioning VNO (29). Surprisingly, we find that selective pressure on the TRPC2 gene, although high in a common ancestor of NW monkeys, is relaxed in several species of extant NW monkeys. It is interesting to note that, although NW monkeys show prominent scent marking behavior, it has not been demonstrated that this form of chemical communication acts through the VNO or that it serves a unique function. Notably, in the marmoset, in which ovulation is suppressed among subordinate females in a social group, visual as well as chemical cues appear to play a role in suppressing ovulation (30). Interestingly, a recent study of V1R pheromone receptor genes in the marmoset has revealed the presence of only pseudogenes (31). This may be because of either extreme sequence divergence of functional V1R sequences (24), as the authors suggest, or a lack of functional significance of the VNO in this species. Our data suggest that, in NW monkeys, signaling through the VNO may, if present, be redundant, leading to a relaxation of selection on all signaling components within the VNO.
Within the lineage leading to humans, our evidence suggests that selective pressure on the TRPC2 gene was relaxed in a common ancestor of OW monkeys and apes. In present-day OW monkeys and apes, we find no evidence for selective pressure on TRPC2. These data correspond with evidence indicating that OW monkeys and apes have not retained a distinct accessory olfactory bulb and that many species of OW monkeys and apes lack a VNO (3). They are also consistent with the observed paucity of intact V1R receptor genes in the human genome; in a comprehensive study, only five V1R receptors with intact ORFs were identified, as compared with ≈200 V1R pseudogenes (18). We presume that, at a certain point in human evolution, ancestral species may have relied more on visual and auditory signals, rather than on chemical signals, for communicating social and reproductive status. Remarkably, it is at the time in evolution when selective pressure on TRPC2 is relaxed that the common ancestor of OW monkeys and apes developed trichromatic color vision through a gene duplication of the green/red opsin gene (32, 33). Indeed, many species of OW monkeys signal sexual and social status via colorful skin pigmentation of the face or genitalia. Thus, it is interesting to speculate that, in the evolution of OW monkeys and apes, an enhanced reliance on vision may have led to a reduced reliance on chemical signaling in mediating social interactions.
Acknowledgments
We thank Don Arnold, David Corey, Magnus Nordborg, Joseph Hacia, Simon Tavare, and Norm Arnheim for helpful discussions and Gina Ochoa and Dan Liu for technical assistance. This work was supported by National Institutes of Health Grant DC02889.
Abbreviations
- VNO
vomeronasal organ
- OW
Old World
- NW
New World
- hTRPC2
human TRPC2
Footnotes
References
- 1.Meredith M. Chem Senses. 2001;26:433–445. doi: 10.1093/chemse/26.4.433. [DOI] [PubMed] [Google Scholar]
- 2.Trotier D, Eloit C, Wassef M, Talmain G, Bensimon J L, Doving K B, Ferrand J. Chem Senses. 2000;25:369–380. doi: 10.1093/chemse/25.4.369. [DOI] [PubMed] [Google Scholar]
- 3.Wysocki C J. Neurosci Biobehav Rev. 1979;3:301–341. doi: 10.1016/0149-7634(79)90015-0. [DOI] [PubMed] [Google Scholar]
- 4.Monti-Bloch L, Diaz-Sanchez V, Jennings-White C, Berliner D L. J Steroid Biochem Mol Biol. 1998;65:237–242. doi: 10.1016/s0960-0760(98)00025-9. [DOI] [PubMed] [Google Scholar]
- 5.Dulac C, Axel R. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 6.Dulac C. Curr Opin Neurobiol. 2000;10:511–518. doi: 10.1016/s0959-4388(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 7.Buck L B. Cell. 2000;100:611–618. doi: 10.1016/s0092-8674(00)80698-4. [DOI] [PubMed] [Google Scholar]
- 8.Liman E R, Corey D P, Dulac C. Proc Natl Acad Sci USA. 1999;96:5791–5796. doi: 10.1073/pnas.96.10.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stowers L, Holy T E, Meister M, Dulac C, Koentges G. Science. 2002;295:1493–1500. doi: 10.1126/science.1069259. [DOI] [PubMed] [Google Scholar]
- 10.Leypold B G, Yu C R, Leinders-Zufall T, Kim M M, Zufall F, Axel R. Proc Natl Acad Sci USA. 2002;99:6376–6381. doi: 10.1073/pnas.082127599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Punta K, Leinders-Zufall T, Rodriguez I, Jukam D, Wysocki C J, Ogawa S, Zufall F, Mombaerts P. Nature. 2002;419:70–74. doi: 10.1038/nature00955. [DOI] [PubMed] [Google Scholar]
- 12.Wes P D, Chevesich J, Jeromin A, Rosenberg C, Stetten G, Montell C. Proc Natl Acad Sci USA. 1995;92:9652–9656. doi: 10.1073/pnas.92.21.9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giorgi D, Friedman C, Trask B J, Rouquier S. Genome Res. 2000;10:1979–1985. doi: 10.1101/gr.10.12.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pantages E, Dulac C. Neuron. 2000;28:835–845. doi: 10.1016/s0896-6273(00)00157-4. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez I, Greer C A, Mok M Y, Mombaerts P. Nat Genet. 2000;26:18–19. doi: 10.1038/79124. [DOI] [PubMed] [Google Scholar]
- 16.Kouros-Mehr H, Pintchovski S, Melnyk J, Chen Y J, Friedman C, Trask B, Shizuya H. Chem Senses. 2001;26:1167–1174. doi: 10.1093/chemse/26.9.1167. [DOI] [PubMed] [Google Scholar]
- 17.Roach J C, Lee I Y, Boysen C, Smit A, Trask B J, Hood L, Kouros-Mehr H. Genome Res. 2002;12:81–87. doi: 10.1101/gr.197901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez I, Mombaerts P. Curr Biol. 2002;12:R409–R411. doi: 10.1016/s0960-9822(02)00909-0. [DOI] [PubMed] [Google Scholar]
- 19.Ina Y. Comput Appl Biosci. 1994;10:11–12. doi: 10.1093/bioinformatics/10.1.11. [DOI] [PubMed] [Google Scholar]
- 20.Jukes T H, Cantor D R. In: Mammalian Protein Metabolism. Munro H N, editor. New York: Academic; 1969. pp. 21–132. [Google Scholar]
- 21.Tajima F. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H S, Montell C. J Cell Biol. 2000;150:1411–1422. doi: 10.1083/jcb.150.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilad Y, Segre D, Skorecki K, Nachman M W, Lancet D, Sharon D. Nat Genet. 2000;26:221–224. doi: 10.1038/79957. [DOI] [PubMed] [Google Scholar]
- 24.Lane R P, Cutforth T, Axel R, Hood L, Trask B J. Proc Natl Acad Sci USA. 2002;99:291–296. doi: 10.1073/pnas.012608399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felsenstein J. Cladistics. 1989;5:164–166. [Google Scholar]
- 26.Messier W, Stewart C B. Nature. 1997;385:151–154. doi: 10.1038/385151a0. [DOI] [PubMed] [Google Scholar]
- 27.Wolfe K H, Sharp P M. J Mol Evol. 1993;37:441–456. doi: 10.1007/BF00178874. [DOI] [PubMed] [Google Scholar]
- 28.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 29.Aujard F. Physiol Behav. 1997;62:1003–1008. doi: 10.1016/s0031-9384(97)00206-0. [DOI] [PubMed] [Google Scholar]
- 30.Barrett J, Abbott D H, George L M. J Reprod Fertil. 1993;97:301–310. doi: 10.1530/jrf.0.0970301. [DOI] [PubMed] [Google Scholar]
- 31.Giorgi D, Rouquier S. Chem Senses. 2002;27:529–537. doi: 10.1093/chemse/27.6.529. [DOI] [PubMed] [Google Scholar]
- 32.Hunt D M, Dulai K S, Cowing J A, Julliot C, Mollon J D, Bowmaker J K, Li W H, Hewett-Emmett D. Vision Res. 1998;38:3299–3306. doi: 10.1016/s0042-6989(97)00443-4. [DOI] [PubMed] [Google Scholar]
- 33.Nathans J. Neuron. 1999;24:299–312. doi: 10.1016/s0896-6273(00)80845-4. [DOI] [PubMed] [Google Scholar]
- 34.Goodman M. Am J Hum Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]