Abstract
Plants contain more DICER-LIKE (DCL) enzymes and double-stranded RNA binding (DRB) proteins than other eukaryotes, resulting in increased small RNA network complexities. Analyses of single, double, triple and quadruple dcl mutants exposed DCL1 as a sophisticated enzyme capable of producing both microRNAs (miRNAs) and siRNAs, unlike the three other DCLs, which only produce siRNAs. Depletion of siRNA-specific DCLs results in unbalanced small RNA levels, indicating a redeployment of DCL/DRB complexes. In particular, DCL2 antagonizes the production of miRNAs and siRNAs by DCL1 in certain circumstances and affects development deleteriously in dcl1 dcl4 and dcl1 dcl3 dcl4 mutant plants, whereas dcl1 dcl2 dcl3 dcl4 quadruple mutant plants are viable. We also show that viral siRNAs are produced by DCL4, and that DCL2 can substitute for DCL4 when this latter activity is reduced or inhibited by viruses, pointing to the competitiveness of DCL2. Given the complexity of the small RNA repertoire in plants, the implication of each DCL, in particular DCL2, in the production of small RNAs that have no known function will constitute one of the next challenges.
Keywords: Arabidopsis , DICER, miRNA, RNA, siRNA
Introduction
Small 20–27-nucleotide (nt) RNAs are regulatory RNAs found in most eukaryotes. The genome of Arabidopsis produces at least four classes of endogenous small RNA: microRNAs (miRNAs), natural antisense transcripts siRNAs (nat-siRNAs), heterochromatin siRNAs (hc-siRNAs) and trans-acting siRNAs (ta-siRNAs), and encodes four DICER-LIKE (DCL) RNase III enzymes (for a review, see Vaucheret, 2006a). Like animal miRNAs, plants miRNAs are processed from stem-loop structures of longer primary transcripts through the action of RNase III enzymes (for a review, see Jones-Rhoades et al, 2006). In animals, miRNA precursors are cut by Drosha in the nucleus and Dicer in the cytoplasm to liberate the mature miRNAs. In Arabidopsis, DCL1 has both Drosha and Dicer functions and produces 21-nt miRNAs in the nucleus (Park et al, 2002; Reinhart et al, 2002; Kurihara and Watanabe, 2004). Because miRNAs are crucial for plant development, mutations in the DCL1 gene result in dramatic defects and dcl1 null alleles are embryo-lethal (Schauer et al, 2002). Hypomorphic weak loss-of-function dcl1 alleles, such as dcl1–7, dcl1–8 and dcl1–9, present less severe phenotypes (Schauer et al, 2002) associated with lower contents of miRNAs as compared to wild-type plants (Vaucheret et al, 2006b). ta-siRNAs have only been described in plants. Primary transcripts encoded by the TAS loci are cleaved by specific miRNAs, processed in double-stranded RNA (dsRNA) by the RNA-dependent RNA polymerase RDR6, and sequentially diced by DCL4 into 21-nt ta-siRNAs (Gasciolli et al, 2005; Xie et al, 2005; Yoshikawa et al, 2005). Like miRNAs, ta-siRNAs act in trans on specific target transcripts, for instance to control some aspects of plant development such as leaf morphology (Adenot et al, 2006). nat-siRNAs derives from pairs of natural antisense transcripts (Borsani et al, 2005). A recent paper describes the case of nat-siRNAs produced from two convergent transcripts expressed in different conditions, one constitutively and the other induced by a salt stress (Borsani et al, 2005). When both transcripts are expressed, DCL2 produces a unique 24-nt nat-siRNA that guides the cleavage of the constitutive transcript and the subsequent production of 21-nt nat-siRNA by DCL1. Like ta-siRNAs, the production of nat-siRNAs is dependent of RDR6 and SGS3, but nat-siRNAs only act in cis. Finally, the large majority of plant endogenous small RNAs derive from transposons, repeated sequences and heterochromatin, and are produced by RDR2 and DCL3, which mostly generates 24-nt siRNA species (Xie et al, 2004).
Dicer and Drosha enzymes are associated with double-stranded RNA-binding (DRB) proteins that serve as essential cofactors. In Caenorhabditis elegans, the unique DICER, DCR-1, is found in a complex that contains rde-4, which encodes a DRB protein required for RNAi (Tabara et al, 2002). Drosophila has one Drosha and two DICER proteins, DCR-1 and DCR-2, involved in various aspect of small RNA biogenesis. Drosha, DCR-1 and DCR-2 are associated with different DRBs, namely Pasha, Loquacious and R2D2 (for reviews, see Hammond, 2005; Kavi et al, 2005). R2D2 binds siRNAs produced by DCR-2 and directs their strand-specific incorporation into the RISC complex. Loquacious and Pasha might have similar roles, but specific of the fly miRNA pathway. Arabidopsis encodes five DRB proteins that contain two DRB motifs in their N-terminal part (Hiraguri et al, 2005). DCL1 interacts with HYPONASTIC LEAVES 1 (HYL1), whereas DCL4 interacts with DRB4 (Hiraguri et al, 2005). Consistently, hyl1 and drb4 mutants exhibit phenotypes similar to dcl1 and dcl4 mutants, and are affected in the production of miRNAs and ta-siRNAs, respectively (Han et al, 2004; Vazquez et al, 2004; Adenot et al, 2006).
In addition to their roles in generating endogenous siRNAs, plant DCL enzymes also participate in defense mechanisms against invading nucleic acids such as viruses or transgenes. DCL4 produces siRNAs from inverted-repeat transgenes (Dunoyer et al, 2005), DCL2 is involved in the production of viral siRNAs derived from the Turnip crinkle virus (TCV) or the Cabbage leaf curl virus (CaLCuV) and DCL3 produces CaLCuV-derived siRNAs (Xie et al, 2004; Akbergenov et al, 2006). Many viruses are counteracting defense mechanisms by suppressing silencing activities by means of various strategies (for a review, see MacDiarmid, 2005). For example, the tombusvirus P19 protein binds siRNAs and sequestrate them to protect viral RNA, the potyvirus P1/HC-Pro protein interacts with plant partners involved in the regulation of RNA silencing, and the potexvirus P25 protein prevents cell-to-cell spreading of silencing signals. Because DCL enzymes are key components of RNA silencing pathways, it is tempting to speculate that some viral suppressors could inhibit their functions. Supporting this hypothesis, P1/HC-Pro was found to reduce the processing of double-stranded RNAs (Mallory et al, 2002; Dunoyer et al, 2004). Suppressors like potexvirus P25 or carmovirus P38 impact only siRNA accumulation, whereas others like furovirus P15, tombusvirus P19, tymovirus P69 or geminivirus AC4 impair both miRNA and siRNA accumulation (Chapman et al, 2004; Chen et al, 2004; Dunoyer et al, 2004; Chellappan et al, 2005; Mlotshwa et al, 2005). Only AC4 has been shown to bind miRNAs directly (Chellappan et al, 2005). Because DCL1 produces miRNAs, inhibition of the miRNA pathway by viruses could result from the direct action of suppressors on DCL1. Alternatively, inhibition of the miRNA pathway could be a side effect of virus infection. Supporting this hypothesis, a recent report showed that the Red clover necrotic mosaic virus (RCNMV) inhibits miRNA biosynthesis but requires DCL1 for its replication (Takeda et al, 2005). As a result, dcl1 mutants are less susceptible than wild-type plants to the infection by RCNMV and do not accumulate viral RNA. It is likely that DCL1 is required by the virus as a host factor to promote its own replication or movement. Whether DCL1 or other DCLs are direct targets of virus counter-defenses remains to be further examined.
Although Dicer enzymes are implicated in specific aspects of silencing, increasing evidences are revealing partially compensatory functions among them. For instance, the two Dicers of Drosophila are involved in miRNA biogenesis and siRNA production, respectively, although some functional redundancy have been observed between the two proteins (Lee et al, 2004). In the filamentous fungus Neurospora crassa, both Dicers are involved in the quelling pathway (Catalanotto et al, 2004). Our previous work pointed toward some redundancies among the four Arabidopsis DCL enzymes (Gasciolli et al, 2005). Indeed, DCL2 and DCL3 can produce RDR6-dependent siRNAs in the absence of DCL4, and some RDR2-dependent siRNAs were produced by DCL2 and DCL4 in the absence of DCL3. Consistent with partial redundancies, double mutants such as dcl3 dcl4 and dcl2 dcl3 exhibit stochastic developmental phenotypes after three generations and dcl1 dcl3 and dcl1 dcl4 had phenotypes more severe than dcl1 mutants. Here, we investigate further the functions of DCL enzymes and their redundancies by examining all double, triple and quadruple dcl mutants. We show that DCL1 is the exclusive producer of miRNAs, whereas RDR6- and RDR2-dependent endogenous siRNAs can be produced by DCL1, DCL2, DCL3 or DCL4. However, in the absence of DCL3 and DCL4, DCL2 antagonizes siRNA production by DCL1. In addition, DCL2 partially antagonizes the production of miRNAs by DCL1 and has deleterious effects on development in dcl1 dcl4 and dcl1 dcl3 dcl4 mutants, whereas the dcl1 dcl2 dcl3 dcl4 mutant is viable. In addition, we reveal the role of DCL4 in generating virus-derived siRNAs. DCL4 is the primary producer of Cucumber mosaic virus (CMV)-derived siRNAs and also produces TCV-siRNAs. DCL2 appears to be the major producer of TCV-derived siRNAs, likely because TCV suppresses the activity of DCL4.
Results and discussion
miRNAs are produced exclusively by DCL1
miRNAs regulate the expression of multigene families involved in key steps of plant development (Jones-Rhoades et al, 2006). In Arabidopsis, miRNAs are processed from double-stranded stem-loop precursors by DCL1. Null dcl1 alleles are embryo-lethal, whereas partial loss-of-function dcl1 mutant plants exhibit less severe developmental defects (Schauer et al, 2002). In the hypomorphic allele dcl1-9, most miRNAs are below detectable levels or accumulate at low levels compared with wild-type plants with the exception of miR165/166 and miR168, which accumulate at near wild-type levels in both leaves (Figure 1A) and flowers (Figure 1B), pointing to the possibility that some miRNAs could be produced by other DICER-LIKE activities in dcl1-9 mutant plants (i.e. DCL2–DCL4). To analyze the contribution of each DCL activity in miRNA production, we generated triple dcl2 dcl3 dcl4 and quadruple dcl1 dcl2 dcl3 dcl4 mutants. In triple dcl2 dcl3 dcl4 mutants, all tested miRNAs accumulated at levels similar to wild-type plants (Figure 1B), and these miRNAs accumulated at similarly reduced levels in dcl1 and dcl1 dcl2 dcl3 dcl4 mutants (Figure 1B). These results rule out the possibility that DCL2, DCL3 or DCL4 produces miRNAs. They also are consistent with the hypothesis that miR165/166 and miR168, which, unlike other miRNAs, are inefficiently loaded onto AGO1 in wild-type plants, accumulate at wild-type levels in dcl1-9 because the reduced production of miRNAs but not AGO1 in dcl1-9 allows a better loading of these miRNAs (Vaucheret et al, 2006b).
Figure 1.
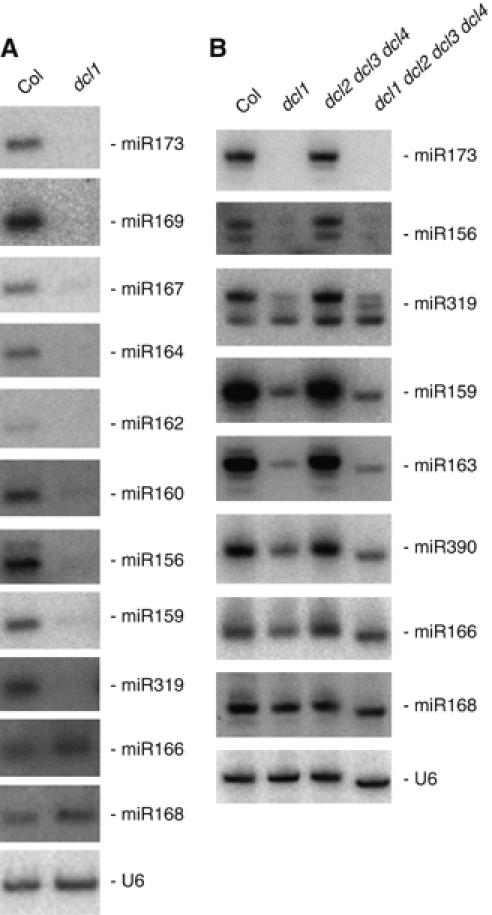
DCL1 is the only DCL that produces miRNAs. RNA gel blots analysis of 10 μg total RNA prepared from rosette leaves of wild-type Col and dcl1 (A), and inflorescences of wild-type Col, dcl1, dcl2 dcl3 dcl4 triple mutants and dcl1 dcl2 dcl3 dcl4 quadruple mutant (B). The blots were successively probed with DNAs complementary to miRNAs and U6, which served as a loading control.
DCL1 can produce RDR6-dependent endogenous siRNAs
Previous analysis of double dcl mutants revealed that DCL2, DCL3 and DCL4 are partially redundant for the production of ta-siRNAs (Gasciolli et al, 2005; Xie et al, 2005). In the absence of DCL4, the accumulation of RDR6-dependent 21-nt siRNAs derived from the TAS loci was reduced, and 22–24-nt siRNAs were produced (see Figure 2B in Gasciolli et al, 2005). Accumulation of 21–24-nt TAS-derived siRNAs was reduced further in dcl3 dcl4 double mutants, indicating that DCL3 can substitute for DCL4 and produce ta-siRNAs (Figure 2). We hypothesized that the production of RDR6-dependent siRNAs also would be impaired in the dcl2 dcl3 dcl4 triple mutant. However, 21-nt siRNAs originating from the three TAS loci were detected in dcl2 dcl3 dcl4 but not in dcl1 dcl2 dcl3 dcl4 plants (Figure 2), indicating that in the absence of DCL2, DCL3 and DCL4, wild-type DCL1 can produce RDR6-dependent 21-nt siRNAs, but the partial-loss-of-function dcl1 mutant does not retain this ability. These results are slightly different from those recently published by Henderson et al (2006), which did not detect ta-siRNAs in a dcl2 dcl3 dcl4 triple mutant. However, these authors generated a triple mutant using dcl2-1, which appeared to behave as a weak allele, transiently impairing the accumulation of siRNAs from TCV (Xie et al, 2004). In contrast, our triple mutant was generated using dcl2–4, which behaves as a strong allele with regards to viral siRNAs (see below).
Figure 2.
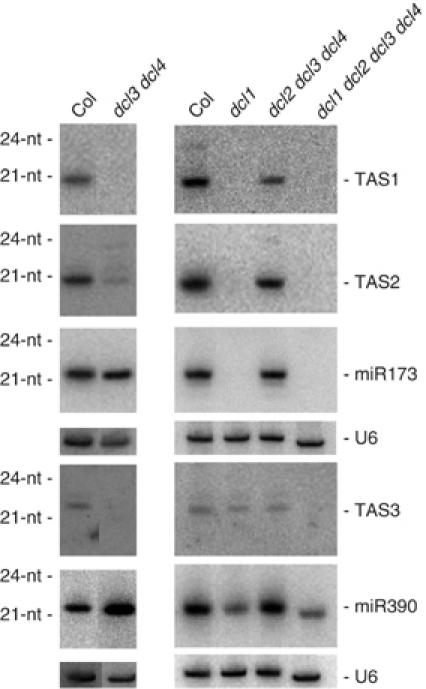
Analysis of RDR6-dependent endogenous ta-siRNAs content in dcl mutants. RNA gel blots analysis of 10 μg total RNA prepared from inflorescence of wild-type Col, and dcl1, dcl3 dcl4, dcl2 dcl3 dcl4, dcl1 dcl2 dcl3 dcl4 mutants. The blots were probed with DNAs complementary to ta-siRNAs, miRNAs and U6, which served as a loading control.
dcl2 and dcl3 single mutants, and dcl2 dcl3 and dcl2 dcl4 double mutants develop normally, whereas dcl4 and dcl3 dcl4 mutants have a mild and severe rdr6 leaf phenotype, respectively, owing to reduced ta-siRNAs accumulation, including TAS3 ta-siRNAs, which are required for proper leaf development (Gasciolli et al, 2005; Adenot et al, 2006). Consistent with the detection of TAS-derived 21-nt siRNAs, dcl2 dcl3 dcl4 plants developed leaves that have less severe rdr6 leaf phenotype than dcl4 or dcl3 dcl4 mutant leaves (Figure 3), indicating that the 21-nt ta-siRNAs produced by DCL1 in the absence of DCL2, DCL3 and DCL4 are, at least partially, functional.
Figure 3.

Rosette leaf morphology of single, double and triple dcl mutants involving dcl4. Pictures of 21-day-old dcl4, dcl2 dcl4, dcl3 dcl4, dcl2 dcl3 dcl4 mutants compared to the sgs3-1 allele and wild-type Col plants. The leaf phenotype was quantified by measuring the average length and width of the fifth rosette leaves of 21-day-old plants, which is indicative of the juvenile-to-adult transition. Numbers under the photographs represent the average length divided by width and the corresponding s.d.'s. Measurements were taken at the widest and longest point of each leaf. Six individual plants were measured for each class.
DCL1 is incapable to produce most RDR2-dependent endogenous siRNAs
Previous analysis of double dcl mutants revealed that DCL2, DCL3 and DCL4 activities are partially redundant for the production of RDR2-dependent 24-nt siRNAs (Gasciolli et al, 2005). In the absence of DCL3, the accumulation of RDR2-dependent 24-nt siRNA02 was abolished, and 21–23-nt siRNAs were detected (Figure 4). 22–23-nt siRNAs were not detected in dcl2 dcl3, whereas 21-nt siRNAs were not detected in dcl3 dcl4 double mutants, indicating that DCL2 and DCL4 can substitute for DCL3 and produce 22–23-nt and 21-nt species, respectively (Figure 4). Although no siRNAs were predicted to accumulate in the dcl2 dcl3 dcl4 triple mutant, 21-nt siRNAs originating from the siRNA02 locus were detected (Figure 4). These siRNAs accumulated at very low level compared with wild-type plants and were not detected in dcl1 dcl2 dcl3 dcl4 plants (Figure 4), indicating that in the absence of DCL2, DCL3 and DCL4, DCL1 can produce some RDR2-dependent 21-nt siRNAs, although very inefficiently.
Figure 4.
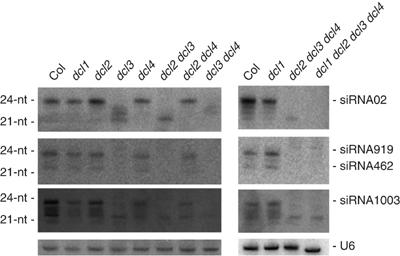
Analysis of RDR2-dependent endogenous siRNAs content in dcl mutants. RNA gel blots analysis of 10 μg total RNA prepared from inflorescence of wild-type Col, and dcl1, dcl2, dcl3, dcl4, dcl2 dcl3, dcl2 dcl4, dcl3 dcl4, dcl2 dcl3 dcl4, dcl1 dcl2 dcl3 dcl4. The blots were probed with DNAs complementary to siRNAs and U6, which served as a loading control.
siRNA02 represents one class of RDR2-dependent siRNA that also depends on DCL3, NRPD1a and NRPD2, but not AGO4, HEN1 and NRPD1b for proper accumulation. hc-siRNAs, another class of RDR2-dependent siRNAs, depend on all six of these proteins for proper accumulation (Xie et al, 2004; Kanno et al, 2005; Pontier et al, 2005). To determine if DCL1 also can produce hc-siRNAs, we examined the accumulation of three hc-siRNAs: siRNA1003, which derives from 5S rDNA repeats, and siRNA462 and siRNA919, which both derive from the SINE retroposon AtSN2 (Gustafson et al, 2005). Results with siRNA1003 were inconclusive because a 21-nt RNA was detected in every genotype (Figure 4). Whether this RNA corresponds to a nonspecific crosshybridization signal or a 21-nt species that is produced in the dcl1 dcl2 dcl3 dcl4 quadruple mutants, owing to residual DCL1 activity, is not known. siRNA 462 is 22-nt long and is internal to siRNA919, which is 24-nt long. Both species accumulated at similar levels in wild-type plants, and were reduced in dcl3 mutants, indicating that they both are produced by DCL3 (Figure 4). There was no 21-nt RNA species detected in any mutant background, suggesting that DCL1 cannot produce the most abundant class of RDR2-dependent siRNA.
In the absence of DCL3 and DCL4, DCL2 antagonizes the production of RDR6- and RDR2-dependent endogenous siRNAs by DCL1
Short-interfering RNAs 21-nt in length deriving from the TAS loci were reduced to low levels in dcl3 dcl4 plants, whereas they accumulated in dcl2 dcl3 dcl4 at levels comparable to wild-type plants (Figure 2), suggesting that in the absence of DCL3 and DCL4, DCL2 antagonized the production of RDR6-dependent siRNAs by DCL1. Similarly, 21-nt siRNAs deriving from the siRNA02 locus were detected in dcl2 dcl3 dcl4 mutants, but were absent in dcl3 dcl4 and dcl1 dcl2 dcl3 dcl4 mutants, although at low levels (Figure 4). Therefore, these 21-nt siRNAs likely result from DCL1 activity and the absence of 21-nt siRNAs in dcl3 dcl4 is likely due to the antagonistic effect of DCL2 on DCL1.
One possible explanation for this antagonism is a competition between DCL1 and DCL2 for the same cofactors or substrates. It is possible that in the dcl3 dcl4 mutant, DCL2 has a higher affinity than DCL1 for cofactors or substrates that normally bind to DCL3 and DCL4. Therefore, DCL1 would not be able to bind these cofactors or substrates in dcl3 dcl4 mutants, and would not produce RDR2- or RDR6-dependent siRNAs. An explanation for the absence of ta-siRNAs in the dcl2 dcl3 dcl4 triple mutant generated by Henderson et al (2006) could be that the dcl2-1 allele present in this triple mutant produces a stable truncated protein that lacks functional DCL2 activity but is able to compete with DCL1 for cofactors or substrates that normally bind to DCL4, therefore antagonizing DCL1 as efficiently as a wild-type DCL2 protein. This dcl2 dcl3 dcl4 triple mutant would therefore behave like a dcl3 dcl4 double mutant with regards to ta-siRNAs.
DCL2 also partially antagonizes the production of miRNAs by DCL1
Mutations in DCL2 have been reported not to affect miRNA accumulation. However, only a few miRNAs have been analyzed, and only RNAs extracted from floral tissues have been examined (Xie et al, 2004). To determine if DCL2 only antagonizes the production of RDR2- and RDR6-dependent siRNAs by DCL1, or if it also impacts the production of miRNAs, we examined the accumulation of several miRNAs in the leaves of single dcl mutants (Figure 5A). The accumulation of miR160, miR164, miR168 and miR173 was unchanged in dcl2. However, miR156, miR159 miR165 and miR319 levels were increased, and miR162 and miR167 level also were increased, although to a lesser extent. A subtle increase in the accumulation of the same miRNAs also was observed in dcl4 mutants (Figure 5A). This contrasts with the results previously observed in flowers where no changes were reported, which was confirmed in our analyses (Figure 5B), but is consistent with a varying dependency on DRB proteins in leaves and flowers. Indeed, the accumulation of TAS3 ta-siRNAs but not TAS1 and TAS2 ta-siRNAs was impaired in drb4 mutant leaves, whereas the accumulation of TAS1 and TAS2 ta-siRNAs but not TAS3 ta-siRNAs was impaired in drb4 flowers (Adenot et al, 2006). These results suggest that DCL2, and to a lesser extent DCL4, could compete with DCL1 for binding to HYL1 in leaves, thus limiting the production of some miRNAs, consistent with the observation that an increase in HYL1 production can increase the accumulation of miRNAs (Han et al, 2004).
Figure 5.
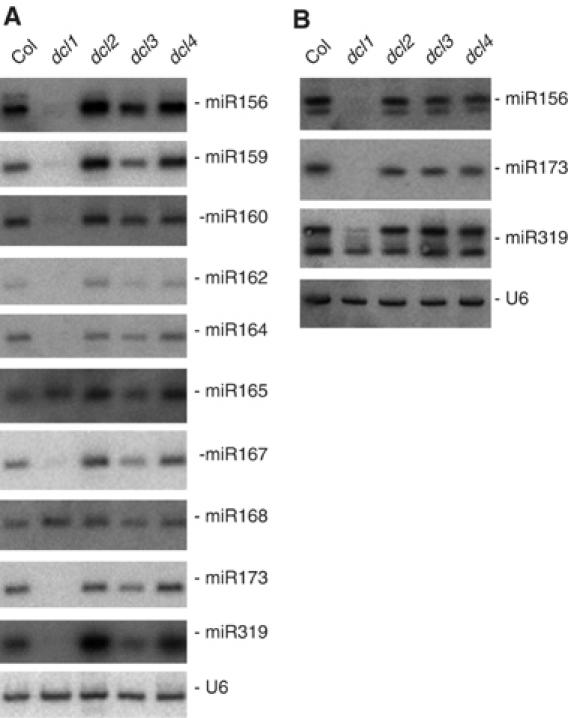
Production of miRNAs in single dcl mutants. RNA gel blot analysis of 10 μg total RNA prepared from rosette leaves (A) or inflorescences (B) of wild-type Col, and dcl1, dcl2, dcl3 and dcl4 single mutants. The blots were successively probed with DNAs complementary to miRNAs and U6, which served as a loading control.
DCL2 has deleterious effects on development in the absence of DCL1 and DCL4
DCL1 is required for plant development as exemplified by the dramatic developmental defects of dcl1 alleles (Schauer et al, 2002). In contrast, dcl2 and dcl3 resemble wild-type plants, and dcl4 has a subtle rdr6-like phenotype due to reduced ta-siRNA accumulation (Gasciolli et al, 2005). All combinations of dcl mutations involving dcl1 were generated to further analyze interactions among the four DCLs. dcl1 dcl2 and dcl1 dcl3 double mutants, dcl1 dcl2 dcl3 and dcl1 dcl2 dcl4 triple mutants and dcl1 dcl2 dcl3 dcl4 quadruple mutants resembled the single dcl1 mutant (Figure 6), although dcl1 dcl3 and dcl1 dcl2 dcl3 flowered later than dcl1 and as such produced more leaves (data not shown). In contrast, dcl1 dcl4 double mutant and dcl1 dcl3 dcl4 triple mutant plants exhibited a severe phenotype associated with a high rate of early mortality (Figure 6). Because dcl1 dcl2 dcl4 and dcl1 dcl2 dcl3 dcl4 mutants did not show such mortality, it is unlikely that the mortality is due to the simultaneous absence of DCL1 and DCL4. It is more likely that DCL2 activity interferes with plant development, when DCL1 and DCL4 are disrupted.
Figure 6.

Phenotype of all combinations of dcl mutations involving dcl1. 65-day-old wild-type dcl1, dcl1 dcl2, dcl1 dcl3, dcl1 dcl4, dcl1 dcl2 dcl3, dcl1 dcl2 dcl4, dcl1 dcl3 dcl4, dcl1 dcl2 dcl3 dcl4 mutants were sowed in vitro, grown for 20 days under standard conditions, and transferred to soil. Photographs were taken 45 days later.
Until recently, it was unclear whether DCL2 produced endogenous siRNAs or if its role was restricted to virus-induced gene silencing (Xie et al, 2004; Akbergenov et al, 2006). A recent report showed that DCL2 processes endogenous siRNAs that derives from pairs of natural cis-antisense transcripts. In the case described in this report, nat-siRNAs are produced in response to salt stress, and the authors suggested that DCL2 could be involved in the production of other stress-related nat-siRNAs (Borsani et al, 2005). Our results point toward the deleterious role of DCL2 when DCL1 and DCL4 are impaired. One explanation could be that DCL2 produces aberrant nat-siRNAs in this context by interacting with DRBs that normally bind to DCL1 or DCL4. Plants would then respond inefficiently to environmental changes resulting in general developmental defects that mimic stressed plants. Supporting this hypothesis, we observed that the mortality rate of dcl1 dcl4 and dcl1 dcl3 dcl4 plants strongly depends on the conditions of growth. Indeed, mortality rates ranged between 20% in growth chambers that have tightly controlled conditions of light, temperature and humidity, and 100% in greenhouses that are more exposed to fluctuating conditions, including heat during the day and cold at night.
DCL4 is the primary producer of CMV-derived siRNAs
In addition to producing endogenous siRNAs, DCL enzymes produce exogenous viral- and transgene-derived siRNAs. The biogenesis of viral siRNAs is not fully understood. In Arabidopsis, DCL2 has been implicated in the production of TCV and CaLCuV 22-nt siRNAs (Xie et al, 2004; Akbergenov et al, 2006). In addition, DCL3 has been implicated in the production of 24-nt CaLCuV siRNAs (Akbergenov et al, 2006). To gain additional insight into the production of viral siRNAs by the various DCLs, we inoculated all combinations of dcl mutants with CMV. Wild-type plants are relatively tolerant to CMV infection, in that CMV-infected plants flower and set seeds normally. In contrast, ago1, hen1, rdr6 and sgs3 mutants are hypersusceptible to CMV infection and die prematurely, suggesting that CMV RNA levels are kept in check in wild-type plants by an siRNA-based immune system (Mourrain et al, 2000). In CMV-inoculated wild-type plants and dcl1, dcl2 and dcl3 mutants, CMV-siRNAs accumulated as 21-nt species (Figure 7A). In dcl4, the accumulation of 21-nt CMV-siRNAs was abolished, but 22–24-nt siRNA species were detected. The accumulation of 22-nt siRNAs was abolished in dcl2 dcl4 mutants, whereas the accumulation of 24-nt siRNAs was reduced in dcl3 dcl4 mutants (Figure 7A), indicating that in the absence of DCL4, these two species are produced by DCL2 and DCL3, respectively. Neither 22- or 24-nt siRNAs accumulated in dcl2 dcl3 dcl4, but a low level of 21-nt siRNAs was detected (Figure 7A). These results indicate that CMV siRNAs mostly behave similar to ta-siRNAs, that is they are produced by DCL4 in wild-type plants, or DCL2 and DCL3 in dcl4 mutants. Very low levels of 21-nt CMV-siRNAs were detected in dcl2 dcl3 dcl4 mutant plants, suggesting that DCL1 could produce 21-nt CMV-siRNAs in the absence of DCL2, DCL3 and DCL4, although much less efficiently than ta-siRNAs (compare Figures 2 and 7A). However, the production of 21-nt CMV-siRNAs was not abolished in dcl1 dcl2 dcl3 dcl4 mutants, suggesting that these siRNAs are DCL1-derived products that behave like miR165/166 and miR168 (Figure 1) or that they are produced by some of the six non-DCL RNAseIII enzymes encoded by the Arabidopsis genome.
Figure 7.
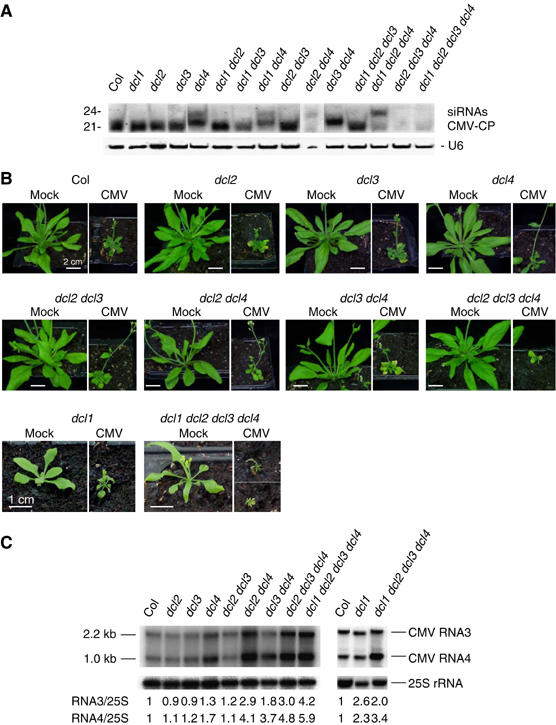
DCL4 is the primary producer of CMV-derived siRNAs. (A) RNA gel blot analysis of 15 μg total RNA prepared from rosette leaves of wild-type Col, dcl1, dcl2, dcl3 and dcl4 single mutants, dcl1 dcl2, dcl1 dcl3, dcl1 dcl4, dcl2 dcl3, dcl2 dcl4, dcl3 dcl4 double mutants, dcl1 dcl2 dcl3, dcl1 dcl2 dcl4, dcl2 dcl3 dcl4 triple mutants and dcl1 dcl2 dcl3 dcl4 quadruple mutant infected with CMV. Plants were grown for 10 days in vitro, transferred to soil, inoculated the day after, and RNAs were extracted from pools of 2–10 plants, 21 days after inoculation. The blots were probed with antisense radiolabeled RNA probes transcripted in vitro complementary to the region coding for the coat protein of CMV. U6 served as a loading control. (B) 31-day-old wild-type dcl2, dcl3, dcl4, dcl2 dcl3, dcl2 dcl4, dcl3 dcl4, dcl2 dcl3 dcl4, dcl1, dcl1 dcl2 dcl3 dcl4 mutants were sowed in vitro, grown for 10 days under standard conditions, transferred to soil, and inoculated the day after with CMV. Photographs were taken 21 days later. (C) High molecular weight RNA blot analysis of 5 μg total RNA prepared from wild-type Col, dcl1, dcl2, dcl3 and dcl4 single mutants, dcl2 dcl3, dcl2 dcl4 and dcl3 dcl4 double mutants, dcl2 dcl3 dcl4 triple mutants and dcl1 dcl2 dcl3 dcl4 quadruple mutant infected with CMV. Total RNA was extracted from a pool of 2–10 CMV-infected plants 21 days after inoculation. This experiment was repeated three times and one representative experiment is shown. Blots were probed with radiolabeled DNAs complementary to the region coding for the coat protein of CMV. 25S RNA served as a loading control.
Decreased levels of CMV-siRNAs correlated with increased levels of CMV RNAs and increased developmental defects and mortality (Figure 7B). Indeed, dcl4 mutants only showed a slight increase in the accumulation of CMV RNAs (Figure 7C), suggesting that 22-nt CMV siRNAs produced by DCL2 in the dcl4 mutant are, at least partially, functional. In contrast, dcl2 dcl4, dcl2 dcl3 dcl4 and dcl1 dcl2 dcl3 dcl4 mutants strongly overaccumulated CMV RNAs (Figure 7C), similar to ago1, hen1, rdr6 and sgs3 mutants (Mourrain et al, 2000; Morel et al, 2002; Boutet et al, 2003), demonstrating the hypothesis that PTGS acts as an antiviral defense to reduce CMV RNA levels in wild-type plants (Mourrain et al, 2000). These results also suggest that, unlike RCNMV, whose replication requires DCL1 (Takeda et al, 2005), CMV replication does not rely on a functional DCL1 (Figure 7C). They also suggest that the 21-nt CMV siRNAs produced by DCL1 or other RNaseIII enzymes in dcl2 dcl3 dcl4 and dcl1 dcl2 dcl3 dcl4 mutant plants are not functional or are not present in sufficient amount to control the proliferation of CMV.
DCL4 also produces TCV siRNAs but likely is inhibited by TCV
Analysis of TCV siRNAs in dcl1, dcl2 and dcl3 single mutants previously suggested that DCL2 was involved in TCV siRNAs production (Xie et al, 2004). We extended this analysis to the single dcl4 mutant and observed that 22-nt TCV siRNAs accumulated at similar levels in wild-type plants and dcl1, dcl3 and dcl4 mutants, whereas their accumulation was reduced in dcl2 mutants (Figure 8A). However, a low amount of 20–21nt TCV-siRNAs was detected in dcl2. Analysis of double dcl mutants revealed that the accumulation of these 20–21-nt TCV siRNAs was abolished in dcl2 dcl4 but not dcl1 dcl2 or dcl2 dcl3, indicating that they are made by DCL4 in the absence of DCL2. Analysis of triple and quadruple dcl mutants confirmed that neither DCL1 nor DCL3 were able to produce TCV siRNAs (Figure 8A).
Figure 8.
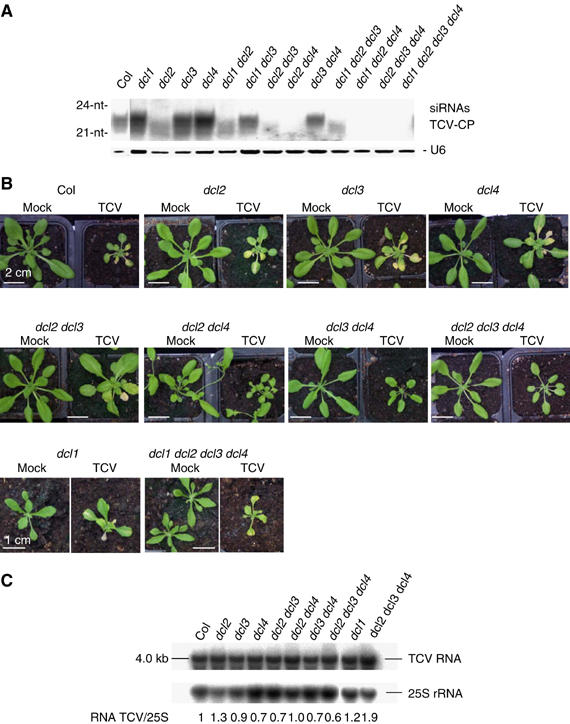
DCL4 and DCL2 produce TCV siRNAs. (A) RNA gel blot analysis of 15 μg total RNA prepared from rosette leaves of wild-type Col, dcl1, dcl2, dcl3 and dcl4 single mutants, dcl1 dcl2, dcl1 dcl3, dcl2 dcl3, dcl2 dcl4, dcl3 dcl4 double mutants, dcl1 dcl2 dcl3, dcl1 dcl2 dcl4, dcl2 dcl3 dcl4 triple mutants and dcl1 dcl2 dcl3 dcl4 quadruple mutant infected with TCV. Plants were grown for 10 days in vitro, transferred to soil, inoculated the day after, and RNAs were extracted from pools of 2–10 plants, 21 days after inoculation. The blots were probed with antisense radiolabeled RNAs probes transcripted in vitro complementary to the region coding for the coat protein of TCV. U6 served as a loading control. (B) 31-day-old wild-type dcl2, dcl3, dcl4, dcl2 dcl3, dcl2 dcl4, dcl3 dcl4, dcl2 dcl3 dcl4, dcl1, dcl1 dcl2 dcl3 dcl4 mutants were sowed in vitro, grown for 10 days under standard conditions, transferred to soil, and inoculated the day after with TCV. Photographs were taken 21 days later. (C) High molecular weight RNA blot analysis of 5 μg total RNA prepared from wild-type Col, dcl1, dcl2, dcl3 and dcl4 single mutants, dcl2 dcl3, dcl2 dcl4 and dcl3 dcl4 double mutants, dcl2 dcl3 dcl4 triple mutants and dcl1 dcl2 dcl3 dcl4 quadruple mutant infected with TCV. Total RNA was extracted from a pool of 2–10 TCV-infected plants 21 days after inoculation. This experiment was repeated three times and one representative experiment is shown. Blots were probed with radiolabeled DNAs complementary to the region coding for the coat protein of TCV. 25S RNA served as a loading control.
These results suggested that DCL2 and DCL4 were the major and minor contributors of TCV siRNAs, respectively, whereas it was the opposite for CMV siRNAs (DCL4 is the major contributor followed by DCL2). However, these results may be interpreted differently considering the different suppressor activities of TCV and CMV. Indeed, TCV encodes a strong suppressor of both S-PTGS and IR-PTGS in Arabidopsis (Béclin et al, 2002; Qu et al, 2003), whereas CMV encodes a weak suppressor of both S-PTGS and IR-PTGS (Mourrain et al, 2000; Béclin et al, 2002). In addition, wild-type plants can tolerate CMV infection, whereas they are hypersusceptible to TCV infection, presumably because TCV is able to inhibit the siRNA-based plant defenses more effectively (Mourrain et al, 2000). Consistently, wild-type plants and dcl mutants were equally susceptible to TCV infection (Figure 8B) and accumulated similar amounts of TCV RNA (Figure 8C), indicating that the siRNA-based plant antiviral defense is ineffective against TCV. We hypothesized that DCL2 could appear to produce TCV siRNAs because DCL4, which also produces TCV-siRNAs, is inhibited by TCV and not because DCL2 has a stronger affinity than DCL4 for TCV dsRNA. To test this hypothesis, we examined the accumulation of DCL4-dependent ta-siRNAs in mock-, CMV- and TCV-inoculated plants (Figure 9). Both TAS1 and TAS2 ta-siRNAs accumulated at low levels in TCV-inoculated plants, whereas they accumulated at similar levels in mock- and CMV-inoculated plants. In TCV-infected plants, we observed a downward shift in mobility for TAS1- and TAS2-derived ta-siRNAs, similar to the mobility shift observed for miRNAs in transgenic plants expressing P19 (Dunoyer et al, 2004). Because the accumulation of miR173, which is required to produce TAS1 and TAS2 ta-siRNAs, was unchanged in TCV-inoculated plants (Figure 9), it is likely that TCV inhibits DCL4 and not DCL1. Therefore, we propose that DCL4 can produce both CMV- and TCV-siRNAs, but that TCV inhibits DCL4 to counteract the plant siRNA-based defense and establish a successful infection. Whether all viral siRNAs also are produced primarily by DCL4 but appear to be produced by other DCLs that substitute to DCL4 due to viral suppressing activity remains to be determined.
Figure 9.
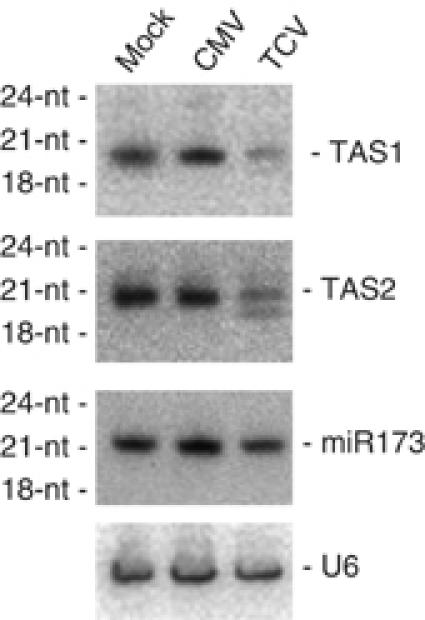
Production of ta-siRNAs in plants infected by TCV or CMV. RNA gel blot analysis of 10 μg total RNA prepared from 1-month-old rosette leaves of wild-type Col mock-inoculated or inoculated with CMV or TCV. RNAs were extracted from pools of 2–10 plants, 21 days after inoculation. The blots were probed with DNAs complementary to ta-siRNAs and miRNAs, and U6, which served as a loading control.
Concluding remarks
In worms, a unique Dicer acts as a core component of all RNAi pathways, and produces different types of small RNAs through its association with distinct DRB cofactors (Duchaine et al, 2006; Lee et al, 2006). These small RNAs can be amplified by different RNA-dependent RNA polymerases and loaded into different Argonaute proteins. Mutation in worm Dicer impacts the accumulation of all types of small RNAs, whereas mutation in specific components can lead to either specific depletion of one type of small RNAs or unbalanced small RNA profiles. For example, mutations in worm RRF-3 or ERI-1 result in decreased levels of siRNAs and tncRNAs, as well as increased levels of miRNAs and exogenous siRNAs, likely because of the redeployment of a limiting core component that is ordinarily shared between RRF-3, ERI-1 and specific cofactors of the miRNA and exogenous siRNA pathways (Duchaine et al, 2006; Lee et al, 2006). Our analyses of Arabidopsis single, double, triple and quadruple dcl mutants point to similar conclusions. DCL1 is able to produce miRNAs and siRNAs, whereas the three other DCLs only produce siRNAs. DCL2, DCL3 and DCL4 exhibit specificity, in that they ordinarily produce different types and sizes of siRNAs, presumably owing to the association of each DCL with a different DRB. Depletion of siRNA-related DCLs usually results in the production of siRNAs of aberrant sizes, indicating a possible redeployment of DRBs towards the available DCLs. The amount of small RNAs produced by a given DCL also can be perturbed when one or more DCLs are missing. In particular, DCL2 antagonizes the production of RDR6- and some RDR2 siRNAs by DCL1 when DCL3 and DCL4 are missing. DCL2 also appears to have deleterious effects during development when DCL1 and DCL4 are reduced. In light of a recent report showing the importance of DCL2 in stress-related pathways (Borsani et al, 2005), our results suggest that DCL2 may take on additional roles when other DCLs are missing. DCL2 partially antagonizes the production of miRNAs by DCL1, at least in leaf, and fully substitutes for DCL4 to produce viral siRNAs when DCL4 is missing or is inhibited by viruses, pointing to the competitiveness and adaptability of DCL2. Given the high number and diversity of small RNAs in plants (Lu et al, 2005), and newly revealed role of DCL2 (Borsani et al, 2005), one may anticipate additional roles for small RNAs and DCL2 in the control of adaptive stress responses.
Materials and methods
Plant materials
dcl1-9 (Jacobsen et al, 1999) was back-crossed five times from Ler to Col. dcl2-4 (FLAG_451B03) and dcl4-1 (FLAG_330A04) were back-crossed two times from Ws to Col. dcl3-1 (Xie et al, 2004) corresponds to the SALK_005512 line in Col. Double dcl mutants have been described before (Gasciolli et al, 2005). Triple and quadruple dcl mutants were generated as follows: a heterozygous dcl1-9/DCL1 plant was crossed to a homozygous dcl4-1/dcl4-1 plant, and a homozygous dcl2-4/dcl2-4 plant was crossed to a homozygous dcl3-1/dcl3-1 plant. The resulting double heterozygous plants were selected and crossed to each other. The resulting quadruple heterozygous plants were selected and allowed to self-fertilize. The following genotypes were identified: dcl1/DCL1 dcl2/dcl2 dcl3/dcl3 DCL4/DCL4, dcl1/DCL1 dcl2/dcl2 DCL3/DCL3 dcl4/dcl4, dcl1/DCL1 DCL2/DCL2 dcl3/dcl3 dcl4/dcl4, DCL1/DCL1 dcl2/dcl2 dcl3/dcl3 dcl4/dcl4, dcl1/DCL1 dcl2/dcl2 dcl3/dcl3 dcl4/dcl4, and allowed to self-fertilize in order to produce sufficient amount of plants for virus infection experiments.
Virus infection assays
Plants were grown in vitro for 10 days, transferred to soil, and mock-inoculated or inoculated with TCV or CMV the day after, as previously described (Mourrain et al, 2000; Béclin et al, 2002). Plants were grown in controlled-environment chambers under the following conditions: 150 μE/m2/s irradiance, 16 h light, 40% relative humidity, 18°C night temperature, 23°C day temperature. Aerial parts of 2–10 plants were pooled 3 weeks after inoculation, and RNA was extracted.
RNA blot analysis
Total RNA was extracted as described before (Gasciolli et al, 2005). Small RNAs were separated by electrophoresis on 15% polyacrylamide gels and blotted on membranes. Oligonucleotide probes corresponding to TAS1 (si-480(+)), TAS2 (si-F) and TAS3 (mixed oligo combinations) have been described before (Gasciolli et al, 2005). 32P-end-labeled oligonucleotide probes were prepared as described before (Gasciolli et al, 2005). Radiolabeled RNA probes transcripted in vitro were used to detect siRNAs corresponding to the regions coding for the coat proteins of the TCV (accession NC 003821) or the CMV (accession Y18137). High molecular weight RNA blots were probed with radiolabeled DNAs complementary to the regions coding for the coat proteins of the CMV or the TCV. RNA hybridization signals were quantified using a Fuji phosphorimager.
Note added in the proof
Work by Deleris et al also implicates DCL4 and DCL2 in the production of viral siRNAs. These authors further demonstrate that DCL4 is inhibited by the TCV coat protein P38 (Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O (2006) Hierarchical action and inhibition of plant dicer-like proteins in antiviral defense. Science Express; doi:10.1126/science.1128214).
Acknowledgments
We thank Allison Mallory and David Bartel for fruitful discussions, and Bruno Letarnec for plant care in the greenhouse. This work was partly supported by a grant from the European Commission (Riboreg program) to HV.
References
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, Gasciolli V, Vaucheret H (2006) Uncoupled production of ta-siRNAs in hypomorphic rdr6 and sgs3 mutants uncovers a role for TAS3 in AGO7-DCL4-DRB4-mediated control of leaf morphology. Curr Biol 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Akbergenov R, Si-Ammour A, Blevins T, Amin I, Kutter C, Vanderschuren H, Zhang P, Gruissem W, Meins F Jr, Hohn T, Pooggin MM (2006) Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res 34: 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béclin C, Boutet S, Waterhouse P, Vaucheret H (2002) A branched pathway for transgene-induced RNA silencing in plants. Curr Biol 12: 684–688 [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet S, Vazquez F, Liu J, Béclin C, Fagard M, Gratias A, Morel JB, Vaucheret H (2003) Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr Biol 13: 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C, Pallotta M, ReFalo P, Sachs MS, Vayssie L, Macino G, Cogoni C (2004) Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol Cell Biol 24: 2536–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC (2004) Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev 18: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan P, Vanitharani R, Fauquet CM (2005) MicroRNA-binding viral protein interferes with Arabidopsis development. Proc Natl Acad Sci USA 102: 10381–10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li WX, Xie D, Peng JR, Ding SW (2004) Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell 16: 1302–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, Yates JR, Mello CC (2006) Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354 [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O (2005) DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet 37: 1356–1360 [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O (2004) Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16: 1235–1250 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD (2005) ASRP: the Arabidopsis Small RNA Project database. Nucleic Acids Res 33: D637–D640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM (2005) Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett 579: 5822–5829 [DOI] [PubMed] [Google Scholar]
- Han MH, Goud S, Song L, Fedoroff N (2004) The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA 101: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38: 721–725 [DOI] [PubMed] [Google Scholar]
- Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T (2005) Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol 57: 173–188 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM (1999) Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126: 5231–5243 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37: 761–765 [DOI] [PubMed] [Google Scholar]
- Kavi HH, Fernandez HR, Xie W, Birchler JA (2005) RNA silencing in Drosophila. FEBS Lett 579: 5940–5949 [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Hammell CM, Ambros V. (2006) Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW (2004) Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ. (2005) Elucidation of the small RNA component of the transcriptome. Science 309: 1567–1569 [DOI] [PubMed] [Google Scholar]
- MacDiarmid R (2005) RNA silencing in productive virus infections. Annu Rev Phytopathol 43: 523–544 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Bartel D, Vance VB, Bowman LH (2002) A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc Natl Acad Sci USA 99: 15228–15233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S, Schauer SE, Smith TH, Mallory AC, Herr JM Jr, Roth B, Merchant DS, Ray A, Bowman LH, Vance VB (2005) Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell 17: 2873–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Remoue K, Sanial M, Vo TA, Vaucheret H (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi M.A, Lerbs-Mache S, Colot V, Lagrange T (2005) Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev 19: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ren T, Morris TJ (2003) The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J Virol 77: 511–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7: 487–491 [DOI] [PubMed] [Google Scholar]
- Tabara H, Yigit E, Siomi H, Mello CC (2002) The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109: 861–871 [DOI] [PubMed] [Google Scholar]
- Takeda A, Tsukuda M, Mizumoto H, Okamoto K, Kaido M, Mise K, Okuno T (2005) A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J 24: 3147–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H (2006a) Posttranscriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20: 759–777 [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Mallory AC, Bartel DP (2006b) AGO1 homeostasis requires co-expression of MIR168 and AGO1, posttranscriptional stabilization of miR168 by AGO1, and miR168-guided AGO1 cleavage. Mol Cell 22: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crété P, Vaucheret H (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]


