Abstract
We describe a comparative mathematical framework for two genome-scale expression data sets. This framework formulates expression as superposition of the effects of regulatory programs, biological processes, and experimental artifacts common to both data sets, as well as those that are exclusive to one data set or the other, by using generalized singular value decomposition. This framework enables comparative reconstruction and classification of the genes and arrays of both data sets. We illustrate this framework with a comparison of yeast and human cell-cycle expression data sets.
Keywords: DNA microarrays, cell cycle, yeast Saccharomyces cerevisiae, human HeLa cell line
Recent advances in high-throughput genomic technologies enable acquisition of different types of molecular biological data, e.g., DNA-sequence and mRNA-expression data, on a genomic scale. Comparative analysis of these data among two or more model organisms promises to enhance fundamental understanding of the universality as well as the specialization of molecular biological mechanisms. It also may prove useful in medical diagnosis, treatment, and drug design. Comparisons of the DNA sequence of entire genomes already give insights into evolutionary, biochemical, and genetic pathways.
Comparative analysis of mRNA-expression data requires mathematical tools that are able to distinguish the similar from the dissimilar among two or more large-scale data sets. These tools should provide mathematical frameworks for the description of the data, where the variables and operations may represent some biological reality. Recently we showed that singular value decomposition (SVD) provides such a framework for genome-wide expression data (refs. 1–3; see also refs. 4–7).
Now we show that generalized SVD (GSVD) (8) provides a comparative mathematical framework for two genome-scale expression data sets. GSVD is a linear transformation of the two data sets from the two genes × arrays spaces to two reduced and diagonalized “genelets” × “arraylets” spaces. The genelets are shared by both data sets. Each genelet is expressed only in the two corresponding arraylets, with a corresponding “angular distance” indicating the relative significance of this genelet, i.e., its significance, in one data set relative to that in the other.
We show that a genelet of equal significance in both data sets may represent a process common to both data sets. The two corresponding arraylets may represent the cellular states in each data set that correspond to this common process. A genelet of no significance in one data set relative to the other may represent a process exclusive to the latter data set. The corresponding arraylet of this data set may represent the cellular state that corresponds to this exclusive process.
We also show that mathematical reconstruction of gene expression in a subset of genelets may simulate experimental observation of only the process that these genelets are inferred to represent. Similarly, reconstruction of array expression in the subset of corresponding arraylets may simulate observation of only the corresponding cellular state. Reconstruction of each data set in two or more subspaces may simulate observation of genome-scale differential expression in the processes, which these subspaces are inferred to span. We demonstrate comparative classification of both sets of genes and arrays based on similarity in their reconstructed rather than overall expression.
We illustrate this framework with a comparison of yeast (9) and human (10) cell cycle-expression data sets.
Mathematical Methods: GSVD
A single microarray probes the relative expression levels of N1 genes in a single sample. A series of M1 arrays probes the genome-scale expression levels in M1 different samples, i.e., under M1 different experimental conditions. Let the matrix ê1, of size N1-genes × M1-arrays, tabulate the full expression data. The vector in the nth row of the matrix ê1, 〈g1,n| ≡ 〈n|ê1, lists the expression of the nth gene across the different samples that correspond to the different arrays.§ The vector in the mth column of the matrix ê1, |a1,m〉 ≡ ê1|m〉, lists the genome-scale expression measured by the mth array. Let the matrix ê2, of size N2-genes × M2-arrays, tabulate the relative expression levels of N2 genes under M2 = M1 ≡ M <max{N1, N2} experimental conditions that correspond one to one to the M1 conditions underlying ê1. This one-to-one correspondence between the two sets of conditions is at the foundation of the GSVD comparative analysis of the two data sets and should be mapped out carefully.
GSVD then is simultaneous linear transformation of the two expression data sets ê1 and ê2 from the two N1-genes × M-arrays and N2-genes × M-arrays spaces to the two reduced M-genelets × M-arraylets spaces (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org, and also at http://genome-www.stanford.edu/GSVD/),
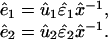 |
In these spaces the data are represented by the diagonal nonnegative matrices ɛ̂1 and ɛ̂2, which satisfy 〈k|ɛ̂1|m〉 ≡ ɛ1,mδkm ≥ 0 and 〈k|ɛ̂2|m〉 ≡ ɛ2,mδkm ≥ 0 for all 1 ≤ k, m ≤ M. The mth genelet is expressed only in the two mth arraylets, each of which corresponds to one of the two data sets. Therefore, each genelet is decoupled from all other genelets in both data sets simultaneously.
The antisymmetric angular distance between the data sets,
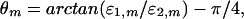 |
indicates the relative significance of the mth genelet, i.e., its significance in the first data set relative to that in the second in terms of the ratio of the expression information captured by this genelet in the first data set to that in the second. An angular distance of 0 indicates a genelet of equal significance in both data sets, with ɛ1,m = ɛ2,m; ±π/4 indicates no significance in the second data set relative to the first, with ɛ1,m ≫ ɛ2,m, or in the first relative to the second, respectively. The angular distances are arranged in decreasing order of significance in the first data set relative to the second such that π/4 ≥ θ1 ≥ ⋯ ≥ θM ≥ −π/4. The “generalized fractions of eigenexpression” of each data set separately indicate the significance of each genelet and its corresponding arraylet in this data set in terms of the fraction of the overall expression information that they capture in this data set alone (see Appendix, Eqs. 4 and 5, and Fig. 6, which are published as supporting information on the PNAS web site).
The transformation matrix x̂−1 defines the M-genelets × M-arrays basis set that is shared by both data sets. The transformation matrices û1 and û2 define the N1-genes × M-arraylets and N2-genes × M-arraylets basis sets that correspond to the first and second data sets, respectively. The vector in the mth row of x̂−1, 〈γm| ≡ 〈m|x̂−1, lists the expression of the mth genelet across the different arrays in both data sets simultaneously. The vectors in the mth columns of û1 and û2, |α1,m〉 ≡ û1|m〉 and |α2,m〉 ≡ û2|m〉, list the genome-scale expression in the mth arraylets of the first and second data sets, respectively. The genelets are normalized, such that 〈γm|γm〉 = 1 for all 1 ≤ m ≤ M, but not necessarily orthogonal superpositions of the genes of the first and, at the same time, the second data set. The arraylets of either data set are orthonormal superpositions of the arrays of this data set such that, in general, x̂−1 is nonorthogonal, whereas û1 and û2 are both orthogonal,
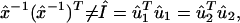 |
where Î is the identity matrix. Therefore, each arraylet of either data set is decoupled and decorrelated from all other arraylets of this data set. The genelets and arraylets are unique, and therefore also data-driven, up to a phase factor of ±1, because each genelet and arraylet capture both parallel and antiparallel gene- or array-expression patterns, respectively, except in degenerate subspaces, defined by subsets of equal angular distances.
GSVD Calculation.
From Eqs. 1 and 3, the M-arrays × M-arrays symmetric correlation matrices â1 = ê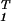 ê1 = (x̂−1)Tɛ̂
ê1 = (x̂−1)Tɛ̂ x̂−1 and â2 = ê
x̂−1 and â2 = ê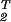 ê2 are represented in the M-genelets × M-genelets space by the simultaneously diagonal matrices ɛ̂
ê2 are represented in the M-genelets × M-genelets space by the simultaneously diagonal matrices ɛ̂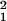 and ɛ̂
and ɛ̂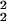 , respectively. In theory, it is possible to calculate the GSVD of the two data sets ê1 and ê2 by (i) diagonalizing â
, respectively. In theory, it is possible to calculate the GSVD of the two data sets ê1 and ê2 by (i) diagonalizing â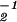 â1 = x̂(ɛ̂
â1 = x̂(ɛ̂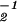 ɛ̂1)2x̂−1 to obtain x̂; (ii) projecting x̂ onto ê1 and ê2 to obtain ɛ̂
ɛ̂1)2x̂−1 to obtain x̂; (ii) projecting x̂ onto ê1 and ê2 to obtain ɛ̂ = (û1ɛ̂1)T(û1ɛ̂1) = (ê1x̂)T(ê1x̂) and ɛ̂
= (û1ɛ̂1)T(û1ɛ̂1) = (ê1x̂)T(ê1x̂) and ɛ̂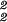 ; and (iii) projecting x̂, ɛ̂1, and ɛ̂2 onto ê1 and ê2 to obtain û1 = ê1x̂ɛ̂
; and (iii) projecting x̂, ɛ̂1, and ɛ̂2 onto ê1 and ê2 to obtain û1 = ê1x̂ɛ̂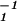 and û2. In practice, we avoid computing the quotient of the correlation matrices, â
and û2. In practice, we avoid computing the quotient of the correlation matrices, â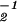 â1, and use the numerically robust GSVD algorithm (8, 9) to obtain x̂.
â1, and use the numerically robust GSVD algorithm (8, 9) to obtain x̂.
Comparative Pattern Inference.
The decorrelation of the arraylets suggests that some of the significant arraylets of each data set, i.e., these with the largest generalized fractions of eigenexpression (see Appendix, Eqs. 4 and 5, and Fig. 6), may represent independent cellular states, where the corresponding genelets represent the corresponding regulatory programs, biological processes, or experimental artifacts that contribute to the overall expression signal in each data set. The one-to-one correspondence between the two sets of experimental conditions that underlie the two data sets suggests that among these genelets, a genelet of equal significance in both data sets with angular distance of ≈0 may represent a process common to both data sets; a genelet of no significance in one data set relative to the other with angular distance of ≈±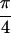 may represent a process exclusive to the latter data set. We infer that a genelet represents a process exclusive to one or common to both data sets when its expression pattern across the corresponding one or both sets of arrays is biologically or experimentally interpretable. We associate this genelet with a biological process when this inference is supported by one or two coherent biological themes, reflected in the functions of the genes of the corresponding one or both data sets, whose coefficients of this genelet in the GSVD expansion, as listed in the corresponding one or both arraylets, are largest in magnitude compared to those coefficients of all other genes. With this we assume that the corresponding one or both arraylets represent the cellular states of this exclusive or common process, respectively. We estimate the probabilistic significance of these associations by annotations using combinatorics (ref. 10; see Appendix, Fig. 7, and Table 1, which are published as supporting information on the PNAS web site).
may represent a process exclusive to the latter data set. We infer that a genelet represents a process exclusive to one or common to both data sets when its expression pattern across the corresponding one or both sets of arrays is biologically or experimentally interpretable. We associate this genelet with a biological process when this inference is supported by one or two coherent biological themes, reflected in the functions of the genes of the corresponding one or both data sets, whose coefficients of this genelet in the GSVD expansion, as listed in the corresponding one or both arraylets, are largest in magnitude compared to those coefficients of all other genes. With this we assume that the corresponding one or both arraylets represent the cellular states of this exclusive or common process, respectively. We estimate the probabilistic significance of these associations by annotations using combinatorics (ref. 10; see Appendix, Fig. 7, and Table 1, which are published as supporting information on the PNAS web site).
Comparative Data Reconstruction.
The decoupling of the genelets and both sets of arraylets allows reconstructing either data set in a given subspace of K-genelets and corresponding arraylets without eliminating genes or arrays, êi → ∑ ɛi,k|αi,k〉〈γk|, where i = 1, 2. For visualization and classification, we set the arithmetic mean of each genelet across the arrays and that of each arraylet across the genes to 0, such that the expression of each gene and array in the reconstructed data set is centered at its array- or gene-invariant level, respectively.
ɛi,k|αi,k〉〈γk|, where i = 1, 2. For visualization and classification, we set the arithmetic mean of each genelet across the arrays and that of each arraylet across the genes to 0, such that the expression of each gene and array in the reconstructed data set is centered at its array- or gene-invariant level, respectively.
Comparative Data Classification.
Inferring that subsets of genelets and arraylets represent independent processes or states, exclusive to one or common to both data sets, allows classifying the genes and arrays of one or simultaneously both data sets by similarity in their expression of these genelets or arraylets, respectively, rather than their overall expression. We least-squares-approximate a subspace spanned by K > 2 genelets with that spanned by the two orthonormal vectors |x〉 and |y〉, which maximize ∑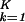 〈γk|(|x〉〈x| + |y〉〈y|)|γk〉. We plot the projection of each gene of either data set 〈gi,n|, where i = 1, 2, from the K-genelets subspace onto |y〉, ∑
〈γk|(|x〉〈x| + |y〉〈y|)|γk〉. We plot the projection of each gene of either data set 〈gi,n|, where i = 1, 2, from the K-genelets subspace onto |y〉, ∑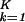 ɛi,k〈n|αi,k〉〈γk|y〉/Ni,n, along the y axis vs. that onto |x〉 along the x axis, normalized by its ideal amplitude, where the contribution of each genelet to the overall projected expression of the gene adds up rather than cancels out, N
ɛi,k〈n|αi,k〉〈γk|y〉/Ni,n, along the y axis vs. that onto |x〉 along the x axis, normalized by its ideal amplitude, where the contribution of each genelet to the overall projected expression of the gene adds up rather than cancels out, N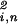 = ∑
= ∑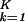 ∑
∑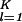 ɛi,kɛi,l|〈n|αi,k〉〈αi,l|n〉〈γk|(|x〉〈x| + |y〉〈y|)|γl〉|. In this plot, the distance of each gene from the origin, ri,n, is the amplitude of its normalized projection. An amplitude of 1 indicates that the genelets add up; 0 indicates that they cancel out. The phase difference of each gene from the x axis, φi,n, is its phase in the progression of expression across the genes from |x〉 to |y〉 and back to |x〉, going through the projections of all K-genelets in this subspace (|x〉〈x| + |y〉〈y|)|γk〉. We sort the genes according to φi,n. Similarly, we plot the projection of each array, |ai,m〉, from the K-arraylets subspace onto ∑
ɛi,kɛi,l|〈n|αi,k〉〈αi,l|n〉〈γk|(|x〉〈x| + |y〉〈y|)|γl〉|. In this plot, the distance of each gene from the origin, ri,n, is the amplitude of its normalized projection. An amplitude of 1 indicates that the genelets add up; 0 indicates that they cancel out. The phase difference of each gene from the x axis, φi,n, is its phase in the progression of expression across the genes from |x〉 to |y〉 and back to |x〉, going through the projections of all K-genelets in this subspace (|x〉〈x| + |y〉〈y|)|γk〉. We sort the genes according to φi,n. Similarly, we plot the projection of each array, |ai,m〉, from the K-arraylets subspace onto ∑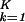 |αi,k〉〈γk|y〉, ∑
|αi,k〉〈γk|y〉, ∑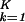 ɛi,k〈y|γk〉〈γk|m〉/Ni,m, along the y axis vs. that onto ∑
ɛi,k〈y|γk〉〈γk|m〉/Ni,m, along the y axis vs. that onto ∑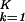 |αi,k〉〈γk|x〉 along the x axis, normalized by its ideal amplitude, N
|αi,k〉〈γk|x〉 along the x axis, normalized by its ideal amplitude, N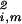 = ∑
= ∑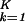 ∑
∑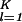 ɛi,k ɛi,l|〈m|γk〉〈γl|m〉〈γk|(|x〉〈x| + |y〉〈y|)|γl〉|. We sort the arrays according to their phase differences from the x axis, φi,m.
ɛi,k ɛi,l|〈m|γk〉〈γl|m〉〈γk|(|x〉〈x| + |y〉〈y|)|γl〉|. We sort the arrays according to their phase differences from the x axis, φi,m.
Biological Results: Comparison of Yeast and Human Cell-Cycle Expression Data Sets
Spellman et al. (11) monitored mRNA levels for 6,113 putative ORFs of the yeast Saccharomyces cerevisiae over two cell-cycle periods in a yeast culture synchronized initially in the cell-cycle stage M/G1 by the pheromone α factor, relative to reference mRNA from an asynchronous culture, at 7-min intervals for 119 min. The data set for the yeast experiments we analyze (see Data Sets 1–4, which are published as supporting information on the PNAS web site and mathematica notebook at http://genome-www.stanford.edu/GSVD/) tabulates the ratios of gene-expression levels for the N1 = 4,523 genes with no missing data in at least 15 of the M1 = 18 arrays. Of these genes, 604 were classified as cell cycle-regulated by Spellman et al., and 77 were classified by traditional methods. Whitfield et al. (12) monitored mRNA levels for 43,198 human gene clones over two and a half cell-cycle periods in a HeLa cell-line culture synchronized initially in S by a double-thymidine block, relative to reference mRNA from an asynchronous HeLa culture, at 2-h intervals for 34 h. The data set for the human experiments we analyze (see Data Sets 5–8, which are published as supporting information on the PNAS web site) tabulates the ratios of gene-expression levels for the N2 = 12,056 clones with no missing data in at least 15 of the M2 = 18 arrays. Of these clones, 750 were classified as cell cycle-regulated by Whitfield et al., and 73 were classified by traditional methods. We estimate the missing data in each data set using SVD (ref. 2; see Appendix and Figs. 8–11, which are published as supporting information on the PNAS web site) and calculate the GSVD of both data sets.
Common Yeast and Human Cell-Cycle Subspace.
The time, i.e., array variations of the third, fourth, and fifth genelets, 〈γ3|, 〈γ4|, and 〈γ5|, that are almost equally significant in both data sets (slightly more in the yeast data), with 0 < θ3, θ4, θ5 < π/16 (Fig. 1), fit normalized cosine functions of two periods and initial phases of π/3, 0, and −π/3, respectively, superimposed on time-invariant expression (Fig. 2). The genelets 〈γ14|, 〈γ15|, and 〈γ16|, which are also almost equally significant in both data sets (slightly more in the human data), with −π/6 < θ14, θ15, θ16 < 0, fit normalized cosines of two and a half periods and initial phases of −π/3, π/3, and 0, respectively. Coherent themes of yeast and human cell-cycle programs emerge from the annotations of the 100 yeast and 100 human genes (13, 14), with largest parallel and separately also antiparallel contributions from each one of these six genelets as listed in the corresponding yeast and human arraylets (see Data Sets 9 and 10, which are published as supporting information on the PNAS web site). We associate all these six genelets with the cell-cycle gene-expression oscillations common to both the yeast and human genomes and manifested in both data sets. We assume that the corresponding six yeast and six human arraylets represent the yeast and human cell-cycle cellular states, respectively. The probabilistic significance of these associations by annotations, estimated using combinatorics, is high: Most of the P values, calculated assuming hypergeometric probability distribution of the annotations among the genes, are orders of magnitude <0.01 (ref. 10; see Appendix, Fig. 7, and Table 1). Following the traditional classifications, the 0-phase genelet 〈γ4| is associated in parallel with the yeast cell-cycle stage M/G1, in which the yeast culture is initially synchronized, and both 0-phase genelets 〈γ4| and −〈γ16| are associated in parallel with the human cell-cycle stage S, in which the human culture is initially synchronized.
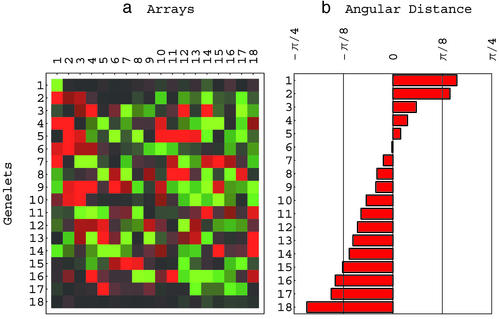
Fig 1.
Yeast and human genelets. (a) Raster display of x̂−1, the expression of 18 genelets in 18 yeast and human arrays simultaneously, centered at their array-invariant levels. (b) Bar chart of the angular distances showing 〈γ1| and 〈γ2| highly significant in the yeast data relative to the human data, 〈γ3|, 〈γ4|, 〈γ5|, 〈γ6|, 〈γ14|, 〈γ15|, and 〈γ16| almost equally significant in both data sets and 〈γ17| and 〈γ18| highly significant in the human data relative to the yeast data. All other genelets are significant in neither the yeast data nor the human data (see Appendix).
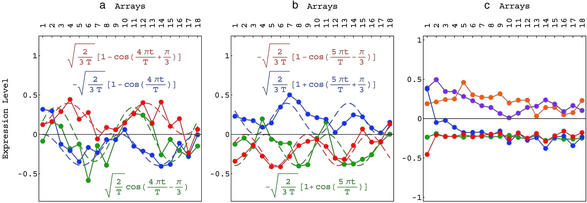
Fig 2.
Line-joined graphs of the expression levels of the genelets. (a) 〈γ3| (red), 〈γ4| (blue), and 〈γ5| (green), which are associated with the common yeast and human cell-cycle gene-expression oscillations, fit dashed graphs of normalized cosines of two periods and initial phases of π/3 (red), 0 (blue), and −π/3 (green), respectively. (b) 〈γ14| (red), 〈γ15| (blue), and 〈γ16| (green), which also are associated with cell-cycle gene-expression oscillations, fit dashed graphs of normalized cosines of two and a half periods and initial phases of −π/3 (red), π/3 (blue), and 0 (green), respectively. (c) 〈γ1| (red) and 〈γ2| (blue) are associated with the exclusive yeast pheromone response, 〈γ17| (orange) and 〈γ18| (green) are associated with the exclusive human stress response, and 〈γ6| (violet) is associated with both the yeast and human transitions from synchronization response into the cell cycle.
Projecting the expression of the 18 yeast arrays from this six-dimensional yeast arraylets subspace onto the two-dimensional subspace that approximates it, ≥50% of the contributions of the six arraylets add up (rather than cancel out) in the overall expression of 16 arrays, the normalized amplitudes of which satisfy 0.5 ≤ r1,m < 1 (Fig. 3). Sorting the arrays according to their phases, {φ1,m}, gives an array order similar to that of the cell-cycle time points measured by the arrays that describes the yeast cell-cycle progression from the M/G1 stage through G1, S, S/G2, and G2/M back to M/G1 twice. Because the projection of the 0-phase arraylets |α1,4〉 and −|α1,16〉, which correspond to the 0-phase genelets, 〈γ4| and −〈γ16|, is correlated with the arrays |a1,1〉, |a1,2〉, and |a1,10〉 and also |a1,9〉 and |a1,18〉, we associate both yeast 0-phase arraylets with the cell-cycle cellular state of transition from G2/M to M/G1, in which the yeast culture is synchronized initially. Projecting the expression of the 18 human arrays from the six-dimensional human arraylets subspace onto the two-dimensional subspace that approximates it, ≥50% of the contributions of the six arraylets add up in the expression of 16 arrays. Sorting the arrays describes the human cell-cycle progression from S through G2, G2/M, M/G1, and G1/S back to S two and a half times. Because the projection of the 0-phase arraylets, |α2,4〉 and −|α2,16〉, is correlated with the arrays |a2,2〉 and |a2,9〉, we associate both human 0-phase arraylets with the cell-cycle stage S, in which the human culture is synchronized.
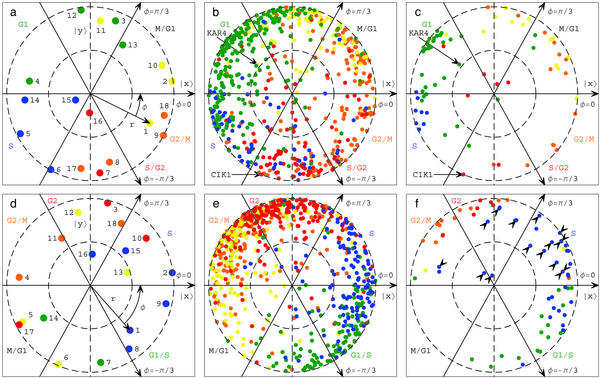
Fig 3.
Yeast (a–c) and human (d–f) expression reconstructed in the six-dimensional cell-cycle subspaces approximated by two-dimensional subspaces. (a) Yeast array expression, projected onto π/2-phase along the y axis vs. that onto 0-phase along the x axis and color-coded according to the classification of the arrays into the five cell-cycle stages: M/G1 (yellow), G1 (green), S (blue), S/G2 (red), and G2/M (orange). The dashed unit and half-unit circles outline 100% and 50% of added-up (rather than canceled-out) contributions of the six arraylets to the overall projected expression. The arrows describe the projections of the −π/3-, 0-, and π/3-phase arraylets. (b) Yeast expression of 603 cell cycle-regulated genes projected onto π/2-phase along the y axis vs. that onto 0-phase along the x axis and color-coded according to the classification by Spellman et al. (11) (c) Yeast expression of 76 cell cycle-regulated genes color-coded according to the traditional classification. (d) Human array expression color-coded according to the classification of the arrays into the five cell-cycle stages: S (blue), G2 (red), G2/M (orange), M/G1 (yellow), and G1/S (green). (e) Human expression of 750 cell cycle-regulated genes color-coded according to the classification by Whitfield et al. (12) (f) Human expression of 73 cell cycle-regulated genes color-coded according to the traditional classification; the arrows point to 16 human histones that were not classified by Whitfield et al. as cell cycle-regulated based on their overall expression.
Projecting the expression of the yeast and human genes from the six-dimensional genelets subspace onto the two-dimensional subspace that approximates it, ≥50% of the contributions of the six genelets add up in the overall expression of 547 of the 604 yeast genes that were classified as cell cycle-regulated by Spellman et al. (11), 709 of the 750 human genes classified by Whitfield et al. (12), and 71 of the 77 yeast and 71 of the 73 human genes classified by traditional methods (including, e.g., 14 of 16 human histones, that were not classified by Whitfield et al. as cell cycle-regulated based on their overall expression). Simultaneous classification of the yeast and human genes into the five cell-cycle stages describes the yeast and human cell cycles' progression along the yeast and human genes, respectively, and is in good agreement with the classifications by Spellman et al. and Whitfield et al. and also the traditional ones. Because the projection of the 0-phase genelets, 〈γ4| and −〈γ16|, is correlated with yeast genes that peak late in G2/M and early in M/G1 and human genes that peak in S, we associate 〈γ4| and −〈γ16| with cell-cycle expression oscillations of yeast at the transition from G2/M to M/G1 and human at S. This simultaneous classification therefore outlines a correspondence between the groups of yeast genes and those of human genes, e.g., yeast genes that peak at M/G1 correspond to human genes that peak at S, the cell-cycle stages in which the yeast and human cultures are synchronized initially, respectively.
With all 4,523 yeast and 12,056 human genes sorted, the gene variations of the six yeast and six human arraylets approximately fit one-period cosines of π/3, 0, and −π/3 initial phases (Fig. 4) such that the initial phase of each arraylet is similar to that of its corresponding genelet. Both sorted and reconstructed yeast and human expressions approximately fit traveling waves of one-period cosinusoidal variation across the genes and of two or two and a half periods across the arrays, respectively.
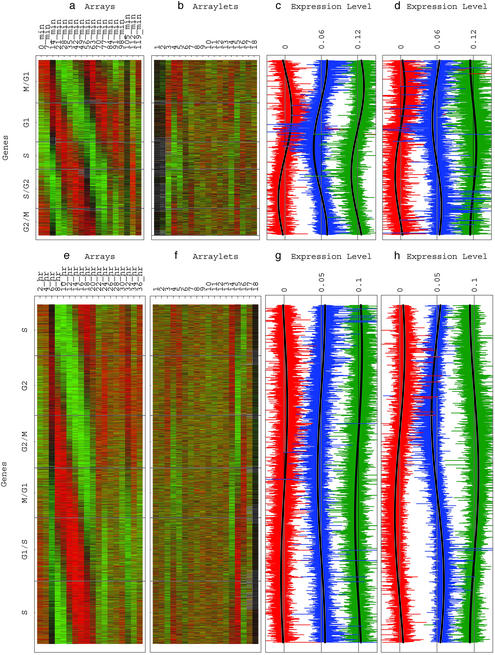
Fig 4.
Yeast (a–d) and human (e–h) expression reconstructed in the six-dimensional cell-cycle subspaces with genes sorted according to their phases in the two-dimensional subspaces that approximate them. (a) Yeast expression of the sorted 4,523 genes in the 18 arrays, centered at their gene- and array-invariant levels, showing a traveling wave of expression. (b) Yeast expression of the sorted 4,523 genes in the 18 arraylets, centered at their array-invariant levels. The expression of the arraylets |α1,3〉, |α1,4〉, |α1,5〉, |α1,14〉, |α1,15〉, and |α1,16〉 displays the sorting. (c) Yeast cell-cycle arraylet expression levels |α1,3〉 (red), |α1,4〉 (blue), and |α1,5〉 (green) fit one-period cosines of π/3 (red), 0 (blue), and −π/3 (green) initial phases. (d) Yeast cell-cycle arraylet expression levels |α1,14〉 (red), |α1,15〉 (blue), and |α1,16〉 (green) fit one-period cosines of −π/3 (red), π/3 (blue), and 0 (green) initial phases. (e) Human expression of the sorted 12,056 genes in the 18 arrays centered at their gene- and array-invariant levels showing a traveling wave of expression. (f) Human expression of the sorted 12,056 genes in the 18 arraylets centered at their array-invariant levels. The expression of the arraylets |α2,3〉, |α2,4〉, |α2,5〉, |α2,14〉, |α2,15〉 and |α2,16〉 displays the sorting. (g) Human cell-cycle arraylet expression levels |α2,3〉 (red), |α2,4〉 (blue), and |α2,5〉 (green) fit one-period cosines of π/3 (red), 0 (blue), and −π/3 (green) initial phases. (h) Human cell-cycle arraylet expression levels |α2,14〉 (red), |α2,15〉 (blue), and |α2,16〉 (green) fit one-period cosines of −π/3 (red), π/3 (blue), and 0 (green) initial phases.
Exclusive Yeast Pheromone-Response Subspace.
The genelets 〈γ1| and 〈γ2|, insignificant in the human data set relative to that of the yeast, with θ1, θ2 > π/7 (Fig. 1), describe initial transient increase and decrease in expression, respectively (Fig. 2). A theme of yeast response to pheromone synchronization emerges from the annotations of those yeast genes with contributions from 〈γ1| and 〈γ2| that are largest in magnitude. The genelet 〈γ6|, equally significant in both data sets with θ6 ∼ 0, describes an initial transient increase in expression superimposed on cosinusidial variation. A theme of transition from pheromone response to cell-cycle progression emerges from the annotations of those yeast genes with contributions from 〈γ6|, as listed in the corresponding yeast arraylet |α1,6〉, that are largest in magnitude (see Data Set 9). We associate these three genelets and corresponding three yeast arraylets with the pheromone response, which is exclusive to the yeast genome. Classification of the yeast genes and arrays into pheromone-response stages in the subspaces spanned by these genelets and arraylets, respectively, is in good agreement with the traditional understanding of this program (ref. 13; Figs. 12–14, which are published as supporting information on the PNAS web site).
Exclusive Human Stress-Response Subspace.
The genelets 〈γ17| and 〈γ18| are insignificant in the yeast data set relative to that of the human, with θ17, θ18 < −π/6. A theme of human synchronization stress response emerges from the annotations of those human genes with contributions from 〈γ17| and 〈γ18| that are largest in magnitude. Also, from the annotations of those human genes with contributions from 〈γ6|, as listed in the corresponding human arraylet |α2,6〉, that are largest in magnitude emerges a theme of transition from stress response to cell-cycle progression (see Data Set 10). We associate these three genelets and corresponding three human arraylets with this human-exclusive stress response. Classification of the human genes and arrays into stress-response stages in the subspaces spanned by these genelets and arraylets, respectively, is in agreement with current understanding of this program (ref. 12; Figs. 15–17, which are published as supporting information on the PNAS web site).
Differential Expression of Yeast Genes in the Exclusive Pheromone-Response and the Common Cell-Cycle Subspaces.
According to their expression in the yeast-exclusive pheromone-response subspace, mRNA expression of both yeast genes KAR4 and CIK1 peak early in the time course (together with that of other genes known to be involved in the α-factor response) (Fig. 3). In the common cell-cycle subspace, KAR4 peaks at the G1 cell-cycle stage, whereas CIK1 peaks almost half a cell-cycle period later (and also earlier) at S/G2 (Fig. 12). This differential expression of CIK1 and KAR4 in the response to pheromone program vs. that of the cell cycle is in agreement with the experimental observation of Kurihara et al. (15), who showed that induction of CIK1 depends on that of KAR4 during mating, and is independent of KAR4 during mitosis.
Differential Expression of Human Genes in the Exclusive Stress-Response and the Common Cell-Cycle Subspaces.
In the human-exclusive stress-response subspace, most human histones reach their expression minima early (Fig. 3). In the common cell-cycle subspace, most histones peak early, together with other genes known to peak in the cell-cycle stage S (Fig. 14). This differential expression of most histones may explain why these histones do not appear to be cell cycle-regulated based on their overall expression.
Conclusions
We have shown that GSVD provides a comparative mathematical framework for two genome-scale expression data sets, in which the variables and operations may represent some biological reality. Using GSVD in a comparison of yeast and human cell-cycle expression data sets, we were able to find (i) biological similarity in these two disparate organisms in terms of their mRNA expression during their cell-cycle programs; (ii) experimental dissimilarity in terms of yeast and human mRNA expression during their different synchronization-response programs; and (iii) differential gene expression in the yeast and human cell-cycle programs vs. their synchronization-response programs, respectively.
Possible additional applications of GSVD include comparison of two genomic data sets, each corresponding to (i) the same experiment repeated, e.g., using different experimental protocols, to separate the biological signal that is similar in both data sets from the dissimilar experimental artifacts; (ii) one of two different types of genomic information (e.g., DNA copy number, mRNA expression, or protein abundance) collected from the same set of samples (e.g., tumor samples) to elucidate the molecular composition of the overall biological signal in these samples; (iii) one of two chromosomes of the same organism to illustrate the relation, if any, between these chromosomes in terms of their, e.g., mRNA expression in a given set of samples; and (iv) one of two interacting organisms, e.g., during infection, to illuminate the exchange of biological information in these interactions.
Supplementary Material
Acknowledgments
We thank G. H. Golub for insightful discussions of matrix computation, M. L. Whitfield for discussions of the human cell-cycle data and careful reading, and G. M. Church, S. R. Eddy, and E. Rivas for thoughtful reviews of this manuscript. This work was supported by National Cancer Institute Grants CA77097 (to D.B.) and CA85129 (to P.O.B.) and National Institute of General Medical Sciences Grant GM46406 (to D.B.). O.A. is a Sloan Foundation/Department of Energy Postdoctoral Fellow in Computational Molecular Biology (DE-FG03-99ER62836) and a National Human Genome Research Institute Individual Mentored Research Scientist Development Awardee in Genomic Research and Analysis (5 K01 HG00038-01). P.O.B. is a Howard Hughes Medical Institute Investigator.
Abbreviations
SVD, singular value decomposition
GSVD, generalized SVD
In this article m̂ denotes a matrix, |v〉 denotes a column vector, and 〈u| denotes a row vector such that m̂|v〉, 〈u|m̂, and 〈u|v〉 all denote inner products, and |v〉〈u| denotes an outer product.
References
- 1.Alter O., Brown, P. O. & Botstein, D. (2000) Proc. Natl. Acad. Sci. USA 97, 10101-10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter O., Brown, P. O. & Botstein, D. (2001) in Microarrays: Optical Technologies and Informatics, eds. Bittner, M. L., Chen, Y., Dorsel, A. N. & Dougherty, E. R. (Int. Soc. Optical Eng., Bellingham, WA), Vol. 4266, pp. 186. [Google Scholar]
- 3.Nielsen T. O., West, R. B., Linn, S. C., Alter, O., Knowling, M. A., O'Connell, J. X., Ferro, M., Sherlock, G., Pollack, J. R., Brown, P. O., et al. (2002) Lancet 359, 1301-1307. [DOI] [PubMed] [Google Scholar]
- 4.Wen X., Fuhrman, S., Michaels, G. S., Carr, D. B., Smith, S., Barker, J. L. & Somogyi, R. (1998) Proc. Natl. Acad. Sci. USA 95, 334-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilsenbeck S. G., Friedrichs, W. E., Schiff, R., O'Connell, P., Hansen, R. K., Osborne, C. K. & Fuqua, S. A. (1999) J. Natl. Cancer Inst. 91, 453-459. [DOI] [PubMed] [Google Scholar]
- 6.Raychaudhuri S., Stuart, J. M. & Altman, R. B. (2000) in Proceedings of the Pacific Symposium on Biocomputing, eds. Altman, R. B., Lauderdale, K., Dunker, A. K., Hunter, L. & Klein, T. E. (World Scientific, Singapore), pp. 455.
- 7.Holter N. S., Mitra, M., Maritan, A., Cieplak, M., Banavar, J. R. & Fedoroff, N. V. (2000) Proc. Natl. Acad. Sci. USA 97, 8409-8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golub G. H. & Van Loan, C. F., (1996) Matrix Computation (Johns Hopkins Univ. Press, Baltimore).
- 9.Paige C. C. & Saunders, M. A. (1981) SIAM J. Numer. Anal. 18, 398-405. [Google Scholar]
- 10.Tavazoie S., Hughes, J. D., Campbell, M. J., Cho, R. J. & Church, G. M. (1999) Nat. Genet. 22, 281-285. [DOI] [PubMed] [Google Scholar]
- 11.Spellman P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown, P. O., Botstein, D. & Futcher, B. (1998) Mol. Biol. Cell 9, 3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitfield M. L., Sherlock, G., Saldanha, A., Murray, J. I., Ball, C. A., Alexander, K. E., Matese, J. C., Perou, C. M., Hurt, M. M., Brown, P. O. & Botstein, D. (2002) Mol. Biol. Cell 13, 1977-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dwight S. S., Harris, M. A., Dolinski, K., Ball, C. A., Binkley, G., Christie, K. R., Fisk, D. G., Issel-Tarver, L., Schroeder, M., Sherlock, G., et al. (2002) Nucleic Acids Res. 30, 69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherlock G., Hernandez-Boussard, T., Kasarskis, A., Binkley, G., Matese, J. C., Dwight, S. S., Kaloper, M., Weng, S., Jin, H., Ball, C. A., et al. (2001) Nucleic Acids Res. 29, 152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurihara L. J., Stewart, B. G., Gammie, A. E. & Rose, M. D. (1996) Mol. Cell. Biol. 16, 3990-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.