Abstract
Telomeres of most animals, plants, and unicellular eukaryotes are made up of tandem arrays of repeated DNA sequences produced by the enzyme telomerase. Drosophila melanogaster has an unusual variation on this theme; telomeres consist of tandem arrays of sequences produced by successive transpositions of two non-LTR retrotransposons, HeT-A and TART. To explore the phylogenetic distribution of these variant telomeres, we have looked for TART homologues in a distantly related Drosophila species, virilis. We have found elements that, despite many differences in nucleotide sequence, retain significant amino acid similarity to TART from D. melanogaster. These D. virilis TART elements have features that characterize TART elements in D. melanogaster: (i) they are found in tandem arrays on chromosome ends, (ii) they are not found in euchromatin, and (iii) they produce both sense and antisense transcripts, with the antisense RNA being in excess. The D. virilis TART elements have one surprising feature: both of the ORFs contain long stretches of the trinucleotide repeat CAX, encoding polyglutamine (with a few interspersed histidines). These long polyglutamine stretches are conserved in the three D. virilis elements sequenced. They do not interrupt any domains of known function in the TART proteins and are not seen in TART proteins from other species. Comparison of the D. virilis and D. melanogaster telomeres suggests that the retrotransposon mechanism of telomere maintenance may have arisen before the separation of the genus Drosophila.
The broad phylogenetic distribution of organisms using telomerase to extend telomeres demonstrates the success of reverse transcription of RNA as a mechanism for maintaining chromosome ends. The repeats that telomerase reverse-transcribes onto chromosomes are surprisingly similar from species to species; most are 6–10 bp long and G/T rich. In contrast to the slight variations in repeat sequences, the length and chromatin structure of the arrays can differ greatly between species (1). For example, macronuclear telomeres of Oxytricha are 20 bp of double-stranded sequence with a terminal single-strand overhang of 16 bp, whereas human telomeres are 5–15 kbp, vary with age and cell type (1), and have a large terminal T loop (2, 3). Repeats in the Drosophila melanogaster telomere arrays (4) consist of two retrotransposons, HeT-A (≈6 kb) and TART (≈10 kb), orders of magnitude larger than repeats made by telomerase. Nevertheless, there are intriguing similarities between the two types of telomeres. The most important similarity is that the D. melanogaster telomeres, like telomerase telomeres, are extended by reverse transcription of an RNA template. HeT-A and TART are non-LTR retrotransposons and transpose by being reverse-transcribed onto the end of the chromosome. Successive transpositions form head-to-tail arrays on chromosome ends. A second similarity is the G/T strand bias of the Drosophila arrays, resembling the strand bias of arrays produced by telomerase.
Because some insects (5) have telomerase, Drosophila must have shared ancestors with species that now have telomerase. To explore the phylogenetic distribution and range of variation of transposon telomeres, we have looked for homologues of TART and HeT-A in other Drosophila species. The search is complicated by rapid sequence change in these elements. Both have long stretches of noncoding sequence, which can differ significantly between copies of the element in the same Drosophila stock. Both elements also encode Gag proteins whose sequence can vary as much as that of the noncoding regions (4). TART has a second ORF (ORF2) encoding a protein with endonuclease and reverse transcriptase (RT) activities. As with retroviruses (6), ORF2 is more conserved than the Gag coding sequence, and the most conserved part, RT, has been the only useful probe for these cross-species homology searches. Although the high sequence divergence makes it difficult to study the telomere elements, it also increases the probability that conserved features are of biological importance.
We have reported studies of telomere arrays in Drosophila yakuba (7). The evolutionary distance between D. yakuba and D. melanogaster is estimated to be only 5–15 million years; nevertheless, it has been long enough for sequences of the telomeric transposons to change significantly, yet their basic features have been conserved. Both HeT-A and TART homologues are found in D. yakuba, where they form head-to-tail arrays on telomeres and are not present in euchromatin. At least in our limited sample, no other elements are present in the telomere arrays. Apparently, these two elements have been evolving together on telomeres since before the separation of the melanogaster and yakuba species complexes.
To look for telomere transposons in much more distant Drosophila species, we chose the most studied distant relative of D. melanogaster, the virilis group (estimated separation from D. melanogaster of ≈60 million years; ref. 8). D. virilis was of special interest because it has been reported to lack telomere transposons and instead to maintain its telomeres by recombination of a 370-bp satellite sequence (9).
We find that D. virilis and Drosophila americana have elements recognizable as TART despite diverged sequence and some structural differences (Fig. 1 and Table 1). TARTvir elements form head-to-tail repeats on telomeres and are not found in euchromatin. One of the arrays cloned contains a 5′ sequence from a second element. This other element may be a HeT-A homologue, but it was truncated and has not yet been identified.
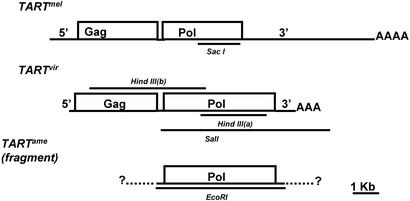
Figure 1.
Diagrams of TART elements in different Drosophila species drawn approximately to scale. 5′, 5′ UTR; 3′, 3′ UTR; Gag, ORF1; Pol, ORF2; AAA, the 3′oligo(A) that characterizes non-LTR retrotransposons. The TARTmel element shows an average of the sizes of the 5′ and 3′ UTRs of the three subfamilies, TARTmelA, TARTmelB, and TARTmelC. The TARTame is the only fragment cloned. Dotted lines represent putative flanking regions. Solid bars above and below elements correspond to probes (see Materials and Methods).
Table 1.
Comparisons of sequence identity, and synonymous and nonsynonymous substitutions
| Species | Nt (%) | AA (%) | Ks | Ka |
|---|---|---|---|---|
| TART Gag | ||||
| mel/vir | 26 (37) | 13 (18) | 1.79 ± 0.36 | 1.09 ± 0.2 |
| TART Pol | ||||
| mel/vir | 45 (52) | 36 (45) | 1.8 ± 0.64 | 0.57 ± 0.17 |
| mel/ame | 44 (50) | 34 (38) | 1.65 ± 0.37 | 0.61 ± 0.03 |
| vir/ame | 57 | 54 | 1.5 ± 0.19 | 0.37 ± 0.02 |
| R1 Pol | ||||
| mel/hyd | 60 | 59 | 1.3 ± 0.16 | 0.32 ± 0.03 |
| Adh | ||||
| mel/vir | 75 | 79 | 1.04 ± 0.25 | 0.12 ± 0.03 |
Nt, % nucleotide identity; AA, % amino acid identity. Values in parentheses are calculated omitting residues in gapped regions. Ks, synonymous substitutions; Ka, replacement substitutions (±SE). Species are indicated by their first three letters. All pairwise sequence alignments were performed with clustalw (11), refined in gendoc (12), and analyzed with mega 2.1; genetic distance is based on Kumar et al. (13). Separation of D. melanogaster and Drosophila hydei is approximately equal to the separation of D. melanogaster and D. virilis. R1mel, X51968; R1hyd, U23196; Adhmel, X98338; Adhvir, AB033640.1. When TARTmel Pol is compared to longer Pol proteins, the extra domain of TARTvir and TARTame is not included in the calculations.
As described below, our studies lead us to suggest that the 370-bp satellite sequence previously reported is a telomere-associated sequence (TAS), similar to the TAS repeats found proximal to telomere arrays in other organisms.
Our findings that telomere transposons have been present >60 million years suggest that elements and host have had time to coevolve complex interactions.
Materials and Methods
Fly Stocks.
D. melanogaster stocks are Oregon R. D. yakuba stocks are described in ref. 7. D. virilis no. S170 and D. americana no. 15010-0951.0 were obtained from the European Drosophila Stock Centre (Umeå, Sweden).
Cross-Hybridization Studies.
Low-stringency Southern hybridization was performed overnight at 55°C in 4× SET (1× SET = 0.15 M NaCl/0.03 M Tris, pH 7.4/2 mM EDTA), 5× Denhardt's solution, 0.5% SDS, and 50 μg/ml of salmon sperm DNA, followed by four 20-min washes at 55°C with 2× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7), and 0.5% SDS. After exposure, filters were washed two times for 20 min at 55°C with 1× SSC and reexposed, rewashed two times for 20 min at 65°C with 0.5× SSC, and reexposed again. A D. virilis genomic library in λ phage was screened overnight at 65°C in the solution described above, washed two times for 20 min at 65°C with 2× SSC, once for 20 min with 1× SSC, and once for 20 min with 0.5× SSC.
Northern Hybridization.
RNA analyses were as described (10). Strand-specific probes were labeled with [32P]UTP by in vitro transcription. Because the strand transcribed for the sense probe has nearly twice the A content, the two probes differ in specific activity. The blot shown in Fig. 4 had equal counts in each hybridization reaction.
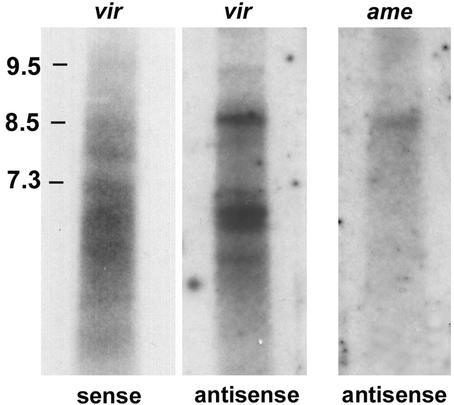
Figure 4.
Northern hybridizations of TARTvir and TARTame. Autoradiograph of total RNA from D. virilis and D . americana probed with the Pol coding region of the homologous TART. Both TARTvir probes detect several bands of RNA in the range of 6–9.5 kb. Sense and antisense RNAs do not migrate at exactly the same position, possibly because of differences in size or differences in conformation of the strands, which differ significantly in base composition. The exposure for the TARTvir sense-strand blot was equivalent to four times that of the antisense blot. Only the antisense blot of TARTame is shown because we have been unable to detect sense-strand RNA for this element.
Probes.
The SacI probe used in the cross-hybridization studies contains nucleotides 434–2,683 from TARTmel (GenBank accession no. U02279). The HindIII(b) probe for library screening contains nucleotides 2,525–7,139 of TARTvir (GenBank accession no. AY219708). The HindIII(a) probe for Northern blots contains nucleotides 9,308–12,428 of TARTvir (GenBank accession no. AY219709). The SalI probe for in situ hybridization contains nucleotides 6,465–12,950 of TARTvir (GenBank accession no. AY219708). The D. americana EcoRI probe contains nucleotides 1–5,055 of TARTame (GenBank accession no. AY219710).
Sequence Analyses.
Sequences were analyzed by blast searches. Alignments were with clustalw (11). Nucleotide alignments from coding regions were corrected by hand with gendoc (12) to agree with protein alignment. Phylogenetic analyses were performed with mega 2.1 (13), by using both neighbor-joining and UPGMA (unweighted pair-group method with arithmetic mean) algorithms.
Results
Isolation of Clones Containing TART Elements from D. virilis and D. americana.
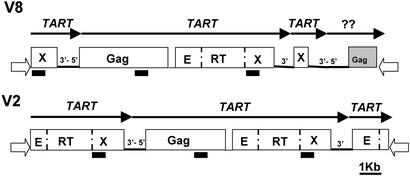
Our strategy to identify diverged TART sequences began with low-stringency hybridization to Southern blots of DNA from D. virilis and D. americana. The probe was the most conserved region of the pol gene, encoding RT from D. melanogaster TART (TARTmel; see Fig. 1). We saw no cross-hybridization to D. virilis DNA but found one band in D. americana DNA. DNA fragments in the size range of this band were cloned and screened with the TARTmel RT sequence. Sequence of the D. americana clone showed that it encoded a TART homologue. This D. americana sequence was then used to probe D. virilis DNA. It identified a band that we cloned and sequenced. The sequence of this D. virilis clone showed it was a TART homologue. The D. virilis clone was used to screen a λ phage library of D. virilis DNA. We isolated five clones and sequenced two clones, V2 and V8, that were clearly different and nonoverlapping (Fig. 2).
Figure 2.
Diagrams of the two D. virilis phage clones, V8 and V2. Arrows above diagram identify different elements and indicate 5′ → 3′ of sense-strand. ?? indicates unidentified element. Regions in each element are marked as in Fig. 1. 3′-5′ indicates apparently complete junctions between elements. Domains in ORF2 are: E, endonuclease; RT; and X, extra domain. Black rectangles under Gag and Pol indicate high content of CAX repeats; white arrowheads mark the phage arms. Diagrams are approximately to scale.
Clones V2 and V8 each had inserts of slightly more than 14 kbp, consisting entirely of head-to-tail TART elements except for what appears to be the 5′ end of a second non-LTR element on clone V8. This second element was truncated near the middle of the Gag coding region by the vector. Enough sequence was present to show that the element is not similar to TART. Unfortunately, the 5′ end is the most variable part of the Gag coding region and does not allow us to identify this second element. It may be a HeT-A homologue.
Clones V2 and V8 each contain an apparently complete junction between TART elements (labeled 3′-5′ in Fig. 2). We assume that the similarity of the two junctions indicates that none of these elements is truncated at the end in the junction.
Both V2 and V8 also contain truncated TART elements. One element in V8 is truncated by the vector, but each clone also has an element truncated at the 5′ end. The 5′ truncations might have resulted from telomere erosion before the next transposition, although other types of sequence rearrangement are also possible.
Chromosome Locations of TARTvir and TARTame.
A major question about the evolution of telomere-specific transposons is whether these elements evolved from more typical non-LTR elements. If so, we might find some species in which TART elements are present in euchromatic regions, whether or not they also transpose onto chromosome ends. However, this scenario is not what we have found. In situ hybridization to polytene chromosomes shows that TART elements are telomere-specific in D. virilis and D. americana, as in other species studied. We have found no euchromatic TART elements in either D. virilis or D. americana.
D. virilis has four pairs of long acrocentric autosomes, a pair of dot chromosomes, and sex chromosomes. The D. americana karyotype is similar except that two of the long autosomes have fused to form a metacentric chromosome. The centromere regions of all chromosomes tend to aggregate to form a chromocenter. TART probes hybridize to the telomeres of the acentric ends of the long chromosomes of D. virilis (Fig. 3a). Although there must be telomeres on the centromeric ends of these chromosomes, we do not detect TART hybridization in the chromocenter. On D. americana polytene chromosomes, the TART probe hybridizes to a telomere on one of the long chromosomes and to two or three discrete spots in the chromocenter (Fig. 3 b and c). These spots may represent telomeres of the centromeric ends; however, the chromatin structure in the chromocenter is too amorphous to allow a definite localization.
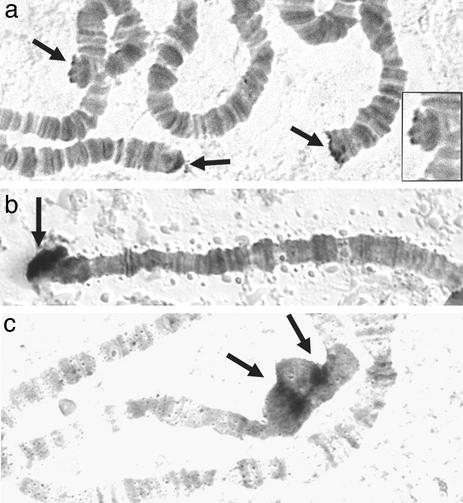
Figure 3.
Hybridization of TARTvir and TARTame to the telomeres in polytene chromosomes. (a) D. virilis. Three telomeres from the same nucleus are shown. Each has probe hybridized to the terminal region (arrows). Note that this region is aggregated into four balls on one telomere (shown enlarged in Inset). There is no hybridization over any banded chromosome regions. (b) The free telomere of D. americana that hybridizes with TARTame. As in D. virilis, there is no hybridization in any banded chromosome regions. (c) Chromocenter from a polytene nucleus of D. americana; two discrete regions of the heterochromatic chromocenter are labeled (arrows). [Hybridized probe detected by alkaline phosphatase activity (7). See Materials and Methods for probes.]
The unlabeled telomeres in D. virilis and D. americana do not necessarily lack transposon telomeres. In different D. melanogaster stocks, TART hybridization is detected on different numbers of ends, frequently fewer than those labeled by HeT-A probes. There are significantly fewer TART elements than HeT-A elements in the D. melanogaster genome. It is likely that TART is also a minor component of telomeres in D. virilis and D. americana. The uncharacterized partial element seen in clone V8 suggests that D. virilis, like D. melanogaster and D. yakuba, may have at least two families of telomere elements. D. americana may also have additional telomere transposons.
The 370-bp Satellite Sequence.
Because the polytene chromosome telomere regions that bind TART probes on polytene chromosomes appear to be indistinguishable from those reported to bind the 370-bp satellite sequence (9), we looked for association between TART and 370-bp repeat sequences. Primers based on the 370-bp database sequence were used to amplify and clone a repeat of the satellite to probe the λ clones isolated with the D. virilis TART probe. None of the clones with TART sequences also contained satellite sequence, suggesting that the two sequences are not intermixed.
To obtain a longer-range comparison of the relation of satellite to TART, we hybridized polytene chromosomes with the satellite probe. We detected satellite on each chromosome end and at a number of sites in euchromatin (data not shown). However, the telomere regions are too compact and amorphous to allow us to distinguish differences in the regions of hybridization of TART and the satellite. Even chromosomes ending in a fringe of balls showed both TART and satellite hybridization covering the balls (Fig. 3a). Satellite is detected at internal euchromatic sites, whereas TART is found only at telomeres. Otherwise, the hybridization patterns are very similar. Thus, evidence to date suggests that the 370-bp satellite is similar to the satellite sequences (TAS) located just proximal to the transposon arrays in other species (14).
Transcription of TARTvir and TARTame.
The sense-strand transcripts of non-LTR retrotransposons serve as both mRNA and transposition template. Many non-LTR elements produce only sense-strand RNA, but in other species TART yields both sense and antisense RNA (10). Surprisingly, for most of the subfamilies of TART studied, antisense RNA is clearly in excess of sense RNA. TART elements of both D. virilis and D. americana show this same pattern of transcription.
The amount of RNA detected by Northern hybridization to RNA from larvae of D. virilis and D. americana (Fig. 4) is significantly less than the amounts found in D. melanogaster or D. yakuba. The lower levels of RNA correlate with the lesser amounts of TART sequence detected on polytene chromosomes (Fig. 3) and by hybridization to genomic DNA (data not shown). In Fig. 4, the lane showing hybridization to TARTvir sense RNA was exposed four times longer than the lanes showing antisense RNA. When probe-specific activity is taken into account (see Materials and Methods), comparison of the blots suggests that there is much more antisense-strand than sense-strand RNA. We assume that TARTame also produces excess antisense RNA because we detect this antisense RNA with difficulty (Fig. 4) and have not been able to detect sense-strand transcripts.
Sequence Evolution of ORF2.
ORF2 encodes the Pol protein with both endonuclease and RT motifs. Consistent with our failure to find cross-hybridization of TARTmel probes to TARTvir DNA, blast comparisons detected no significant similarity in the nucleotide sequences. However, when sequences were aligned, we could detect a low level of sequence identity (Table 1). Surprisingly, we see more sequence similarity between TARTvir and TARTmel than between TARTame and TARTmel, although we did not detect hybridization between TARTmel and D. virilis DNA on Southern blots. We suppose the lack of detectable hybridization is because of a more heterogeneous population of restriction fragments with the RT sequence in D. virilis than D. americana.
Amino acid identity between TARTmel and TARTvir is also low (Table 1). Nevertheless, all the complete ORFs in the D. virilis clones are open. When compared as the amino acid sequences, blast reports highly significant similarity to the other TART ORF2 proteins (E = 0.0). There is also lower similarity to ORF2 proteins of several other Drosophila non-LTR retrotransposons (X, jockey, F, Bs, Doc, and strider) and, with lower significance, to proteins from elements in other insects (E = e−142 to 6e−71 for Drosophila and 3e−77 to 2e−24 for other insects).
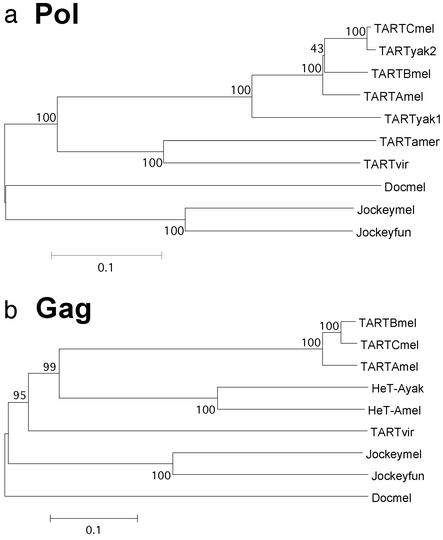
Comparisons of the D. americana sequence produce similar results (Table 1). The available TART ORF2 sequences give a tree that is consistent with the species relationship (Fig. 5a). The sole exception is one TART subfamily in D. yakuba that is surprisingly similar to a subfamily in D. melanogaster (7). The nontelomeric elements tested, jockey and Doc, group separately from the TART elements.
Figure 5.
Phylogenetic relationships of TART, HeT-A, Doc, and jockey elements in different Drosophila species. Neighbor-joining trees for the protein sequences are shown. (The UPGMA trees yield the same relationships as do the nucleotide trees.) Bootstrap tests were performed with 500 replications and a cut-off value of 50% for the consensus tree. Numbers indicate bootstrap values of >40% in the corresponding node. Scale bar corresponds to the P value (number of differences normalized by the number of total residues). Elements are indicted by the first three letters of the species (fun, Drosophila funebris). Note that TARTyak2 groups with TARTmelC as proposed (7). Gag from TARTyak was not included because all of the fragments cloned to date are 5′ truncated. GenBank accession numbers: jockeymel ORF1, M22874; ORF2, AAA28675; jockeyfun, PIR:B38418; Doc, CAA35587.
To compare the rate of evolution of TART with that of other sequences, we chose the well studied alcohol dehydrogenase (Adh) gene and the pol gene of the non-LTR element R1. (The R1 sequences were from D. melanogaster and D. hydei, an evolutionary distance (8) about equal to that between D. melanogaster and D. virilis; Table 1.) Even though ORF2 is the more conserved ORF in retroelement evolution (6), the TART ORF2 shows more rapid sequence change than the Adh gene and the pol gene of R1. The TART protein is also more tolerant of amino acid change, as shown by the increased proportion of amino acid replacements (nonsynonymous substitutions, Ka) compared with synonymous substitutions (Ks).
Comparisons of the ORF2 proteins from D. virilis and D. americana produced two surprises. Both proteins are much more glutamine rich than their equivalents in D. melanogaster or D. yakuba. TARTvir has 13.3% glutamine and TARTame has 8.6%, whereas the ORF2 protein of TARTmelB has only 4% glutamine. In D. virilis, this increased content of glutamine is primarily in long runs of CAX trinucleotide repeats. The triplets are predominantly CAA and CAG, encoding glutamine, with some scattered CAC and CAT trinucleotides encoding histidine. None of these runs interrupts motifs known to be important for the enzymatic activities of the ORF2 protein (15).
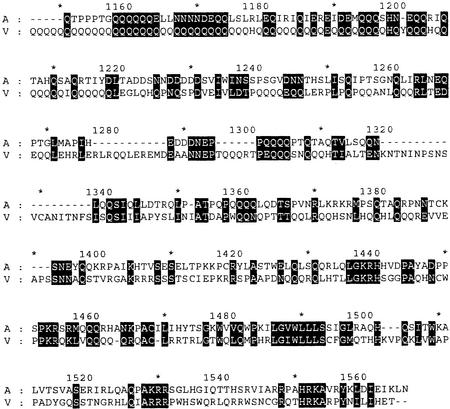
A second surprise is the size of the ORF2 proteins in these two species. Both proteins extend ≈400 aa beyond the C terminus of the proteins from the other two species. This extension (labeled X in Fig. 2) does not show obvious motifs that might suggest its function. It is tempting to suggest that this extension is an RNase H domain similar to that near the C terminus of ORF2 of I factor (16); however, we have not been able to detect strong similarities. The most obvious feature of the X domain is the long stretches of polyglutamine in TARTvir. The TARTame domain does not have similar long regions of CAX codons, but a comparison of the amino acid sequences (Fig. 6) shows groups of amino acids with similar properties throughout the extensions, suggesting that these domains have similar functions.
Figure 6.
Alignment of the X domain of TARTvir and TARTame. Sequence of the TARTvir X domain is residues 1,121–1,538, and TARTame is residues 1,146–1,518 of the Pol protein. In both cases, the sequence shown here begins just after the residue that aligns with the last residue on the TARTmel and TARTyak protein. Numbers on top indicate residues of the TARTame protein. Residues of the same chemical property groups are shaded in black. A, TARTame; V, TARTvir; −, gaps in alignment.
Sequence Evolution of ORF1.
ORF1 encodes a Gag protein (4) with both the amino acid motifs that characterize retroviral Gags, the CCHC or zinc knuckle region and the major homology region (MHR). Our results (Table 1) show that ORF1 has undergone more change than ORF2 in the TART phylogeny, as seen for retroviruses and other retrotransposons (6). In addition, ORF1 has an even higher ratio of amino acid replacements to synonymous changes than does ORF2. These conclusions are based on the two complete TARTvir Gag coding sequences. We have not yet isolated this region from TARTame.
blast comparisons of the ORF1 nucleotide sequences yield no significant matches. Both the D. virilis ORF1 sequences were open, and conceptual translation yields an amino acid sequence with similarity to Gags from several insect non-LTR retrotransposons, most significantly with Gags from TART and HeT-A (E = 3e−49 to 2e−43). The similarity is in the region of the MHR and zinc knuckle, following a pattern seen in studies of HeT-A Gag from D. yakuba. The nucleotide sequence of TARTmel Gag has 38% similarity to HeT-Ayak Gag and 41% similarity to HeT-Amel Gag. This similarity is concentrated in the region of the MHR and zinc knuckles (10). The close relationship between HeT-A and TART Gags is clearly reflected in the phylogenetic relationship of the proteins (see Fig. 5b).
The D. virilis ORF1 sequence is even more enriched for CAX repeats than is ORF2. In ORF1, the polyglutamine stretches do not increase the length of the protein. They tend to be concentrated in the C-terminal regions of the protein and do not interrupt the known motifs.
Discussion
TART Elements Are Telomere-Specific in Both D. virilis and D. americana.
D. virilis chromosomes all have the centromere near one end. In polytene nuclei, the centromeric ends tend to associate closely to form an amorphous chromocenter. TART hybridization can be detected on the nonchromocentral telomeres, but not within the chromocenter. This difference could indicate a bias in the distribution of TART sequence at the two ends of each chromosome. Alternatively, it could indicate some underreplication of telomere sequences in the chromocenter. There is massive underreplication of pericentric satellite sequences in these polytene chromosomes (17), which might also affect nearby telomere sequences. In theory, this question could be resolved by hybridization to mitotic chromosomes. In practice, telomere sequences have been detected on D. melanogaster mitotic chromosomes from the Tel mutant, which has greatly enlarged telomeres, but not on wild-type chromosomes (18). This technical limitation prevents a direct experimental approach. However, telomere sequences are detected on both ends of the Tel X mitotic chromosome, yet we do not detect telomere sequences on the chromocentral end of the D. melanogaster X in polytenes. This finding suggests that there may be underreplication of these telomeres in D. melanogaster, and possibly in D. virilis.
In D. americana, TART is detected on one of the noncentromeric ends and on two, possibly three, regions of the chromocenter. We believe that the chromocentral hybridization represents telomeres that have undergone more polytenization than chromosomes in D. virilis. A similar difference in chromocentral hybridization was seen with D. melanogaster and D. yakuba (19).
TARTvir Has a Significant Resemblance to TARTmel.
TART and HeT-A are the first retrotransposons with a defined role in chromosome maintenance. They transpose only to chromosome ends, although analyses of their coding regions suggest that they are related to a group of non-LTR retrotransposons that transpose generally throughout the euchromatic regions of D. melanogaster chromosomes (15). This study shows that TART has remained telomere-specific over the ≈60 million years separating the melanogaster and virilis groups.
In addition to its telomere-specific transposition, TARTvir has another of the unusual characteristics of TART from other species. TARTvir produces an excess of antisense RNA. Antisense production has been reported for a subgroup of non-LTR retrotransposons that are also characterized by a 5′ sequence that is perfectly repeated near the 3′ end. Studies of two of these elements, DRE, now called TRE5-A (20, 21), and TOC1 (22), led to a model in which the antisense RNA was needed for replication of the element, although questions remain about this subclass of elements. TARTmel has similar perfect nonterminal repeats longer than 1 kb (10). It is attractive to propose that the antisense RNA is also involved in TART replication, although the two apparently complete TARTvir elements studied do not show these perfect repeats. This finding raises a question about the significance of the repeats in replication unless the two elements in our clones are not transpositionally competent. (In D. melanogaster, the telomere transposons show many defects we suppose are caused by erosion, recombination, and sequence decay.)
This study also provides evidence of a second non-LTR element in the telomere arrays of D. virilis. We began our study of virilis group telomeres with TART because RT sequence has the most sequence conservation of the telomere array. Because telomere arrays in other species are mixtures of TART and HeT-A, we had hoped to identify any partner element(s) in D. virilis by their association with TART. The uncharacterized partial element in clone V8 shows that this strategy has worked. Although TART is much less abundant than HeT-A in D. melanogaster, all stocks studied have both elements, suggesting that the elements have different roles and that TART requires a partner. Studies of D. yakuba show that these elements have been evolving together (7). It is notable that HeT-A does not possess its own RT and that HeT-Amel Gag provides telomere targeting for TARTmel Gag (23). These findings suggest that the elements have coevolved to cooperate.
TARTvir Has Some Differences from TARTmel and TARTyak.
The two telomere retrotransposons in D. melanogaster and D. yakuba differ from other retroelements in the relatively large proportion of their sequence found in the 3′ UTR (4). The 3′ UTRs of HeT-A elements show a conserved pattern of A-rich regions, whereas TART 3′ UTRs show less conservation, even among subfamilies in the same Drosophila species. We have suggested that the 3′ UTR sequences are involved in heterochromatin structure. The TARTvir elements studied here have much shorter 3′ UTRs (650 bp for TARTvir, in contrast to 3–5 kb for TARTmel). These short 3′ UTRs raise questions about the importance of the 3′ UTR of TART but do not eliminate the possibility that other telomere sequences supply this function in D. virilis.
TARTvir and TARTame have another structural difference from TARTmel and TARTyak, the extra domain of ORF2, the X domain. The similarities of this sequence in D. virilis and D. americana suggest that this region has function. There is evidence that another non-LTR element, I factor, has an RNase H domain in this part of the sequence (16), and it will be interesting to see whether the extra domain of TART has a similar function.
The TARTvir Coding Regions Have Long CAX Repeats.
A dramatic difference between TART elements of D. virilis and those of the other species is the abundance of triplet CAX nucleotide repeats in TARTvir elements. Both ORF1 and ORF2 contain significant lengths of CAX repeats, encoding polyglutamine with infrequent histidine residues. It is striking that all of the long repeat segments in the coding strand are CAX, rather than its GTX′ complement. Random events should give a less biased strand distribution. Thus, the strand distribution suggests that there has been selection for glutamine codons (or against the valine codons of the complement). Additionally, the repeats show the same CA prevalence that characterizes the coding strand of all studied telomeric transposons. This strand bias mimics the CA bias of the template used by telomerase, an interesting similarity because the coding strand is the template for Drosophila telomeres. Thus, strand sequence bias may also influence the coding potential.
The interest of the long CAX repeats is enhanced by their resemblance to sequences causing human polyglutamine repeat diseases; Huntington's disease, Kennedy's disease, and six of the spinocerebellar ataxias are characterized by expanded sequence encoding polyglutamine (24). Both of the TARTvir proteins have stretches of polyglutamine of lengths in the range that characterizes disease alleles in humans, although the TART regions are infrequently broken by one or two nonglutamine residues. Disease-associated alleles of human triplet repeat disease genes have produced deleterious phenotypic effects when expressed in D. melanogaster (25, 26). It will be interesting to see whether these D. virilis sequences can also produce deleterious phenotypes in similar situations.
The CAX repeats are open and completely conserved in the sequenced TARTvir elements, suggesting that they have roles in the protein. Perutz et al. (27) have pointed out that polyglutamine regions resemble polar zippers and might be involved in protein–protein associations. This suggestion is consistent with the placement of the long polyglutamine segments in ORF1. These are most concentrated in the C-terminal half of the coding region. This part of both TART and HeT-A Gag from D. melanogaster is implicated in both homologous and heterologous interactions between these Gag proteins (ref. 23 and S. Rashkova, A. Athanasiades, and M.-L.P., unpublished data). Thus, the repeats in ORF1 might play a role in such interactions for TART Gag. It is more difficult to speculate about the role of CAX in TART Pol because we know little about the role of the extra domain.
Although the CAX repeats are a striking characteristic of the TARTvir proteins, the rest of the protein sequences and the characteristics of the element clearly show that these proteins are homologues of TART proteins in other species, yet TART proteins in other species do not have long polyglutamine repeats. This finding presents an interesting analogy to proteins involved in human triplet-repeat disease genes. Some of these human proteins have homologues in other species that do not have the shorter CAX repeats that undergo disease-producing expansions (24).
Conclusions
Our identification of TARTvir and TARTame has shown conservation of a number of characteristics despite very significant changes in both nucleotide and protein sequence. These characteristics strongly suggest that the special role of this element at the telomere is widely distributed in Drosophila, raising the possibility that TART also maintains telomeres in other genera.
Acknowledgments
We thank Ron Blackmun and Thom Kaufman for the D. virilis library. We thank T. Eickbush, K. Lowenhaupt, Josep M. Casacuberta, P. G. DeBaryshe, and the members of the Pardue laboratory for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health Grant GM50315.
Abbreviation
- RT
reverse transcriptase
Footnotes
References
- 1.Greider C W. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 2.Fang G, Cech T. In: Telomeres. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 69–105. [Google Scholar]
- 3.Griffith J D, Comeay L, Rosenfeld S, Stansel R M, Bianchi A, Moss H, de Lange T. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 4.Pardue M-L, DeBaryshe P G. In: Mobile DNA II. Craig N, Craigie R, Gellert M, Lambowitz A, editors. Washington, DC: Am. Soc. Microbiol.; 2002. pp. 870–887. [Google Scholar]
- 5.Okazaki S, Tsuchida K, Maekawa H, Ishikawa H, Fijiwara H. Mol Cell Biol. 1993;13:1424–1432. doi: 10.1128/mcb.13.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClure M A, Johnson M S, Feng D-F, Doolittle R F. Proc Natl Acad Sci USA. 1988;85:2469–2473. doi: 10.1073/pnas.85.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casacuberta E, Pardue M-L. Genetics. 2002;161:1113–1124. doi: 10.1093/genetics/161.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beverly S M, Wilson A C. J Mol Evol. 1984;21:1–13. doi: 10.1007/BF02100622. [DOI] [PubMed] [Google Scholar]
- 9.Biessmann H, Zurovcova M, Yao J G, Lozovskaya E, Walter M F. Chromosoma. 2000;109:372–380. doi: 10.1007/s004120000094. [DOI] [PubMed] [Google Scholar]
- 10.Danilevskaya O N, Traverse K L, Hogan N C, DeBaryshe P G, Pardue M-L. Mol Cell Biol. 1999;19:873–888. doi: 10.1128/mcb.19.1.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholas K B, Nicholas H B, Jr, Deerfield D W, II. EMBNEWNEWS. 1997;4:14. [Google Scholar]
- 13.Kumar S, Tamura K, Jakobsen I B, Nei M. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 14.Karpen G H, Spradling A C. Genetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malik H S, Burke W D, Eickbush T H. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 16.Malik H S, Eickbush T H. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- 17.Gall J G, Cohen E H, Polan M L. Chromosoma. 1971;33:319–344. doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- 18.Siriaco G M, Cenci G, Haoudi A, Champion L E, Zhou C, Gatti M, Mason J M. Genetics. 2002;160:235–245. doi: 10.1093/genetics/160.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danilevskaya O N, Tan C, Wong J, Alibhai M, Pardue M-L. Proc Natl Acad Sci USA. 1998;95:3770–3775. doi: 10.1073/pnas.95.7.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shumann G, Zündorf I, Hofmann J, Marschalek R, Dingermann T. Mol Cell Biol. 1994;14:3074–3084. doi: 10.1128/mcb.14.5.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beck P, Dingermann T, Winckler T. J Mol Biol. 2002;318:273–286. doi: 10.1016/S0022-2836(02)00097-9. [DOI] [PubMed] [Google Scholar]
- 22.Day A, Schirmer-Rahire M R, Kuchka M, Mayfield S P, Rochaix J-D. EMBO J. 1988;7:1967–1972. doi: 10.1002/j.1460-2075.1988.tb03029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rashkova S, Karam S E, Kellum R, Pardue M-L. J Cell Biol. 2002;159:397–402. doi: 10.1083/jcb.200205039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cummings C J, Zoghbi H Y. Annu Rev Genomics Hum Genet. 2000;1:281–328. doi: 10.1146/annurev.genom.1.1.281. [DOI] [PubMed] [Google Scholar]
- 25.Warrick J M, Paulson H L, Gray-Board G L, Bui Q T, Fischbeck K H, Pittman R N, Bonini N M. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 26.Jackson G R, Salecker I, Dong X, Yao X, Arnheim N, Faber P W, MacDonald M E, Zipursky S L. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 27.Perutz M F, Johnson T, Suzuki M, Finch J T. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]