Abstract
Voltage-gated Na+ channels are composed of pore-forming α and auxiliary β subunits. The majority of Na+ channels in the heart contain tetrodotoxin (TTX)-insensitive Nav1.5 α subunits, but TTX-sensitive brain-type Na+ channel α subunits are present and functionally important in the transverse tubules of ventricular myocytes. Sinoatrial (SA) nodal cells were identified in cardiac tissue sections by staining for connexin 43 (which is expressed in atrial tissue but not in SA node), and Na+ channel localization was analyzed by immunocytochemical staining with subtype-specific antibodies and confocal microscopy. Brain-type TTX-sensitive Nav1.1 and Nav1.3 α subunits and all four β subunits were present in mouse SA node, but Nav1.5 α subunits were not. Nav1.1 α subunits were also present in rat SA node. Isolated mouse hearts were retrogradely perfused in a Langendorff preparation, and electrocardiograms were recorded. Spontaneous heart rate and cycle length were constant, and heart rate variability was small under control conditions. In contrast, in the presence of 100 nM TTX to block TTX-sensitive Na+ channels specifically, we observed a significant reduction in spontaneous heart rate and markedly greater heart rate variability, similar to sick-sinus syndrome in man. We hypothesize that brain-type Na+ channels are required because their more positive voltage dependence of inactivation allows them to function at the depolarized membrane potential of SA nodal cells. Our results demonstrate an important contribution of TTX-sensitive brain-type Na+ channels to SA nodal automaticity in mouse heart and suggest that they may also contribute to SA nodal function and dysfunction in human heart.
Voltage-gated sodium channels are responsible for the initiation of action potentials in excitable cells. They are composed of pore-forming α subunit and auxiliary β subunits (1). Ten genes encoding α subunits have been identified, and 9 have been functionally expressed (2, 3). Isoforms preferentially expressed in the central nervous system (Nav1.1, -1.2, -1.3, and -1.6) are inhibited by nanomolar concentrations (KD ≈ 1–10 nM) of tetrodotoxin (TTX), as is the isoform present in adult skeletal muscle (Nav1.4). In contrast, the primary cardiac isoform (Nav1.5) requires micromolar concentrations of TTX (KD ≈ 2–6 μM) for inhibition (4, 5). Na+ channel α subunits are associated with one or two auxiliary β subunits, named β1 to β4. These auxiliary subunits modulate channel gating, interact with extracellular matrix, and function as cell adhesion molecules (6, 7). Although TTX-insensitive Nav1.5 channels are the predominant Na+ channel type in the heart, we recently found that TTX-sensitive brain-type Na+ channels are localized in the transverse tubules of ventricular myocytes where they are required for the normal synchronous coupling of cell surface depolarization to contraction of the myocyte (8).
The heart beat is initiated by slow diastolic depolarization of the sinoatrial (SA) node, which drives the membrane potential after action potential toward the threshold for firing the next action potential. Multiple ionic currents with complex interactions are involved in spontaneous diastolic depolarization (reviewed in refs. 9 and 10), but the contribution of Na+ channels is controversial. Early studies suggested that the action potential in SA node is insensitive to TTX (11), but subsequent work showed that Na+ channels are present but are largely inactivated at diastolic potentials in rabbit and mouse SA node (12–21, ¶, ‖). Connexin 40 and connexin 43 are expressed in myocytes of mouse atrial muscle, with a sharp transition to connexin 45 expression in the SA node. In addition, a sharp transition from atrial to nodal action potential waveforms was observed over ≈100 μm (22). In light of previous work detecting Nav1.1 channels in neonatal rabbit SA node (23), we have examined the localization and function of brain-type Na+ channel subtypes in adult SA node. Our results reveal specific localization of brain-type Na+ channels in mouse and rat SA nodes and show that the function of these Na+ channels is required for normal control of beat rate and heart rate variability in the mouse heart.
Experimental Procedures
Antibodies.
The specifications and the peptide sequences against which the antibodies are directed have been described (8). Antibodies recognizing Nav1.1, Nav1.2, and Nav1.3 were purchased from Chemicon (Temecula, CA) (anti-RI, anti-RII, and anti-RIII). The antibody recognizing Nav1.6 (anti-Scn8a) was from Alomone Laboratories (Jerusalem). The antibodies against Nav1.5 (8) and the β subunits (24, 25) have been described. The mouse monoclonal antibody against connexin 43 was purchased from Chemicon.
Microdissection and Immunocytochemistry of Mouse SA Node.
Adult male (8–10 wk) B6/129F1 mice were preinjected with 300 I.E. heparin i.p. and anesthetized with halothane, and microdissection of the SA node was performed as described (26), following procedures approved by the Institutional Animal Care and Use Committee of the University of Washington. Spontaneously contracting tissue containing the SA node was then fixed in 4% paraformaldehyde at 4°C for 4 h, rinsed in 0.1 M phosphate buffer (pH 7.4) for 5 min and then cryoprotected in buffer containing 30% sucrose for 24 h. Frozen 10-μm sections were cut and incubated with specific antibodies, as described (8). Tissue was viewed by using a Bio-Rad MRC 600 confocal microscope. For control sections, primary antibodies were preincubated with their antigenic peptides, or no primary antibody was used.
Microdissection and Immunocytochemistry of Rat SA Node.
Adult male rats (250–350 g) were studied in accordance with the regulations of the United Kingdom Animals (Scientific Procedures) Act of 1986. The SA node was dissected and analyzed as described (27). Ten-micrometer cryosections were cut perpendicular to the crista terminalis close to the main branch from the crista. Sections were fixed with 4% paraformaldehyde for 30 min at room temperature, treated with 0.1% Triton X-100 for 10 min at room temperature, blocked with 10% normal donkey serum for 60 min at room temperature, incubated with the primary antibody at a dilution of 1:100 overnight at 4°C, and incubated with appropriate secondary antibody (from Jackson ImmunoResearch) for 1 h at room temperature. Tissue was viewed using Leica TCS SP and Zeiss LSM 5 Pascal confocal microscopes. For control sections, primary antibodies were preincubated with their antigenic peptides, or no primary antibody was used.
Electrocardiography.
Mice were anesthetized, and hearts were removed and Langendorff-perfused retrogradely by using a modified Krebs–Henseleit buffer (KHB) containing (in mM) 118 NaCl, 4.7 KCl, 1.2 MgSO4, 1.2 KH2PO4, 11 glucose, 2.0 CaCl2, 2.0 pyruvate, 25 NaHCO3, and 5 units/liter insulin plus 1 μM propranolol and 100 nM atropine to block autonomic responses (28–30), and bubbled with 95% O2 and 5% CO2 at 37°C. Perfusion pressure was 60 mm Hg. Horizontal ECGs were recorded between pairs of thin platinum electrodes attached to the atrial appendages, and data were sampled at 5 kHz with windaq software (Dataq Instruments, Akron, OH) on a personal computer and analyzed with igor (WaveMetrics, Lake Oswego, OR) software. After initial perfusion, the preparation was allowed to stabilize for 10 min, followed by a 4-min ECG recording period (ECG 1). Then the heart was perfused for an additional 10 min with either control KHB or KHB containing 100 nM TTX, followed by a second 4-min recording period (ECG 2) in control KHB or TTX-containing KHB.
Analysis of ECGs.
Custom-written ECG-measurement macros were imbedded in igor for semiautomatic analysis. Waveforms were variable from heart to heart, but the shapes of the P wave and the QRS complex were stable within each experiment. Heart rates and R-R intervals were determined over a 4-min period, whereas P-P and P-R intervals were measured over 60 successive beats. The P-P interval was measured from the first sign of atrial excitation in two successive cycles, and the P-R interval was measured from the first sign of atrial excitation to the first sign of ventricular excitation as conventionally defined (31). Interval durations were measured by aligning the segment from the end of the P wave to the end of the QRS complex by using a least squares approach with the unknown interval duration as the fit parameter, and assignments were verified by inspection. Interval variability was assessed by comparing the standard deviation of intervals in successive cardiac cycles of spontaneously beating hearts (32). All measurements are presented as mean ± SEM.
Results
Immunocytochemical Localization of Na+ Channel α Subunits in SA Node.
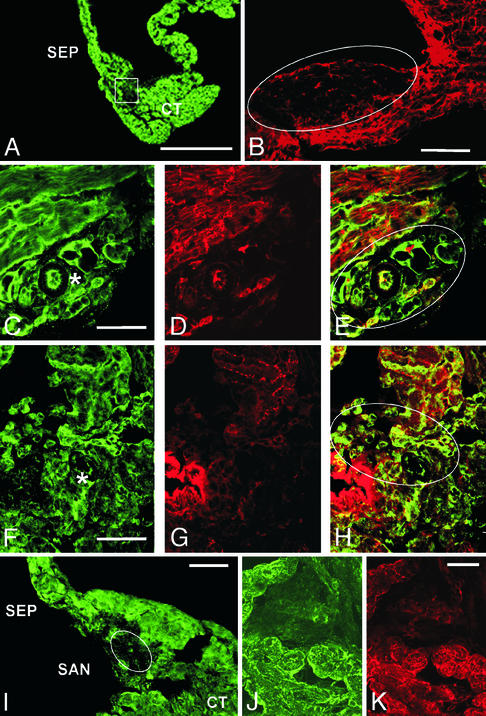
The mouse SA node was isolated in a strip of tissue bounded by the crista terminalis and the atrial septum (Fig. 1A). SA nodal cells were recognized (33, 34) by their characteristic architecture, small size, dense nuclei, and spindle or ring shape (9, 13, 22, 35–38). To confirm identification of SA nodal cells, sections were labeled with antibodies recognizing connexin 43, which stain atrial and ventricular muscle cells but not SA nodal cells (refs. 22 and 39–42; Fig. 1B, circled).
Figure 1.
Localization of α subunits of Na+ channels in the mouse SA node. (A) Section stained with anti-Nav1.5 antibodies to illustrate the region of the crista terminalis (CT), the SA node (boxed unstained region), and the interatrial septum (SEP). (B) Section labeled with anti-connexin 43 to illustrate the lack of expression in the SA node (oval unstained region). (C) Section labeled with anti-Nav1.1 to illustrate staining in the cells of the SA node. An asterisk marks the SA nodal artery as a landmark. (D) Same section shown in C, double-labeled with anti-connexin 43 to label atrial muscle fibers. (E) Merged images from C and D to illustrate specific labeling of SA nodal cells with anti-Nav1.1. Outlined region indicates the area of the node. (F) Section labeled with anti-Nav1.3 illustrating staining of SA nodal cells. Asterisk denotes SA nodal artery as a landmark. (G) Same section as in F, double-labeled with anti-connexin 43. (H) Merged image of G and H illustrating specific staining of SA nodal cells with anti-Nav1.3. Oval indicates region of SA node. (I) Lower magnification image of section labeled with anti-Nav1.5 illustrating staining in surrounding CT and SEP but not in the SA node. (J) Higher magnification of the SA node region illustrating lack of anti-Nav1.5 staining. (K) Same section as J, stained for connexin 43. Scale bars: A, 125 μm; B, 100 μm; C–K, 50 μm.
Na+ channels were labeled with subtype-specific antibodies against Nav1.1, Nav1.2, Nav1.3, Nav1.5, and Nav1.6 α subunits and examined under the confocal microscope. The SA node stains with antibodies against the brain-type Na+ channel Nav1.1 (Fig. 1C), and this staining is observed in myocytes that are not stained for connexin 43 (Fig. 1D), as illustrated in the merged image (Fig. 1E, circled). Mouse SA nodal cells are also stained specifically for Nav1.3 (Fig. 1 F–H). In contrast, Nav1.2 and Nav1.6 channels were not detected (data not shown), although comparatively high background staining of the SA node with the anti-Nav1.6 may have obscured a low level of Nav1.6 channels. Surprisingly, we also did not detect Nav1.5 (Fig. 1 I and J), the principal TTX-sensitive Na+ channel isoform of the myocardium (4, 5), although it is highly expressed in the surrounding atrial muscle cells. Thus, Nav1.1 and Nav1.3 are the principal Na+ channel α subunit isoforms of the SA node, and the “cardiac” Nav1.5 channel is strikingly absent.
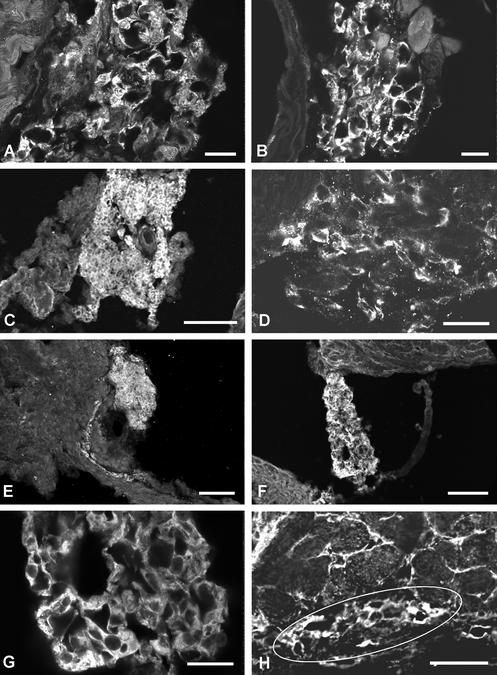
In ventricular muscle, Nav1.5 is concentrated in intercalated disks, whereas Nav1.1, Nav1.3, and Nav1.6 display a t-tubular localization (8). SA nodal cells do not have well developed transverse tubules, and no distinct subcellular localization of Nav1.1 and Nav1.3 channels is evident when examined at high resolution in the light microscope (Fig. 2 A and B).
Figure 2.
Localization of α and β subunits of the Na+ channel in the mouse SA node. (A) Higher magnification of anti-Nav1.1 staining in the SA node, illustrating staining of individual cells. (B) Higher magnification of anti-Nav1.3 staining in the SA node, illustrating staining of individual cells. (C) Lower magnification of anti-β1 staining in the SA node. (D) Higher magnification staining of anti-β1 staining in individual cells of the node. (E) Section stained with anti-β2 antibodies illustrating localization of this subunit in the node. (F and G) Sections labeled with anti-β3, illustrating localization in cells of the SA node. (H) Section labeled with anti-β4, illustrating staining in cells of the SA node (outlined region). Scale bars: A and B, 50 μm; C, 100 μm; D, 25 μm; E and F, 100 μm; G, 25 μm; H, 50 μm.
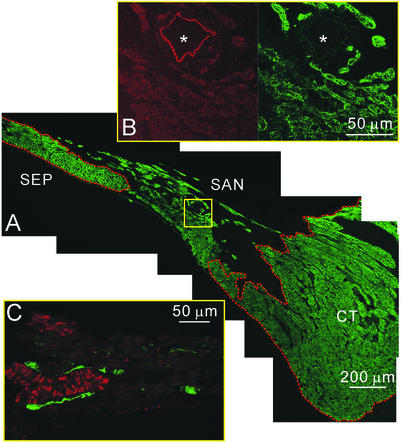
In rat heart, where the dissection to identify the SA node is easier and more precise, sections were also labeled with anti-connexin 43 to identify the SA node between the crista terminalis and the atrial septum (Fig. 3A). SA nodal cells as well as the cells of the crista terminalis and atrial septum labeled positively for Nav1.1 (Fig. 3A). The labeling was present primarily in the plasma membrane (Fig. 3B). As in mouse, labeling of Nav1.2 and Nav1.6 was not observed in the SA node from rat heart. In contrast to mouse, no labeling of Nav1.3 in SA nodal cells was observed in rat (Fig. 3C). However, there was labeling of Nav1.3 in nerve fibers and cell bodies within and near the SA node, which is known to be highly innervated (9).
Figure 3.
Localization of the α subunits of Na+ channels in rat SA node. (A) Section through the crista terminalis and intercaval region labeled for Nav1.1. The section was also labeled for Cx43 (labeling not shown), and the red lines surround the Cx43-positive atrial muscle of the crista terminalis (CT) and atrial septum (SEP) on either side of the Cx43-negative SA node (SAN). Labeling of Nav1.1 in the cell membrane of all cells throughout the section is evident. (B) High-magnification images of labeling of Cx43 (Left) and Nav1.1 (Right) in the area of SA node indicated by the yellow box in A. There is no specific labeling of Cx43 above background in the SA node cells, whereas, for comparison, specific labeling is present in the SA nodal artery (*). In contrast, there is labeling of Nav1.1 in the cell membrane of the SA node cells but no labeling in the artery. (C) Section through the SA node double-labeled for Cx43 (red) and Nav1.3 (green). Cx43 labeling (in red) is largely absent, confirming that the tissue is SA node. There is no labeling of Nav1.3 in the SA node cells, but labeling is present in nerve fibers and cell bodies.
Immunolocalization of Na+ Channel β Subunits in SA Node.
Auxiliary Na+ channel β subunits have been identified in cardiac tissue (25) but not localized to the SA node. Using similar methods as for α subunits, we detected β1, β2, β3, and β4 subunits in SA nodal cells (Fig. 2 C–H). Thus, in contrast to α subunits, we do not find subtype-specific expression of β subunits in the SA node.
Functional Role of Brain-Type Na+ Channels in the SA Node.
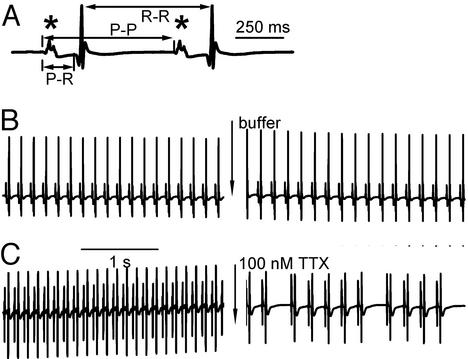
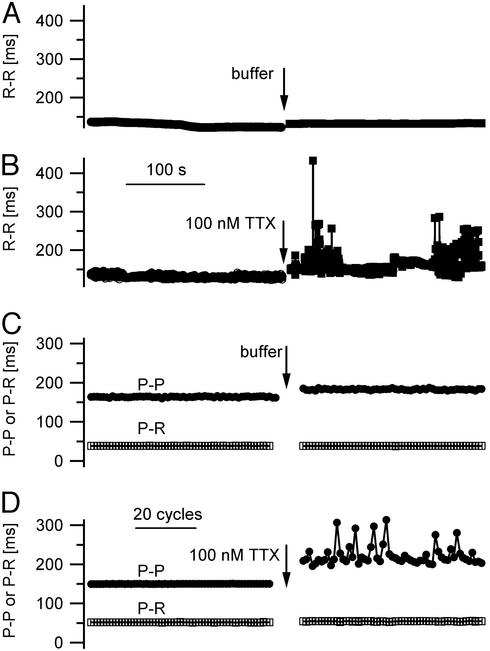
To investigate the functional role of brain-type Na+ channels in the SA node, we used an isolated Langendorff heart preparation and recorded ECGs from spontaneously beating hearts. These studies were performed in the presence of atropine and propranolol to avoid effects of TTX on neurotransmitter release from autonomic nerve terminals. Measurements of the P-P and R-R intervals, which provide estimates of the cycle length of the heart beat, and the P-R interval, which measures the time from atrial excitation to ventricular excitation, are illustrated in Fig. 4A. We used 100 nM TTX to selectively block the TTX-sensitive Na+ channels (Nav1.1 and Nav1.3) that were detected in SA nodes of mice and thereby test their role in cardiac rhythm. This concentration blocks at least 90% of the TTX-sensitive brain-type Na+ channels, as their IC50 values for block by TTX are in the range of 1 to 10 nM (3), but only ≈1.5% of Nav1.5 channels, assuming an IC50 of 6.3 μM as reported for multicellular cardiac preparations (43). ECGs were recorded during a 4-min period while the hearts were perfused with KHB (ECG 1). Perfusion was then continued with KHB (control group; n = 5) or KHB containing 100 nM TTX (TTX group; n = 6) for 10 min. Then, ECGs were recorded for an additional 4 min in control KHB or KHB containing 100 nM TTX (ECG 2). During ECG 1 in KHB, cycle length measured as R-R intervals was ≈138 ms in the control and TTX groups (Table 1). All other measured variables were also similar between these groups (Table 1; P > 0.05). After perfusion for an additional 10 min, cycle length was increased in both groups (Fig. 4 B and C; Table 1). However, cycle length during the following 4 min in ECG 2 was 161 ± 10 ms in the control group vs. 227 ± 34 ms in the TTX group (P = 0.04, control vs. TTX group, unpaired Student's t test), an increase in cycle length of 17.5% in the control group and 64.5% in the TTX group.
Figure 4.
Effect of TTX on electrocardiograms of spontaneously beating Langendorff-perfused mouse hearts. (A) A typical ECG tracing in expanded form illustrating the P wave (*), the P-R interval, the P-P interval, and the R-R interval. (B and C) Example ECGs taken from the control (B) and the TTX group (C) before (ECG 1) and after (ECG 2) a 10-min perfusion with control buffer or buffer containing 100 nM TTX.
Table 1.
Effects of TTX on heart rate and its variability
| Parameter | Control group
|
100 nM TTX group
|
||
|---|---|---|---|---|
| ECG 1 | ECG 2 | ECG 1 | ECG 2 | |
| R-R, ms | 137 ± 6 | 161 ± 10* | 138 ± 7 | 227 ± 34*† |
| SDR-R, ms | 4.8 ± 1.6 | 3.2 ± 1.1 | 2.3 ± 0.4 | 35.3 ± 13.3*† |
| P-P, ms | 140.7 ± 8.2 | 159.8 ± 9.8* | 141.1 ± 6.1 | 222.9 ± 25.8* |
| SDP-P, ms | 0.5 ± 0.20 | 1.7 ± 0.87 | 0.6 ± 0.28 | 32.5 ± 9.8*† |
| P-R, ms‡ | 44.2 ± 2.6 | 41.9 ± 1.4 | 44.9 ± 1.9 | 42.8 ± 2.7 |
| SDP-R, ms | 0.21 ± 0.03 | 0.26 ± 0.11 | 0.3 ± 0.10 | 0.6 ± 0.23 |
Significantly different (P < 0.05) between ECG 1 and ECG 2 within a group.
Significantly different between control and TTX groups during ECG 2 recording period. SD, standard deviation.
P-R as defined in Experimental Procedures.
Application of 100 nM TTX also led to obvious irregularities in heart beat when compared with control (Fig. 4 B and C). To assess the irregularity of the cycle length quantitatively, we measured the variability of the R-R intervals during the 4-min recording periods by using the standard deviation of R-R intervals, SDR-R (32). In the control group, the cycle length variability in ECG 2 was not statistically different from ECG 1 (Table 1). In contrast, treatment with 100 nM TTX increased heart rate variability more than 10-fold when compared with pretreatment values (SDR-R = 2.3 ± 0.4 ms in ECG 1 vs. 35.3 ± 13.3 ms in TTX in ECG 2, P = 0.04, paired Student's t test). These measurements show that treatment with 100 nM TTX causes a slower and more irregular heart beat.
The R-R intervals mirror the beat rate of the ventricles, which can be slowed by delayed or blocked conduction from the atrium to the ventricle under pathophysiological conditions. To further investigate the origin of the slowing and irregularity of the heart beat due to block of TTX-sensitive Na+ channels, we analyzed both P-P and P-R intervals from the ECGs (see definition in Fig. 4A). The P-P interval measures the entire cardiac cycle, like the R-R interval, using the time of initiation of the atrial action potential as an index, whereas the P-R interval corresponds to the time between the atrial action potential and the beginning of the ventricular action potential. In the control group, perfusion with normal KHB for 10 min led to a significant increase in P-P interval from 140.7 ± 0.08 ms in ECG 1 to 159.8 ± 0.23 ms in ECG 2, equivalent to a P-P interval prolongation of 13.6% (P < 0.01, paired Student's t test; Fig. 5C; Table 1), in accordance with the observed 17.7% prolongation of cycle length derived from assessment of R-R intervals. In contrast, P-R interval did not changed significantly after 10 min of perfusion with control KHB (P = 0.9, paired Student's t test; Fig. 5C; Table 1), suggesting stable atrial excitation, AV nodal conduction, and ventricular excitation. Perfusion with 100 nM TTX for 10 min led to a very prominent increase in P-P interval from 141.1 ± 0.14 ms during ECG 1 to 223.0 ± 4.3 ms after TTX treatment during ECG 2, equivalent to a P-P interval prolongation of 58.0% (P = 0.02, paired Student's t test; Fig. 5D; Table 1), in accordance with the observed 65.9% prolongation of cycle length determined by assessing the R-R intervals. As for the control group, P-R intervals changed insignificantly after 10-min perfusion with 100 nM TTX (Fig. 5D; Table 1), suggesting stable AV conduction even after block of TTX-sensitive Na+ channels.
Figure 5.
Effects of TTX on P-P, P-R, and R-R intervals from spontaneously beating Langendorff-perfused mouse hearts. (A and B) R-R intervals representing beat-to-beat intervals and therefore heart rate measured during 4-min recording periods before (ECG 1) and after (ECG 2) a 10-min perfusion with either control buffer (A) or buffer containing 100 nM TTX (B). (C and D) P-P and P-R intervals measured in 60 consecutive cycles before and after a 10-min perfusion with either control buffer (C) or buffer containing 100 nM TTX (D).
To assess the variability of the P-P and P-R intervals quantitatively, we calculated their standard deviation and named these variables SDP-P and SDP-R. As expected, the SDP-P and SDP-R were similar within the control group in ECG 1 and ECG 2 (Table 1). However, in the TTX group, SDP-P increased significantly from 0.6 ± 0.28 ms during ECG 1 to 32.5 ± 9.8 ms after 10 min of perfusion with 100 nM TTX in ECG 2 (P = 0.02, paired Student's t test). SDP-R was affected by TTX to a much lesser extent than SDP-P (0.3 ms vs. 0.6 ms, P = 0.07). Comparison of the two variables in ECG 2 after wash-in of either control KHB or 100 nM TTX shows a significant increase of SDP-P due to TTX treatment from 1.7 to 32.5 ms (P = 0.02, unpaired Student's t test), whereas SDP-R was unaffected (0.26 ms vs. 0.6 ms, P = 0.2, unpaired Student's t test). These measurements of P-P and P-R intervals and their variability confirm that specific block of brain-type Na+ channels with 100 nM TTX slows heart rate and substantially increases its variability.
Discussion
Na+ Channel Expression in the SA Node.
TTX-insensitive Nav1.5 channels are primarily expressed in the heart, and they are the most highly expressed Na+ channels in cardiac tissue (4, 5). Therefore, they have been widely assumed to fulfill all of the functions of Na+ channels in the heart. Recent work has now identified two distinct functional roles for brain-type, TTX-sensitive Na+ channels–Nav1.1, Nav1.3, and Nav1.6. In ventricular myocytes, Nav1.1, Nav1.3, and Nav1.6 are specifically localized in the transverse tubules (8). Block of these channels with low concentrations of TTX reduces the synchrony and efficiency of coupling of cell surface depolarization to contraction (8). Previous studies detected TTX-sensitive Na+ currents and localized expression of Nav1.1 mRNA in neonatal rabbit SA node (23). Here, we have shown that Nav1.1 channels are localized in adult rat SA nodal cells and that both Nav1.1 and Nav1.3 channels are localized in adult mouse SA node, along with the auxiliary β subunits of Na+ channels. Surprisingly, the major cardiac Na+ channel subtype, Nav1.5, is not present in the SA node. The specific expression of the brain-type Na+ channels in SA node strongly suggests that they have a specific functional role there.
Functional Role of Brain-Type Na+ Channels in the SA Node.
The automaticity of the SA node is thought to be derived primarily from the actions of the hyperpolarization-activated, cyclic nucleotide-regulated channels of the HCN family that conduct Ih (44–46), the Cav1.2 and Cav1.3 channels that conduct L-type calcium currents (26, 47), and the Cav3 channels that conduct T-type calcium currents (reviewed in refs. 10 and 48). Nevertheless, many previous studies (see Introduction) have shown that there are significant Na+ currents in the SA node. Here we provide evidence that these Na+ currents are conducted primarily by brain-type Nav1.1 and Nav1.3 channels by showing that these subtypes are specifically expressed in SA node, whereas Nav1.5 channels are not. Moreover, we find that specific block of these TTX-sensitive brain-type Na+ channels reduces the spontaneous heart rate and greatly increases its variability in mouse heart. These functional effects of 100 nM TTX show that TTX-sensitive Nav1.1 and Nav1.3 channels are required for normal regulation of the heart beat in mouse.
What properties of the brain-type Nav1.1 and Nav1.3 might make them well-suited for a functional role in the SA node in comparison with Nav1.5 channels? An important difference between SA nodal cells and ventricular or atrial myocytes is their comparatively positive maximum diastolic membrane potential of approximately −60 mV (10, 13, 45, 48). Comparison of the voltage dependence of gating of the most prevalent brain-type Na+ channel, Nav1.2, shows that its half-maximal steady state inactivation requires a 15- to 25-mV more positive membrane potential compared with Nav1.5, when analyzed side-by-side in cell lines expressing these Na+ channels endogenously or by expression of cDNAs in heterologous mammalian cells (5, 49). In contrast, Nav1.1 and Nav1.3 channels have similar voltage dependence of inactivation to Nav1.2 channels when expressed in the same heterologous mammalian cells (ref. 50; M. Mantegazza, W.A.C., and T.S., unpublished results). The principal Na+ current conducted by Nav1.5 in ventricular myocytes is half-inactivated at approximately −82 mV and essentially completely inactivated at −60 mV (51, 52). At the diastolic membrane potential of approximately −60 mV in SA nodal cells, Nav1.5 channels would be nearly completely inactivated whereas approximately half of brain-type Na+ channels would remain in the resting state. Therefore, the more positive voltage dependence of inactivation of brain-type Na+ channels allows them to function in SA node at a membrane potential that would inactivate Nav1.5. In addition to their more positive voltage dependence of inactivation, Nav1.1 and Nav1.3 channels conduct larger persistent Na+ currents than Nav1.5 after repolarization (refs. 53 and 54; M. Mantegazza, W.A.C., and T.S., unpublished results). These persistent inward Na+ currents after opening and inactivation of Na+ channels during the SA node action potential may be important in supporting diastolic depolarization in the SA node.
The highly variable heart rate we have observed after blocking TTX-sensitive Na+ channels resembles sick-sinus syndrome in man, suggesting that dysfunction of Na+ channels in the SA node may be involved in that arrhythmia. Brain-type Na+ channels are inhibited by neurotransmitters acting through the cAMP-dependent protein kinase and protein kinase C signaling pathways (55). Therefore, over-stimulation of the SA node by the many neurotransmitters and hormones that activate these signaling pathways might reduce Na+ currents in the SA node and cause or exacerbate sick-sinus syndrome. Some clinical evidence suggests that overactivity of the parasympathetic nervous system may indeed cause sick-sinus syndrome (56, 57).
Although our studies of the effects of TTX on heart rate and its variability gave clear evidence of a role for TTX-sensitive Na+ channels in SA nodal function, we did not observe a major effect on the P-R interval, which is a sensitive measure of conduction through the AV node. Therefore, at least under the experimental conditions used here, we did not find evidence for a major role for TTX-sensitive brain-type Na+ channels in conduction through the AV node. Consistent with this finding, Nav1.5 channels are expressed in the AV node (58), and both mice lacking Nav1.5 channels and humans with mutations in Nav1.5 channels have altered AV nodal conduction (59, 60). Thus, Nav1.5 channels may be predominant in the AV node whereas the brain-type Nav 1.1 and Nav1.3 channels are predominant in SA node.
Regional Specialization of the SA Node.
From studies on rabbit tissue, it is known that the SA node is not uniform and that the action potential is first initiated in the center and is conducted to the surrounding atrial muscle via the peripheral regions (36, 61). In intact SA node, the center is the leading pacemaker site because of its greater slope of diastolic depolarization (62). The leading pacemaker current in the center of the SA node is the L-type Ca2+ current whereas the Na+ current plays a major role in pacemaking in the periphery (63). These functional specializations within the SA node suggest that brain-type Na+ channels may be differentially distributed throughout the node, which may contribute to the functional differences that have been observed. High-resolution analysis of the distribution of Na+ channels and correlation with physiological measurements in individual cells will likely reveal new aspects of the role of brain-type Na+ channels in the SA node.
Acknowledgments
We thank Thuy Vien (University of Washington) and Shin Yoo and Mitsuru Yamamoto (University of Leeds) for help in immunocytochemical experiments and Eric Feigl (University of Washington) for making his laboratory available for the electrocardiographic experiments and for critical comments on the manuscript. Research at the University of Washington was supported by Research Grant P-01 HL44948 from the National Institutes of Health (to W.A.C.).
Abbreviations
- TTX
tetrodotoxin
- SA
sinoatrial
- KHB
Krebs–Henseleit buffer
Footnotes
Lei, M., Cooper, J. P. & Kohl, P. (2002) Biophys. J. 82, 609a (abstr.).
Lei, M., Lancaster, S. A., Jones, S. A. & Boyett, M. R. (2002) J. Physiol. (London) 544, 39P (abstr.).
References
- 1.Catterall W A. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 2.Goldin A L, Barchi R L, Caldwell J H, Hofmann F, Howe J R, Hunter J C, Kallen R G, Mandel G, Meisler M H, Netter Y B, et al. Neuron. 2000;28:365–368. doi: 10.1016/s0896-6273(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 3.Goldin A L. Annu Rev Physiol. 2001;63:871–894. doi: 10.1146/annurev.physiol.63.1.871. [DOI] [PubMed] [Google Scholar]
- 4.Satin J, Kyle J W, Chen M, Bell P, Cribbs L L, Fozzard H A, Rogart R B. Science. 1992;256:1202–1205. doi: 10.1126/science.256.5060.1202. [DOI] [PubMed] [Google Scholar]
- 5.Fozzard H A, Hanck D A. Physiol Rev. 1996;76:887–926. doi: 10.1152/physrev.1996.76.3.887. [DOI] [PubMed] [Google Scholar]
- 6.Isom L L, De Jongh K S, Catterall W A. Neuron. 1994;12:1183–1194. doi: 10.1016/0896-6273(94)90436-7. [DOI] [PubMed] [Google Scholar]
- 7.Isom L L. Neuroscientist. 2001;7:42–54. doi: 10.1177/107385840100700108. [DOI] [PubMed] [Google Scholar]
- 8.Maier S K, Westenbroek R E, Schenkman K A, Feigl E O, Scheuer T, Catterall W A. Proc Natl Acad Sci USA. 2002;99:4073–4078. doi: 10.1073/pnas.261705699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyett M R, Honjo H, Kodama I. Cardiovasc Res. 2000;47:658–687. doi: 10.1016/s0008-6363(00)00135-8. [DOI] [PubMed] [Google Scholar]
- 10.Schram G, Pourrier M, Melnyk P, Nattel S. Circ Res. 2002;90:939–950. doi: 10.1161/01.res.0000018627.89528.6f. [DOI] [PubMed] [Google Scholar]
- 11.Narahashi T. Physiol Rev. 1974;54:813–889. doi: 10.1152/physrev.1974.54.4.813. [DOI] [PubMed] [Google Scholar]
- 12.Muramatsu H, Zou A R, Berkowitz G A, Nathan R D. Am J Physiol. 1996;270:H2108–H2119. doi: 10.1152/ajpheart.1996.270.6.H2108. [DOI] [PubMed] [Google Scholar]
- 13.Mangoni M E, Nargeot J. Cardiovasc Res. 2001;52:51–64. doi: 10.1016/s0008-6363(01)00370-4. [DOI] [PubMed] [Google Scholar]
- 14.Kreitner D. J Mol Cell Cardiol. 1975;7:655–662. doi: 10.1016/0022-2828(75)90142-x. [DOI] [PubMed] [Google Scholar]
- 15.Maylie J, Morad M. J Physiol (London) 1984;355:215–235. doi: 10.1113/jphysiol.1984.sp015415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noma A, Yanagihara K, Irisawa H. Pflügers Arch. 1977;372:43–51. doi: 10.1007/BF00582205. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama T, Kurachi Y, Noma A, Irisawa H. Pflügers Arch. 1984;402:248–257. doi: 10.1007/BF00585507. [DOI] [PubMed] [Google Scholar]
- 18.Nathan R D. Am J Physiol. 1986;250:H325–H329. doi: 10.1152/ajpheart.1986.250.2.H325. [DOI] [PubMed] [Google Scholar]
- 19.Oei H I, Van Ginneken A C, Jongsma H J, Bouman L N. J Mol Cell Cardiol. 1989;21:1137–1149. doi: 10.1016/0022-2828(89)90691-3. [DOI] [PubMed] [Google Scholar]
- 20.Denyer J C, Brown H F. J Physiol (London) 1990;428:405–424. doi: 10.1113/jphysiol.1990.sp018219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muramatsu H, Zou A-R, Berkowitz G A, Nathan R D. Am J Physiol. 1996;270:H2108–H2119. doi: 10.1152/ajpheart.1996.270.6.H2108. [DOI] [PubMed] [Google Scholar]
- 22.Verheijck E E, van Kempen M J, Veereschild M, Lurvink J, Jongsma H J, Bouman L N. Cardiovasc Res. 2001;52:40–50. doi: 10.1016/s0008-6363(01)00364-9. [DOI] [PubMed] [Google Scholar]
- 23.Baruscotti M, Westenbroek R, Catterall W A, DiFrancesco D, Robinson R B. J Physiol (London) 1997;498:641–648. doi: 10.1113/jphysiol.1997.sp021889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ratcliffe C F, Qu Y, McCormick K A, Tibbs V C, Dixon J E, Scheuer T, Catterall W A. Nat Neurosci. 2000;3:437–444. doi: 10.1038/74805. [DOI] [PubMed] [Google Scholar]
- 25.Malhotra J D, Chen C, Rivolta I, Abriel H, Malhotra R, Mattei L N, Brosius F C, Kass R S, Isom L L. Circulation. 2001;103:1303–1310. doi: 10.1161/01.cir.103.9.1303. [DOI] [PubMed] [Google Scholar]
- 26.Bohn G, Moosmang S, Conrad H, Ludwig A, Hofmann F, Klugbauer N. FEBS Lett. 2000;481:73–76. doi: 10.1016/s0014-5793(00)01979-7. [DOI] [PubMed] [Google Scholar]
- 27.Dobrzynski H, Rothery S M, Marples D D, Coppen S R, Takagishi Y, Honjo H, Tamkun M M, Henderson Z, Kodama I, Severs N J, Boyett M R. J Histochem Cytochem. 2000;48:769–780. doi: 10.1177/002215540004800606. [DOI] [PubMed] [Google Scholar]
- 28.Nishimaru K, Tanaka Y, Tanaka H, Shigenobu K. Life Sci. 2000;66:607–615. doi: 10.1016/s0024-3205(99)00633-5. [DOI] [PubMed] [Google Scholar]
- 29.Ng G A, Brack K E, Coote J H. Exp Physiol. 2001;86:319–329. doi: 10.1113/eph8602146. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka H, Nishimaru K, Kobayashi M, Matsuda T, Tanaka Y, Shigenobu K. Naunyn-Schmiedeberg's Arch Pharmacol. 2001;363:577–582. doi: 10.1007/s002100100411. [DOI] [PubMed] [Google Scholar]
- 31.Katz A M. Physiology of the Heart. New York: Raven; 1992. [Google Scholar]
- 32.Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Circulation. 1996;93:1043–1065. [PubMed] [Google Scholar]
- 33.Opthof T, de Jonge B, Jongsma H J, Bouman L N. J Mol Cell Cardiol. 1987;19:1221–1236. doi: 10.1016/s0022-2828(87)80532-1. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen P M, Le Grice I J, Smaill B H, Hunter P J. Am J Physiol. 1991;260:H1365–H1378. doi: 10.1152/ajpheart.1991.260.4.H1365. [DOI] [PubMed] [Google Scholar]
- 35.Alings A M, Abbas R F, de Jonge B, Bouman L N. J Mol Cell Cardiol. 1990;22:1453–1466. doi: 10.1016/0022-2828(90)90988-e. [DOI] [PubMed] [Google Scholar]
- 36.Bleeker W K, Mackaay A J, Masson-Pevet M, Bouman L N, Becker A E. Circ Res. 1980;46:11–22. doi: 10.1161/01.res.46.1.11. [DOI] [PubMed] [Google Scholar]
- 37.James T N, Sherf L, Fine G, Morales A R. Circulation. 1966;34:139–163. doi: 10.1161/01.cir.34.1.139. [DOI] [PubMed] [Google Scholar]
- 38.Opthof T, de Jonge B, Mackaay A J, Bleeker W K, Masson-Pevet M, Jongsma H J, Bouman L N. J Mol Cell Cardiol. 1985;17:549–564. doi: 10.1016/s0022-2828(85)80024-9. [DOI] [PubMed] [Google Scholar]
- 39.Coppen S R, Kodama I, Boyett M R, Dobrzynski H, Takagishi Y, Honjo H, Yeh H I, Severs N J. J Histochem Cytochem. 1999;47:907–918. doi: 10.1177/002215549904700708. [DOI] [PubMed] [Google Scholar]
- 40.Davis L M, Kanter H L, Beyer E C, Saffitz J E. J Am Coll Cardiol. 1994;24:1124–1132. doi: 10.1016/0735-1097(94)90879-6. [DOI] [PubMed] [Google Scholar]
- 41.Davis L M, Rodefeld M E, Green K, Beyer E C, Saffitz J E. J Cardiovasc Electrophysiol. 1995;6:813–822. doi: 10.1111/j.1540-8167.1995.tb00357.x. [DOI] [PubMed] [Google Scholar]
- 42.Oosthoek P W, Viragh S, Mayen A E, van Kempen M J, Lamers W H, Moorman A F. Circ Res. 1993;73:473–481. doi: 10.1161/01.res.73.3.473. [DOI] [PubMed] [Google Scholar]
- 43.Antoni H, Bocker D, Eickhorn R. J Physiol (London) 1988;406:199–213. doi: 10.1113/jphysiol.1988.sp017376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Irisawa H, Brown H F, Giles W. Physiol Rev. 1993;73:197–227. doi: 10.1152/physrev.1993.73.1.197. [DOI] [PubMed] [Google Scholar]
- 45.DiFrancesco D. Annu Rev Physiol. 1993;55:455–472. doi: 10.1146/annurev.ph.55.030193.002323. [DOI] [PubMed] [Google Scholar]
- 46.Moosmang S, Stieber J, Zong X, Biel M, Hofmann F, Ludwig A. Eur J Biochem. 2001;268:1646–1652. doi: 10.1046/j.1432-1327.2001.02036.x. [DOI] [PubMed] [Google Scholar]
- 47.Platzer J, Engel J, Schrott-Fischer A, Stephan K, Bova S, Chen H, Zheng H, Striessnig J. Cell. 2000;102:89–97. doi: 10.1016/s0092-8674(00)00013-1. [DOI] [PubMed] [Google Scholar]
- 48.Kaupp U B, Seifert R. Annu Rev Physiol. 2001;63:235–257. doi: 10.1146/annurev.physiol.63.1.235. [DOI] [PubMed] [Google Scholar]
- 49.Mantegazza M, Yu F H, Catterall W A, Scheuer T. Proc Natl Acad Sci USA. 2001;98:15348–15353. doi: 10.1073/pnas.211563298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clare J J, Tate S N, Nobbs M, Romanos M A. Drug Discovery Today. 2000;5:506–520. doi: 10.1016/s1359-6446(00)01570-1. [DOI] [PubMed] [Google Scholar]
- 51.Bean B P, Cohen C J, Tsien R W. J Gen Physiol. 1983;81:613–642. doi: 10.1085/jgp.81.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen C J, Bean B P, Colatsky T J, Tsien R W. J Gen Physiol. 1981;78:383–411. doi: 10.1085/jgp.78.4.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joho R H, Moorman J R, VanDongen A M J, Kirsch G E, Silberberg H, Schuster G, Brown A M. Mol Brain Res. 1990;7:105–113. doi: 10.1016/0169-328x(90)90087-t. [DOI] [PubMed] [Google Scholar]
- 54.Smith R D, Goldin A L. J Neurosci. 1998;18:811–820. doi: 10.1523/JNEUROSCI.18-03-00811.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cantrell A R, Catterall W A. Nat Rev Neurosci. 2001;2:397–407. doi: 10.1038/35077553. [DOI] [PubMed] [Google Scholar]
- 56.Turakhia D P, Shah B P, Gandhi M J. J Assoc Physicians India. 1985;33:535–536. [PubMed] [Google Scholar]
- 57.Santinelli V, Chiariello M, Clarizia M, Condorelli M. Int J Cardiol. 1984;5:532–535. doi: 10.1016/0167-5273(84)90093-7. [DOI] [PubMed] [Google Scholar]
- 58.Petrecca K, Amellal F, Laird D W, Cohen S A, Shrier A. J Physiol (London) 1997;501:263–274. doi: 10.1111/j.1469-7793.1997.263bn.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan H L, Bink-Boelkens M T, Bezzina C R, Viswanathan P C, Beaufort-Krol G C, van Tintelen P J, van den Berg M P, Wilde A A, Balser J R. Nature. 2001;409:1043–1047. doi: 10.1038/35059090. [DOI] [PubMed] [Google Scholar]
- 60.Papadatos G A, Wallerstein P M, Head C E, Ratcliff R, Brady P A, Benndorf K, Saumarez R C, Trezise A E, Huang C L, Vandenberg J I, et al. Proc Natl Acad Sci USA. 2002;99:6210–6215. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kodama I, Boyett M R. Pflügers Arch. 1985;404:214–226. doi: 10.1007/BF00581242. [DOI] [PubMed] [Google Scholar]
- 62.Opthof T, de Jonge B, Jongsma H J, Bouman L N. Eur Heart J. 1987;8:1249–1259. doi: 10.1093/oxfordjournals.eurheartj.a062200. [DOI] [PubMed] [Google Scholar]
- 63.Kodama I, Nikmaram M R, Boyett M R, Suzuki R, Honjo H, Owen J M. Am J Physiol. 1997;272:H2793–H2806. doi: 10.1152/ajpheart.1997.272.6.H2793. [DOI] [PubMed] [Google Scholar]