Abstract
Methylating agents are ubiquitous in the environment, and central in cancer therapy. The 1-methyladenine and 3-methylcytosine lesions in DNA/RNA contribute to the cytotoxicity of such agents. These lesions are directly reversed by ABH3 (hABH3) in humans and AlkB in Escherichia coli. Here, we report the structure of the hABH3 catalytic core in complex with iron and 2-oxoglutarate (2OG) at 1.5 Å resolution and analyse key site-directed mutants. The hABH3 structure reveals the β-strand jelly-roll fold that coordinates a catalytically active iron centre by a conserved His1-X-Asp/Glu-Xn-His2 motif. This experimentally establishes hABH3 as a structural member of the Fe(II)/2OG-dependent dioxygenase superfamily, which couples substrate oxidation to conversion of 2OG into succinate and CO2. A positively charged DNA/RNA binding groove indicates a distinct nucleic acid binding conformation different from that predicted in the AlkB structure with three nucleotides. These results uncover previously unassigned key catalytic residues, identify a flexible hairpin involved in nucleotide flipping and ss/ds-DNA discrimination, and reveal self-hydroxylation of an active site leucine that may protect against uncoupled generation of dangerous oxygen radicals.
Keywords: alkylation, crystal structure, DNA repair, hABH3, oxidative demethylation
Introduction
Humans are exposed to alkylating agents produced endogenously and exogenously, such agents are also frequently used in cancer therapy. Alkylating agents may react with oxygen and nitrogen atoms in DNA (Singer and Grunberger, 1983), causing cytotoxic and mutagenic lesions (Drabløs et al, 2004; Ringvoll et al, 2006). The global response to methylating agents may be comprehensive and similar to those needed for vital cellular functions, as observed in yeast (Said et al, 2004). Depending on the type of alkylation, DNA repair may take place by base excision repair, nucleotide excision repair, mismatch repair or recombination repair (Hoeijmakers, 2001). In an important addition to these repair pathways, certain alkylations may be removed by direct repair by the methyltransferase Ada, the demethylase AlkB and their eukaryote homologues (Drabløs et al, 2004).
The Escherichia coli alkB gene was discovered more than 20 years ago (Kataoka et al, 1983). An indication on the nature of the substrates came from the observation that AlkB specifically reactivated methylated single-stranded DNA bacteriophages, where 1-meA and 3-meC are major cytotoxic lesions (Dinglay et al, 2000). A clue towards the mechanism of repair came from a bioinformatics study in which AlkB was demonstrated to be a member of the Fe(II)/2-oxoglutarate (2OG) dioxygenase superfamily (Aravind and Koonin, 2001). Oxidative demethylation of 1-meA and 3-meC was subsequently verified experimentally for E. coli AlkB (Falnes et al, 2002; Trewick et al, 2002) and for the human homologues hABH2 and 3 (Duncan et al, 2002; Aas et al, 2003). Putative relatives of AlkB were found in several RNA viruses, and it was predicted that AlkB and its homologues may act in RNA demethylation (Aravind and Koonin, 2001). RNA repair was later proven experimentally for AlkB and hABH3 in vitro and in vivo (Aas et al, 2003; Ougland et al, 2004), although the significance of RNA repair remains elusive. AlkB and hABH3 prefer ssDNA and RNA substrates, whereas hABH2 works best with dsDNA (Aas et al, 2003; Falnes et al, 2004). Six other human homologues of AlkB have been identified (Kurowski et al, 2003; Drabløs et al, 2004), but demethylase activity has only been experimentally confirmed for hABH2 and hABH3. In addition to 1-meA and 3-meC, AlkB also repairs 1-meG, 1-etA, 3-etC, ɛA and ɛC in DNA (Duncan et al, 2002; Delaney and Essigmann, 2004; Delaney et al, 2005), 1-methyldeoxyadenosine mono- and triphosphates, 3-meT, and the larger hydroxyethyl, propyl, and hydroxypropyl lesions (Koivisto et al, 2003). The substrate preference of hABH3 is more restricted. Besides 1-meA and 3-meC, only a low level of repair of 3-meT and ɛA has been demonstrated (Mishina et al, 2005).
AlkB belongs to the superfamily of Fe(II)/2OG-dependent dioxygenases, which catalyse a remarkable diversity of enzymatic reactions. These include protein side-chain modification, biosynthesis of antibiotics and plant products, metabolism of lipids and repair of alkylated DNA/RNA (Hausinger, 2004). Common for most enzymes in this superfamily is the coupling of substrate oxidation to the conversion of 2OG to succinate and CO2. The crystal structures that have been determined within this superfamily show a β-strand jelly-roll fold that coordinates a catalytically active iron-centre using a highly conserved His1-X-Asp/Glu-Xn-His2 motif (Hausinger, 2004). During revision of this manuscript, the crystal structure of AlkB in complex with a 1-meA-containing trinucleotide was published (Yu et al, 2006), showing that AlkB shares several features with structures of other superfamily members.
Here, we present the high-resolution structure of the human ABH3 catalytic core domain in complex with iron and 2OG together with activity analysis of site-directed mutants of full-length protein. The combined results reveal new features of this class of repair enzymes, including previously unassigned key catalytic residues, a flexible hairpin involved in nucleotide flipping and ss/ds-DNA discrimination, and self-hydroxylation of an active site leucine that may have a protective function by inhibiting production of dangerous oxygen radicals.
Results and discussion
Overall structure of hABH3
The N-terminally truncated hABH3Δ69 used for crystallisation displayed ∼36% catalytic activity compared to the full-length enzyme (averaged by using 0.3, 1, 3 and 10 pmol enzyme). Crystals of hABH3Δ69 were grown under aerobic conditions at room temperature, and the structure was determined by multiwavelength anomalous dispersion (MAD) with a single Pt derivative using data from 50 to 2.8 Å. A high-resolution X-ray diffraction data set was collected on a crystal of hABH3Δ69 in complex with Fe and 2OG from 50 to 1.5 Å. The model built from MAD data was used to obtain phases for the high-resolution data set. Crystallographic data are presented in Supplementary Table I.
hABH3Δ69 is composed of 14 β-strands and two α-helices (Figure 1). The N-terminal His-tag, a flexible loop between β10 and β11 and the last seven C-terminal residues, were not visible in the electron density maps and were therefore omitted from the structure. Eight of the β strands (β7–β14) form a central jelly-roll fold consisting of two anti-parallel β-sheets characteristic of many metal-binding and nonmetal-binding proteins, including the Fe(II)/2OG-dependent dioxygenase family (Dunwell et al, 2004). β1 is located at the end of the smaller β-sheet in the jelly-roll fold, and is the only strand that is parallel to its adjacent strand. β2 and β3 at one end and β6 at the other end extend the larger sheet of the jelly-roll while the two helices buttress the larger sheet. The active site iron is coordinated by His191, Asp193, His257 and 2OG and likely exists in the inactive Fe(III) state in the aerobically grown crystal. β4 and β5 form a hairpin that creates a lid over the active site (Figure 1). This hairpin and a corresponding loop in AlkB (Yu et al, 2006) (and putatively in hABH2) are absent in non-AlkB proteins of the Fe(II)/2OG-dependent dioxygenase family (Supplementary Figure 1). A positively charged groove traverses the active site between the β4–β5 hairpin and the jelly-roll core, and must constitute the DNA/RNA binding cleft.
Figure 1.
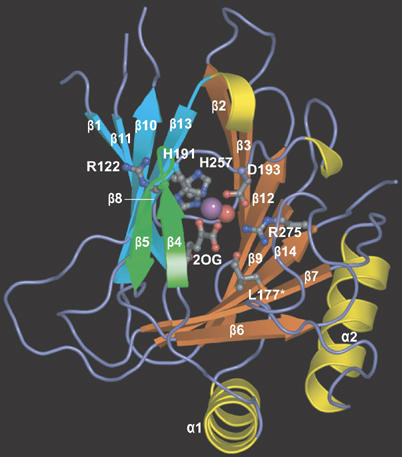
hABH3 overall structure and active site location. Ribbon representation of hABH3Δ69 with secondary structure assignments. The β-strand jelly roll is made up by two antiparallel β-sheets coloured orange and blue, respectively. The β4–β5 hairpin is coloured green. Helices are coloured yellow, and key residues as well as the co-substrate 2OG are shown as balls and sticks. The iron (violet) and the iron bound water (red) are shown as spheres.
Key regions in hABH3 and AlkB are structurally different
To characterise differences associated with greater substrate specificity of hABH3 compared to AlkB, we superimposed the human enzyme structure onto the E. coli structure. hABH3Δ69 superimposes onto the structure of AlkB (Yu et al, 2006) with 114 Cα atoms to an rms deviation of 1.28 Å. The overall similarity is particularly evident in the central jelly-roll core (Figure 2A), where the iron-binding triad as well as 2OG adapts nearly identical geometry in the two enzymes. Structure-based sequence alignment (Figure 2B) reveals relatively low sequence conservation with 18% overall identity (34% similarity) between hABH3 and AlkB, rising to 21% (39%) in their C-terminal halves (Supplementary Table II).
Figure 2.
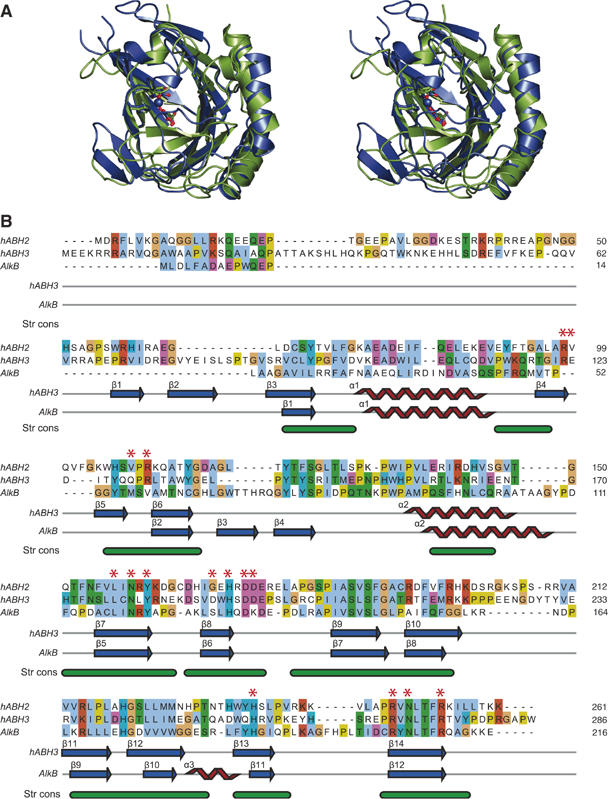
The overall hABH3 β-strand jelly roll core resembles AlkB. (A) Stereo pair ribbon representation of hABH3Δ69 (blue) superimposed upon AlkB (2FDJ) (green). The side chains in the His1-X-Asp-Xn-His2 motif and the 2OG (hABH3) and succinate (AlkB) are shown as balls and sticks, whereas the iron ions are presented as spheres. (B) Sequence alignment and secondary structure assignment of AlkB, hABH2 and hABH3. The alignment of hABH3 and AlkB was generated with DeepView (Guex and Peitsch, 1997) based on a structural alignment of the 3D structures. The hABH2 sequence was added to the alignment by a combination of profile alignment of hABH2 versus the structure-based hABH3/AlkB alignment, using ClustalX (Thompson et al, 1997), as well as hABH2/hABH3 and hABH2/AlkB alignments from iterative PSI-Blast searches (Altschul et al, 1997) starting from hABH2. The final alignment was made with JalView (Clamp et al, 2004). The bottom annotation line (Str cons) shows structurally conserved regions (in green) identified by DeepView. Individual residues are coloured according to ClustalX colour coding, and residues selected for site-directed mutation analysis are indicated by red asterisks.
Notably, significant differences occur in the substrate-binding pockets and in other key regions near the active sites of the two enzymes. The substrate binding pocket of AlkB is predominantly hydrophobic and 1-meA is sandwiched between His131 and Trp69 (Yu et al, 2006). In hABH3, the substrate-binding pocket is considerably more polar (Figure 3A), and lacks a potentially stacking side chain at the position corresponding to Trp69. Thus, tryptophan at this position is not invariant among the AlkB proteins, as suggested by the authors (Yu et al, 2006). Their proposal was, however, based on a sequence alignment where 50 internal residues in the hABH3 sequence were left out. Our structure-based alignment furthermore indicates that a substrate-stacking tryptophan is also absent in hABH2 (Figure 2). Thus, the stacking interactions and the more hydrophobic substrate binding pocket in AlkB may in fact partly explain the broader substrate specificity of AlkB compared to hABH3 (Mishina and He, 2006). Apparently, bulky adducts at the 1- and 3-positions of A and C, respectively, may disrupt position-specific interactions in hABH3 and formation of a productive complex.
Figure 3.
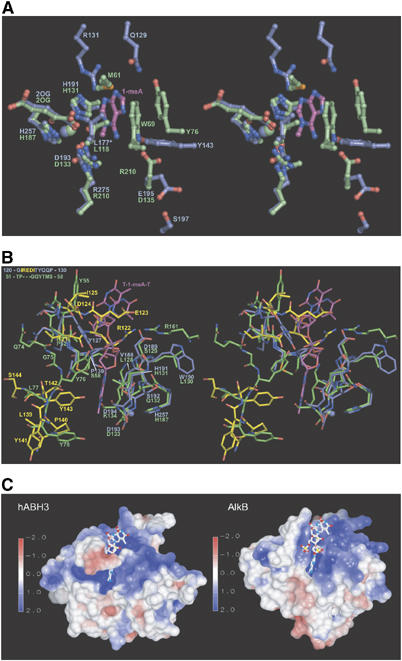
Key regions in hABH3 and their differences from AlkB. (A) Stereo pair ball and stick representation of active site residues in hABH3 (blue) and AlkB (green). The 1-meA lesion from the AlkB complex is coloured magenta. The substrate-binding pocket of hABH3 is dominated by polar residues, while the observed substrate interactions in AlkB are predominantly hydrophobic. The oxidised Leu177 is denoted L177*. (B) Stereo pair representation of the active site entrances of hABH3 (blue) and AlkB (green). The structurally nonconserved residues in the β4–β5 loop (upper left) and the β6–α2 region (lower left) of hABH3 are in yellow. (C) Electrostatic surface presentations of hABH3 and AlkB contoured at ±2 kT/e, where red and blue describe negative and positive potentials, respectively. Electrostatic potentials were calculated by UHBD (Gilson et al, 1993), and are mapped on the molecular surface using AVS (Advanced Visual Systems Inc.). The T-1-meA-T (balls and sticks) was positioned in hABH3 by superimposition of the AlkB/trinucleotide complex upon hABH3.
In AlkB, Asp135 hydrogen bonds to N6 of 1-meA. Interestingly, Glu195 that replaces Asp135 in hABH3, hydrogen bonds to Ser197 and points away from the position of the methylated base (Figure 3A). When hABH3Δ69 is superimposed upon the AlkB/T-1-meA-T structure, hABH3 Tyr143 is apparently positioned to make a hydrogen bond to 1-meA N6. Arg131 in hABH3 is conserved in hABH2, and is positioned to interact with 1-meA N3. Interestingly, the side chain of Arg131 is positioned in the region that likely would be occupied by the 5-methyl group of 3-meT. In contrast, the corresponding Val59 (Figure 2B) in AlkB likely provides room for the 5-methyl group of 3-meT, and may thus explain the higher activity of AlkB towards this lesion compared to hABH3 (Falnes, 2004; Koivisto et al, 2004).
Surprisingly, very few of the residues in AlkB that make contact with the sugar-phosphate backbone of T-1-meA-T are structurally conserved in hABH3 (Figure 3B). This striking variation in contact residues therefore supports different modes of binding to the DNA/RNA backbone by the two enzymes. Distinct binding modes are also implicated by the considerably more electropositive floor of the DNA/RNA-binding groove in hABH3 compared to AlkB (Figure 3C), as well as by significant structural differences in the wall of the grooves. Such structurally implied substrate recognition differences are especially evident in the flexible β4–β5 hairpin that forms the outer wall of the DNA/RNA binding groove and contains some of the key residues in hABH3. This hairpin is extended by three residues (Arg122, Glu123, Asp124) in hABH3 relative to the corresponding AlkB loop, creating a more negatively charged entrance to the DNA/RNA binding groove with resulting implications for substrate recognition as discussed in more detail below. The subsequent β6-strand superimposes well with AlkB β2, whereas the region between this strand and the next α-helix makes different turns and forms different secondary structure elements in the two demethylases (Figure 2A and B). A loop segment between β10 and β11 is not visible in the crystal structure of hABH3. The corresponding, but considerably shorter loop region in AlkB folds towards the DNA/RNA binding cleft, and holds arginine that binds to a substrate phosphate (Arg161 in AlkB) (Figure 3B). hABH3 lacks an arginine corresponding to AlkB Arg161. However, three consecutive prolines encompass the same region in hABH3 and a proline-rich pinch has been shown to play a role in DNA damage scanning and backbone compression in the human uracil-DNA glycosylase UNG (Parikh et al, 1998).
The trinucleotide T-1-meA-T bound to AlkB adopts a rather unusual conformation compared to all other known oligonucleotide structures bound to nucleobase-flipping enzymes (Slupphaug et al, 1996; Barrett et al, 1998; Lau et al, 1998; Daniels et al, 2004). At the 3′-side of 1-meA, the conformation of the phosphate resembles that observed in the substrate bound to the alkyladenine-DNA glycosylase AAG (Lau et al, 1998), while the 5′-phosphate resembles that of the substrate bound to the O6-methylguanine-DNA methyltransferase AGT (Daniels et al, 2004). Thus, in T-1-meA-T bound to AlkB, the 5′-and 3′-phosphates are rotated in such a way that the thymidines adjacent to 1-meA stack with each other separated by a distance of only 3.4–3.6 Å (Figure 3B). This unique AlkB substrate conformation is, however, not easily reconciled with the structure of the hABH3 DNA/RNA binding groove. When the AlkB/T-1-meA-T structure is superimposed upon hABH3Δ69, the DNA both 3′- and 5′- to 1-meA sterically interferes with the groove. The rigid Pro130 (Cδ) in the loop preceding β6 as well as Asp189 (Cβ) and the aromatic ring of Tyr127 (χ1 rotated ∼180° relative to that of the corresponding Y55 in AlkB) all make close contacts to the 3′-phosphate of 1-meA. At the 5′-side of 1-meA the flexible β4–β5 hairpin sterically interferes with the trimer (Figure 3B). Interestingly, these steric hindrances in hABH3 are alleviated if the 5′- to 3′-direction of the T-1-meA-T is rotated 180° (Supplementary Figure 2). Moreover, such rotation would still allow binding of 1-meA with the 1-methyl group in the same position as observed in the AlkB/T-1-meA-T complex. To our knowledge, there are no reports of two alternative DNA-binding orientations in other pairs of related DNA-repair proteins. We therefore propose that hABH3 and AlkB may bind to its DNA/RNA substrate in the opposite direction to that observed in the crystal structure of AlkB complexed with the three nucleotide oligomer used for crystallisation. Alternatively, the sugar-phosphate backbone of longer, biologically relevant oligonucleotides may adopt a significantly different conformation due to a more restricted rotational flexibility. The answers to these questions must, however, await crystallisation of complexes containing such oligonucleotides coupled to mutational analyses validating the crystallographically observed interactions, as performed for UNG (Slupphaug et al, 1996).
Residues involved in iron- and 2OG-coordination are essential for hABH3 activity
To test implications for activity and substrate interactions based on the sequence alignment and structural analyses noted above, selected residues in the full-length hABH3 protein were mutated to alanine and the catalytic activity of the purified mutants analysed (Figure 4). In the crystal structure, His191, Asp193 and His257 bind at one face of the iron ion. Among these, His191 or Asp193 both appear essential. His257 appeared less important, since the His257Ala mutant retained about 35% activity (Figure 4A and E).
Figure 4.
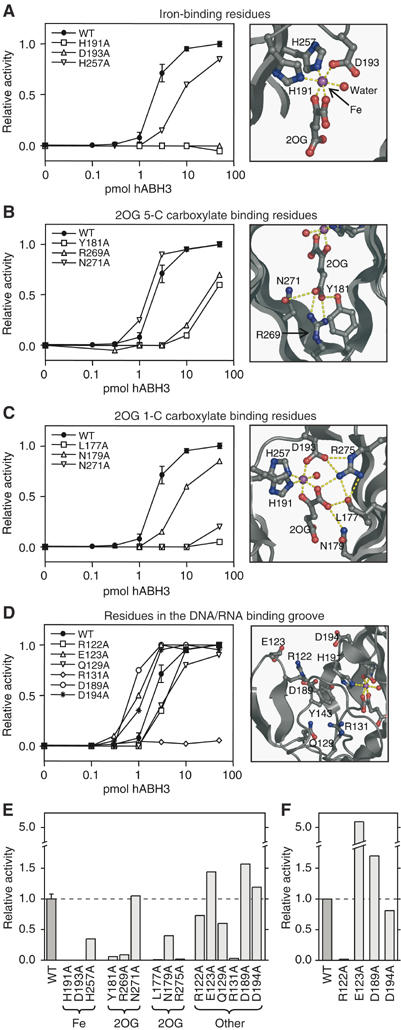
Active site residue structure–function relationships and enzyme activity shown by WT and mutant hABH3-mediated release of radioactivity from [3H]methyl DNA. Left: (A–D) Relative 1-meA repair activity for full-length WT hABH3 and alanine mutants thereof at 0.1, 0.3, 1, 3, 10 and 50 pmol enzyme. Mutants are grouped according to the functions of different residues. Assay reproducibility was verified by including the WT in each experiment comprising three mutants, so every activity curve data point for WT is based on four independent experiments each in duplicate, with standard deviation indicated. Each point in the graphs for mutants represents the mean of duplicates. Right: (A–D) Closeup presentations of the residues in hABH3 analysed by site-directed mutation. The iron ion (violet) and iron-bound water (red) are shown as spheres. (E) Activity of hABH3 mutants involved in iron-binding, 2OG C-5 carboxylate binding, 2OG C-1 carboxylate binding, and residues in the DNA/RNA binding cleft relative to WT on 1-meA-containing ssDNA using 0.3, 1, 3 and 10 pmol enzyme. Bars represent average activity from the four enzyme concentrations relative to WT. (F) Average activity of WT and mutants Arg122Ala, Glu123Ala, Asp189Ala and Asp194Ala, acting upon 1-meA-containing dsDNA substrate using 0.3, 1, 3 and 10 pmol enzyme.
hABH3 uses two co-substrates, O2 and 2OG. 2OG chelates the iron ion in a bi-dentate manner (Figure 4A). The C-1 carboxyl is trans to His257 and the C-2 keto group is trans to Asp193. This ‘off-line' binding mode (Hausinger, 2004) is also observed in the AlkB/T-1-meA-T complex (Yu et al, 2006), as well as in CarC (Clifton et al, 2003) and ANS (Wilmouth et al, 2002). The C-5 carboxylate of 2OG forms a salt-bridge with Arg269 (AlkB Arg204) located at the start of β14 (Figure 4B), an interaction that is conserved in the Fe(II)/2OG-dependent dioxygenases. In addition, the C-5 carboxylate hydrogen bonds to Tyr181 (in β7) (AlkB Tyr122) and Asn271 (in β14) (AlkB Asn206) side chains, residues that are located in the conserved D/NxxLxNxY and RxNLTFR motifs, respectively. These motifs are shared by nucleobase demethylases in the AlkB family. Consistent with the importance of these interactions, both Tyr181Ala and Arg269Ala showed markedly reduced activities (Figure 4E). The enzymatic activity of Asn271Ala resembled wild-type (WT) and Asn271 thus appears less important in 2OG-coordination. The C-1 carboxylate oxygen of 2OG that is not chelating iron lies within hydrogen bonding distances of Asn179 (AlkB Asn120) and Arg275 (AlkB Arg210) (Figure 4C). Of these, Arg275 appears to be essential for activity, while the Asn179Ala mutant displays about 40% activity (Figure 4E). Interestingly, a post-translationally oxidised leucine (Leu177) observed in our crystal structure also lies within hydrogen bonding distance to the C-1 carboxylate of 2OG. Leu177 is conserved in AlkB (Leu118), hABH2 and hABH6, but not in the other human AlkB family members. Notably, the Leu177Ala mutant is virtually inactive against 1-meA (Figure 4E). Potential roles of the Leu177 residue in catalysis will be discussed in more detail below.
Charged residues in the β4–β5 loop mediate base flipping and discrimination between double-stranded and single-stranded substrates
The distance between the hairpin loop and the opposing edge of the cleft is only ∼9 Å (measured between Arg122 Cα and His191 Cα) (Figures 3B and 4D). This is too narrow even for ssDNA or ssRNA to fit between the rims of the putative binding cleft. However, high B-values for residues in the hairpin suggest that it is flexible and may thus change to a more open conformation upon DNA/RNA binding, as observed in the corresponding, but shorter hairpin loop in the AlkB complex (Yu et al, 2006). A striking feature of hABH3 is three negatively charged residues (Glu123, Asp189 and Asp194) strategically located at the entrance of the DNA/RNA-binding groove and absent in AlkB. When the 1-AlkB/T-1-meA-T complex is superimposed upon hABH3Δ69, Glu123 sterically clashes with the 5′-phosphate of the trinucleotide, while Asp189 is in close contact with the phosphate 3′ to 1-maA (Figure 3B). Mutation of either Glu123 or Asp189 to alanine resulted in about 1.5-fold increased activity against ssDNA (Figure 4E), likely caused by increased accessibility of the substrate to the active site. Asp194 is located further away from the trinucleotide, in agreement with the minor effect of the Asp194Ala mutation upon activity. Interestingly, the activity of Glu123Ala against the dsDNA substrate was about five-fold higher than that of the WT enzyme (Figure 4F), suggesting that this glutamate has an important function in discrimination against dsDNA substrates. Conversely, a valine at the corresponding position in hABH2 may contribute to the dsDNA preference of this demethylase (Figure 2B). The above results support our previous findings that hABH3 is not able to reactivate MMS-treated dsDNA bacteriophages (Aas et al, 2003), and may indicate that dsDNA is not a physiologically important substrate for hABH3. This is also sustained by earlier results from our laboratory demonstrating that hABH3 is localised both in the cytoplasm as well as in the nucleus, in contrast to hABH2 that is strictly nuclear. Furthermore, hABH3 is not located to replication foci during the S-phase (Aas et al, 2003).
Nucleotide flipping in DNA repair was first shown for the human uracil-DNA glycosylase UNG (Slupphaug et al, 1996), and has later been found in several co-crystal structures of DNA-repair enzymes and their DNA substrates (Huffman et al, 2005), including AlkB (Yu et al, 2006). Intercalation generally plays an important role in flipping of the damaged base, and in the 3-methyladenine-DNA glycosylase AAG this function is served by an antiparallel β hairpin (Lau et al, 1998). Surprisingly, intercalation is not observed in the crystal structure of AlkB with trinucleotide; however, the hABH3 β4–β5 antiparallel hairpin is ideally positioned to serve such a function. Furthermore, Arg122 located at the top of the hairpin may occupy the subsequent empty position in the base stack, as supported by superimposition of hABH3 upon the AlkB/T-1-meA-T structure (Figure 3B). This arginine is conserved in hABH2, whereas AlkB lacks this region in its shorter flexible loop (Figure 2). In fact, an analogous arginine role in nucleotide flipping has also been observed for SMUG1 (Wibley et al, 2003), O6-alkylguanine DNA transferase (AGT) (Daniels et al, 2004) and vaccinia and cytomegalovirus UNG (Parikh et al, 1998). Notably, whereas the activity of Arg122Ala against ssDNA substrate was about 75% compared to the WT, the activity against dsDNA substrate was essentially abolished (Figure 4E and F). This likely reflects the higher energy barrier for nucleotide flipping in dsDNA, and supports a role of Arg122 in hABH3 base flipping.
Potential functions of Leu177 in substrate binding and catalysis
Leu177 occupies a central position in the hABH3 active site (Figures 4C and 5A). Although it lacks any obvious direct function, Leu177 is however essential for demethylation activity against 1-meA, and is conserved in AlkB, hABH2 and hABH6. Unexpectedly, Leu177 was clearly modified in the experimental electron density and in the refined crystal structure of hABH3 (Figure 5A). A peptide mass shift of +14 and +16 Da found in MALDI-TOF MS analyses of tryptic peptides (Figure 5B) indicated that this modification was a mixture of hydroxyl and carbonyl at Leu177 Cδ. To our knowledge, leucine side chain carbonylation has not been reported to occur in proteins, although it has been proposed to be an intermediate in the formation of 4-methylproline from free L-leucine (Fu and Dean, 1997).
Figure 5.
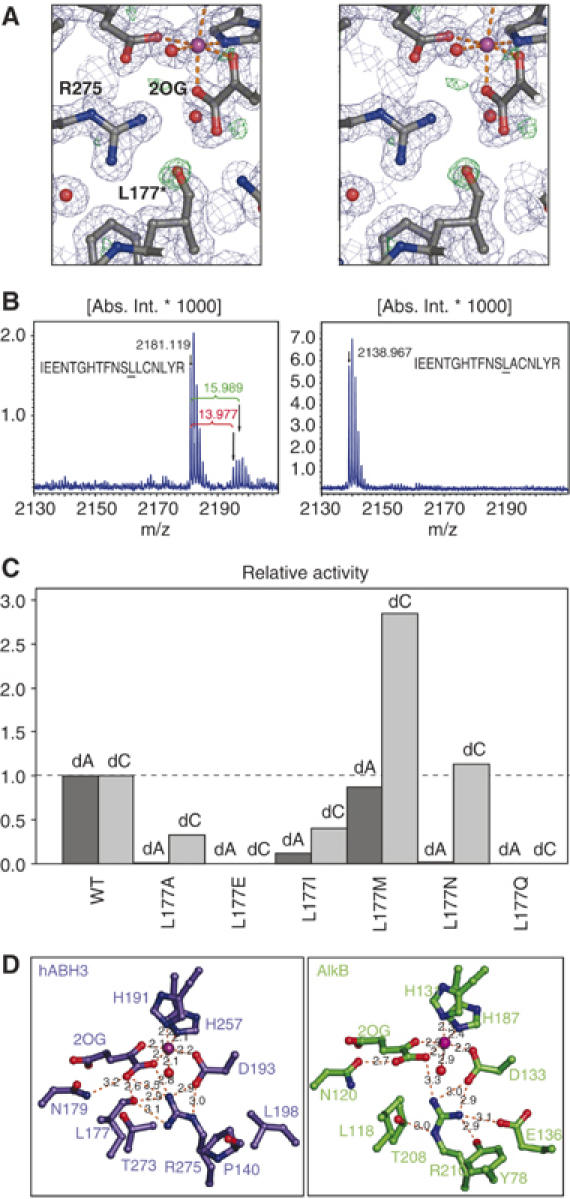
Analysis of Leu177 oxidation and activity of Leu177 mutants. (A) Stereo view of the electron density around the hABH3 active site residues, 2OG and iron. 2Fo−Fc (contoured at 1σ, in blue) and Fo−Fc (contoured at 3σ, in green) were calculated after omitting the Leu177 modification (assigned Oɛ). In the crystal structure, the modification was modelled as carbonyl (L177*) with a refined distance of 1.23 Å between the Leu177 Cδ2 and Oɛ refined to a Cγ-Cδ2-Oɛ angle of 119.6°. (B) The modification gave mass-shifts of +14 and +16 Da in MALDI-TOF MS, revealing a mixture of carbonyl and hydroxyl at Cδ2. (C) Relative activity of Leu177 mutants using [3H]methylated poly(dA) and poly(dC) ssDNA substrates. Activity assays were performed in duplicate using 5 pmol of each hABH3 mutant. Data for Leu177Ala are the same as presented in Figure 4E. (D) Closeup view of the 2OG-coordinations in hABH3 and AlkB. The essential Arg275 in hABH3 adapts a different conformation than the corresponding Arg210 in AlkB, and makes a hydrogen bond to the oxidised Leu177.
To investigate whether a leucine at this position is oxidised in other proteins in the AlkB family, we expressed and purified AlkB, hABH2 and hABH6 proteins in E. coli. Notably, mass shifts of +14 and +16 Da was observed in the corresponding tryptic peptides from these enzymes (data not shown), confirming the presence of the oxidation. Importantly, the mass shift was abolished when the leucine was mutated to alanine in hABH3 (Figure 5B), further confirming Leu177 as the target residue for oxidation. Leu177 is located 6.8 Å away from the iron centre, almost perpendicular to the plane formed by the side chains of His191 and Asp193, the 2-keto group of 2OG, and a water molecule in the crystal structure (Figure 4C). It is thus likely that oxidation of Leu177, and equivalent modifications in other AlkB proteins, results from attack by a hydroxyl radical formed by uncoupled decarboxylation of 2OG in the absence of primary substrate. Self-hydroxylation of Leu177 was supported by lack of oxidation in the catalytically dead His191Ala mutant (data not shown).
The Leu177 oxygen modification was modelled as a carbonyl atom in the crystal structure and refined to occupancy of ∼69%, which is an indication of the ratio of oxidised and unoxidised protein in the crystals. Although equivalent oxidation was not observed in the crystal structures of AlkB (Yu et al, 2006), it is possible that their crystallisation conditions selected for the unoxidised protein bound to the short oligo, as the DNA may not fit sterically with the modified protein.
To examine possible functional implications of this modification, we analysed whether this oxidation in hABH3 can occur in vivo. We found that a mixture of both oxidised and unoxidised forms of Leu177 exist in hABH3 (and of the corresponding leucines in AlkB, hABH2 and hABH6) even after purification under strictly reducing conditions, suggesting that the oxidation indeed occurs in vivo in E. coli. Due to their apparently identical chromatographic and electrophoretic behaviour, we have not been able to separate the two forms of the enzyme to monitor their relative activities. Moreover, even if the peptide encompassing Leu177 could readily be monitored in MALDI-MS, it could not be detected using electrospray ionisation techniques. Thus, we have shown that the Leu modification likely occurs in vivo in E. coli but have been unable to correlate changes in relative abundance of each of the forms to activity. Therefore, to further analyse the function of Leu177 and the potential implications of its oxidation, a series of additional mutants of this residue were made, and their activities analysed against both 1-meA- and 3-meC-containing ssDNA substrates (Figure 5C). Interestingly, the various amino-acid substitutions at the 177 position had striking effects upon the relative activity of hABH3 against 1-meA and 3-meC. Mutations to alanine and isoleucine markedly reduced the activity against 1-meA, while the activity against 3-meC was less affected. Moreover, the Leu177Met had little effect on the activity against 1-meA, while the activity against 3-meC actually increased nearly three-fold (Figure 5C). Leu177 Cδ is located at the bottom of the catalytic pocket only 3.6 Å from the N1-methyl group of 1-meA from the superimposed AlkB complex. The different effects of the mutations upon the activity against the two substrates may thus be explained by Leu177 serving as a ‘buffer stop' to prevent penetration of alkylated purines too deep into the catalytic pocket. This function would not be equally important for the smaller alkylated pyrimidines, since too deep penetration would be restricted by steric contacts between the enzyme and the sugar-phosphate backbone. A peculiar effect was observed with Leu177Asn, in which the activity against 3-meC was similar to that of the WT, whereas the activity against 1-meA was essentially abolished. This mutant may thus be exploited for in vivo complementation assays to specifically address effects of 3-meC repair in the absence of 1-meA repair. Notably, an asparagine is also found at this position in hABH4 (Drabløs et al, 2004).
To mimic the size and (at least partially) the polarity of an oxidised leucine, Leu177 was mutated to glutamine and glutamate. Notably, both mutations abolished catalytic activity, suggesting that oxidation of Leu177 inactivates hABH3. Self-hydroxylation of aromatic residues in the vicinity of the active site has been observed with several Fe(II)/2OG-dependent dioxygenases in the absence of their primary substrates. It has been suggested that this inactivates the enzymes to avoid generation of reactive oxygen species and potentially harmful side-reactions in the cell (Liu et al, 2001; Ryle et al, 2003). It cannot be excluded that self-hydroxylation serves a similar function in the AlkB-family of dioxygenases, and enzymatic inactivation has in fact been observed following hydroxylation of an active-site tryptophan in AlkB (Henshaw et al, 2004). Common for the above inactivation mechanisms are that they are caused by uncoupled decarboxylation of 2OG, and that the newly oxidised aromatic residue binds the iron ion and inhibits damaging reactivity of the enzyme. A similar inactivation mechanism is not likely in hABH3, since the oxidised Leu177 is not bound to the active site iron (Figure 5A). A more likely explanation is that the oxidation negatively affects coordination of 2OG (Figure 5D), thereby preventing the insertion of an oxygen atom into the C1–C2 bond of 2OG and formation of a carbonate-succinate mixed anhydride.
Taken together, the results presented here on the human damage reversal enzyme hABH3 crystal structure coupled to structure-based mutational analysis and comparisons to the recently published structure of the E. coli AlkB yield new insights into the catalytic mechanism and divergent substrate preferences of DNA direct damage reversal enzymes. This work furthermore discovers a novel self-hydroxylation of an active site leucine that may serve an inhibitory function in the absence of primary substrate and protect the cell from harmful oxidative side reactions. In general, these results provide the framework for characterising hABH-proteins in complex with their prime substrates, which could aid the design of small-molecule antagonists useful as adjuvants in cancer therapy that relies upon simple alkylating agents.
Materials and methods
Purification of hABH3Δ69 protein
Full-length hABH3 and mutant proteins were expressed and purified as previously described (Aas et al, 2003). A truncated construct encoding residues 70–286 of hABH3 was cloned into pET28a (Novagen). His-tagged hABH3Δ69 protein was expressed in E. coli BL21 (DE3) RIL (Stratagene). Bacterial pellets were resuspended in buffer A (50 mM Na2PO4, 300 mM NaCl, 10 mM imidazole, 10 mM β-mercaptoethanol, pH 8), sonicated on ice, and the clarified extract was purified by Ni-NTA chromatography (Qiagen). The purified fraction (containing >95% hABH3Δ69) was concentrated to 4–7 mg/ml, and stored at −80°C.
Crystal growth and flash-cooling
Initial crystals were grown at room temperature in 1+1 μl hanging drops over reservoirs of 24-well plates containing reservoir solution of 0.1 M Bis-Tris pH 5.5, 0.1 M ammonium acetate, 17% PEG 10000. Data-quality crystals grew with a reservoir solution of 0.1 M Bis-Tris pH 5.5, 0.1 M ammonium acetate, 4–9% PEG 8000 or PEG 10000. The crystals grew as rods up to 0.1 × 0.1 × 0.7 mm3 in size in 2–4 days. Crystals were soaked for 1 min in reservoir solution containing 20% ethylene glycol before flash-cooling in liquid nitrogen. Heavy atom derivatives for MAD experiments were prepared by soaking crystals for 1 min in 3.3 mM K2PtCl4, backsoaking in reservoir solution for 1 min, and then soaking in the cryoprotectant solution as described above. Iron and 2OG containing crystals were obtained by adding 1 mM fresh FeSO4 and 1 mM 2OG to the concentrated protein prior to crystallisation.
Data collection, phasing, and model refinement
The enzyme crystallised in space group I41 with cell parameters of a=b=120.0 Å and c=40.5 Å. A three-wavelength MAD data set was collected at the Advanced Light Source (ALS) beamline 8.3.1 for data from 50.0 to 2.8 Å, using λ=1.07206, 1.07227, and 1.123051. All X-ray intensity data were integrated using HKL2000 and scaled with Scalepack or HKL2000 (Otwinowski and Minor, 1997). Two Pt sites were located, and phases were calculated by the program SOLVE (Terwilliger and Berendzen, 1999). Density modification was carried out using the program RESOLVE (Terwilliger, 2000) and a solvent content of 52%. The electron density map was easily interpretable, and a polypetide model was built using the XtalView package (McRee, 1999). A high-resolution data set was collected on a crystal of hABH3Δ69 in complex with iron and 2OG at the Stanford Synchrotron Radiation Laboratory (SSRL) beamline 11.1 for data from 50.0 to 1.5 Å. Phases for the high-resolution data set were determined by molecular replacement using the model built into the heavy-atom data and observed amplitudes. Difference electron density and omit maps were manually fitted with the XtalView program package (McRee, 1999) and refined in either CNS 1.1 (Brunger et al, 1998) or SHELX-97 (Sheldrick and Schneider, 1997) in iterative cycles. Five percent of the data were used for Rfree calculations (Brunger et al, 1998).
Oxidative demethylation assay
[3H]methylated poly(dA) (Bjelland et al, 1993), as single-stranded substrate or annealed with equal amount of complementary poly(dT) (typically containing 3000 dpm label, specific activity 599 GBq mmol−1), was incubated in a 50 μl reaction mixture for 30 min at 37°C in the presence of various amounts of hABH3 protein (from 0.1 to 50 ρmol enzyme) in the following buffer: 50 mM Tris (pH 7.5), 30 mM KCl, 4 mM dithiothreitol, 100 μg ml−1 BSA, 100 μM 2OG, 100 μM Fe(NH4)2(SO4)2. In some experiments, hABH3 and mutants thereof were also analysed in the presence of [3H]methylated poly(dC). DNA was precipitated by ethanol, and the radioactivity in the supernatant was measured by scintillation counting.
In-gel enzymatic digestion and mass spectrometry analysis
Proteins were excised from 1D Coomassie-blue stained gels (Invitrogen) and digested in-gel with sequencing grade modified trypsin (Promega). Tryptic peptides were concentrated with Poros C18 stage-tips and 1 μl of the peptide solution was mixed with 1 μl α-cyano-4-hydroxycinnamic acid matrix in an anchorship 600-384 plate. Protein digests were analysed by matrix-assisted laser desorption/ionisation on a Bruker Reflex IV time-of-flight (MALDI-TOF) mass spectrometer equipped with a scout 384 ion source (Bruker Daltonics). Theoretical masses of peptides after trypsin digestion were obtained from a MS-digest web-server (http://prospector.ucsf.edu) for comparison.
Coordinates
The atomic coordinates of the hABH3Δ69 complex have been deposited in the Protein Data Bank with the accession number 2iuw.
Supplementary Material
Supplementary Figures and Tables
Acknowledgments
We thank the late EC Seeberg along with A Arvai, DP Barondeau, L Hagen, PB Aslaksen, E Feyzi and especially JL Huffman for helpful discussions, comments and technical advice. This research was supported by the European Union, the Research Council of Norway, the National Programme for Research in Functional Genomics in Norway (FUGE) in the Research Council of Norway, the Norwegian Cancer Association, the Cancer Fund at St Olav's Hospital, Trondheim, the Svanhild and Arne Must Fund for Medical Research to OS, CBV, MMLS, PAA, FD, HEK, and GS. JAT and his crystallographic efforts are supported by a grant from National Institutes of Health and OS in part by a fellowship from the Skaggs Institute of Chemical Biology. We also like to thank the staff at Stanford Synchrotron Laboratory and the Advanced Light Source, especially J Holton at ALS 8.3.1 and SIBYLS, for assistance in X-ray data collection.
Competing interests statement The authors declare that they have no competing financial interests.
References
- Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, Krokan HE (2003) Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421: 859–863 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV (2001) The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol 2: 7.1–7.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett TE, Savva R, Panayotou G, Barlow T, Brown T, Jiricny J, Pearl LH (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell 92: 117–129 [DOI] [PubMed] [Google Scholar]
- Bjelland S, Bjoras M, Seeberg E (1993) Excision of 3-methylguanine from alkylated DNA by 3-methyladenine DNA glycosylase I of Escherichia coli. Nucleic Acids Res 21: 2045–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54 (Part 5): 905–921 [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20: 426–427 [DOI] [PubMed] [Google Scholar]
- Clifton IJ, Doan LX, Sleeman MC, Topf M, Suzuki H, Wilmouth RC, Schofield CJ (2003) Crystal structure of carbapenem synthase (CarC). J Biol Chem 278: 20843–20850 [DOI] [PubMed] [Google Scholar]
- Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA (2004) DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat Struct Mol Biol 11: 714–720 [DOI] [PubMed] [Google Scholar]
- Delaney JC, Essigmann JM (2004) Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci USA 101: 14051–14056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney JC, Smeester L, Wong C, Frick LE, Taghizadeh K, Wishnok JS, Drennan CL, Samson LD, Essigmann JM (2005) AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat Struct Mol Biol 12: 855–860 [DOI] [PubMed] [Google Scholar]
- Dinglay S, Trewick SC, Lindahl T, Sedgwick B (2000) Defective processing of methylated single-stranded DNA by E. coli AlkB mutants. Genes Dev 14: 2097–2105 [PMC free article] [PubMed] [Google Scholar]
- Drabløs F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE (2004) Alkylation damage in DNA and RNA-repair mechanisms and medical significance. DNA Repair (Amst) 3: 1389–1407 [DOI] [PubMed] [Google Scholar]
- Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B (2002) Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci USA 99: 16660–16665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell JM, Purvis A, Khuri S (2004) Cupins: the most functionally diverse protein superfamily? Phytochemistry 65: 7–17 [DOI] [PubMed] [Google Scholar]
- Falnes PO (2004) Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res 32: 6260–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PO, Bjoras M, Aas PA, Sundheim O, Seeberg E (2004) Substrate specificities of bacterial and human AlkB proteins. Nucleic Acids Res 32: 3456–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PO, Johansen RF, Seeberg E (2002) AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419: 178–182 [DOI] [PubMed] [Google Scholar]
- Fu SL, Dean RT (1997) Structural characterization of the products of hydroxyl-radical damage to leucine and their detection on proteins. Biochem J 324 (Part 1): 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson M, Davis M, Luty B, McCammon J (1993) Computation of electrostatic forces on solvated molecules using the Poisson–Boltzmann equation. J Phys Chem 97: 3591–3600 [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Hausinger RP (2004) FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol 39: 21–68 [DOI] [PubMed] [Google Scholar]
- Henshaw TF, Feig M, Hausinger RP (2004) Aberrant activity of the DNA repair enzyme AlkB. J Inorg Biochem 98: 856–861 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374 [DOI] [PubMed] [Google Scholar]
- Huffman JL, Sundheim O, Tainer JA (2005) DNA base damage recognition and removal: new twists and grooves. Mutat Res 577: 55–76 [DOI] [PubMed] [Google Scholar]
- Kataoka H, Yamamoto Y, Sekiguchi M (1983) A new gene (alkB) of Escherichia coli that controls sensitivity to methyl methane sulfonate. J Bacteriol 153: 1301–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivisto P, Duncan T, Lindahl T, Sedgwick B (2003) Minimal methylated substrate and extended substrate range of Escherichia coli AlkB protein, a 1-methyladenine-DNA dioxygenase. J Biol Chem 278: 44348–44354 [DOI] [PubMed] [Google Scholar]
- Koivisto P, Robins P, Lindahl T, Sedgwick B (2004) Demethylation of 3-methylthymine in DNA by bacterial and human DNA dioxygenases. J Biol Chem 279: 40470–40474 [DOI] [PubMed] [Google Scholar]
- Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM (2003) Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics 4: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T (1998) Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell 95: 249–258 [DOI] [PubMed] [Google Scholar]
- Liu A, Ho RY, Que L Jr, Ryle MJ, Phinney BS, Hausinger RP (2001) Alternative reactivity of an alpha-ketoglutarate-dependent iron(II) oxygenase: enzyme self-hydroxylation. J Am Chem Soc 123: 5126–5127 [DOI] [PubMed] [Google Scholar]
- McRee DE (1999) XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol 125: 156–165 [DOI] [PubMed] [Google Scholar]
- Mishina Y, He C (2006) Oxidative dealkylation DNA repair mediated by the mononuclear non-heme iron AlkB proteins. J Inorg Biochem 100: 670–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y, Yang CG, He C (2005) Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins. J Am Chem Soc 127: 14594–14595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (eds.) (1997) Processing of X-ray Diffraction Data Collected in Oscillation Mode. New York: Academic Press [DOI] [PubMed] [Google Scholar]
- Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, Remme J, Falnes PO (2004) AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol Cell 16: 107–116 [DOI] [PubMed] [Google Scholar]
- Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J 17: 5214–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringvoll J, Nordstrand LM, Vågbø CB, Talstad V, Reite K, Aas PA, Lauritzen KH, Liabakk NB, Bjørk A, Doughty RW, Falnes PØ, Krokan HE, Klungland A (2006) Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryle MJ, Liu A, Muthukumaran RB, Ho RY, Koehntop KD, McCracken J, Que L Jr, Hausinger RP (2003) O2- and alpha-ketoglutarate-dependent tyrosyl radical formation in TauD, an alpha-keto acid-dependent non-heme iron dioxygenase. Biochemistry 42: 1854–1862 [DOI] [PubMed] [Google Scholar]
- Said MR, Begley TJ, Oppenheim AV, Lauffenburger DA, Samson LD (2004) Global network analysis of phenotypic effects: protein networks and toxicity modulation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 101: 18006–18011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM, Schneider TR (eds.) (1997) SHELXL: High Resolution Refinement. Academic Press: New York [PubMed] [Google Scholar]
- Singer B, Grunberger D (1983) Molecular Biology of Mutagens and Carcinogens. Plenum Press: New York [Google Scholar]
- Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA (1996) A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature 384: 87–92 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2000) Maximum-likelihood density modification. Acta Crystallogr D 56 (Part 8): 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D 55 (Part 4): 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B (2002) Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419: 174–178 [DOI] [PubMed] [Google Scholar]
- Wibley JE, Waters TR, Haushalter K, Verdine GL, Pearl LH (2003) Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol Cell 11: 1647–1659 [DOI] [PubMed] [Google Scholar]
- Wilmouth RC, Turnbull JJ, Welford RW, Clifton IJ, Prescott AG, Schofield CJ (2002) Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure (Camb) 10: 93–103 [DOI] [PubMed] [Google Scholar]
- Yu B, Edstrom WC, Benach J, Hamuro Y, Weber PC, Gibney BR, Hunt JF (2006) Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature 439: 879–884 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Tables


