Abstract
It is poorly understood why a particular plant species is resistant to the vast majority of potential pathogens that infect other plant species, a phenomenon referred to as “nonhost” resistance. Here, we show that Arabidopsis NHO1, encoding a glycerol kinase, is required for resistance to and induced by Pseudomonas syringae isolates from bean and tobacco. NHO1 is also required for resistance to the fungal pathogen Botrytis cinerea, indicating that NHO1 is not limited to bacterial resistance. Strikingly, P. s. pv. tomato DC3000, an isolate fully virulent on Arabidopsis, actively suppressed the NHO1 expression. This suppression is abolished in coi1 plants, indicating that DC3000 required an intact jasmonic acid signaling pathway in the plant to suppress NHO1 expression. Constitutive overexpression of NHO1 led to enhanced resistance to this otherwise virulent bacterium. The presence of avrB in DC3000, which activates a cultivar-specific “gene-for-gene” resistance in Arabidopsis, restored the induction of NHO1 expression. Thus, NHO1 is deployed for both general and specific resistance in Arabidopsis and targeted by the bacterium for parasitism.
Keywords: glycerol‖lipids‖parasitism‖JA‖Pseudomonas
Nonhost disease resistance refers to the phenomenon that most plant species are typically resistant to the pathogens of other plant species (1). It is the most prevalent form of disease resistance in plants, because it is effective against the vast majority of phytopathogens. The nature of nonhost resistance implies that it is broad spectrum and durable; thus, it is of great interest to agriculture. Unraveling nonhost resistance mechanisms is also crucial for our understanding of host-specificity and the pathogenesis of phytopathogens.
Although of fundamental importance, nonhost resistance remains a poorly investigated problem in plant pathology. What are the constituents of nonhost resistance? Is nonhost resistance an induced resistance? What is the relationship between nonhost resistance and gene-for-gene resistance? Why is nonhost resistance not effective against the specialized virulent pathogen? An answer to these questions requires the isolation and detailed characterization of nonhost resistance genes.
Previously (2), we have established Arabidopsis-Pseudomonas syringae pv. phaseolicola as a model genetic system for nonhost resistance studies. As a result, NHO1 was identified as a key general resistance gene for resistance to multiple nonhost pathovars of P. syringae bacteria. NHO1 is also required for gene-for-gene resistance to P. syringae. The nho1 mutant is caused by a recessive mutation that nonspecifically supports the multiplication of both nonhost and avirulent Pseudomonas bacteria. However, NHO1 is ineffective against virulent bacteria, and the nho1 mutant shows no increased disease susceptibility (2). We suggested that the NHO1-mediated resistance is overcome by virulent Pseudomonas bacteria (2). Here, we report the isolation of NHO1 that defines an active defense mechanism for general resistance. Furthermore, we demonstrate that the virulent P. s. pv. tomato DC3000 strain actively suppresses the NHO1 gene expression through a jasmonic acid (JA)-dependent pathway. Gene-for-gene resistance negated this suppression. Ectopic expression of NHO1 increased plant resistance to DC3000 bacteria.
Materials and Methods
Mapping.
Approximately 2,000 nho1 × Ler F2 plants were inoculated with P. s. pv. tabaci, and a bacterial growth assay was used to score susceptible plants. Simple sequence length polymorphism (3) markers F18B13-71K and F5I6-115K were PCR-amplified by using the following primer pairs: 5′-TTTCGTTCTGCTTCCGAGCTTAG-3′ and 5′-ACCTGAAGCATCGTCACATTTATG-3′; 5′-GATAACAATCCCAAGCATTTGAAG-3′ and 5′-ACACCTTACATTACCACATACAAC-3′.
Inoculation and RNA Gel Blot Analysis.
Five-wk-old plants were vacuum-infiltrated with P. s. pv. phaseolicola NPS3121, P. s. pv. tomato DC3000, P. s. pv. tabaci R11528 race 0, P. s. pv. syringae 3525 (4), or P. s. pv. tomato DC3000 (avrB) bacteria at the indicated concentrations. RNA was isolated from plants at the indicated times postinoculation. RNA gel blot was hybridized with an Arabidopsis EST that encodes a glycerol kinase (GenBank accession no. T43219) as a probe.
Complementation by Gene Transformation.
A 7-kb XbaI fragment containing At1g80460 was blunt-ended and inserted into a modified pBI121 (CLONTECH) lacking the Cauliflower Mosaic Virus 35S promoter and the UID gene. The construct was mobilized into Agrobacterium tumefaciens strain GV3101, and the nho1 mutant plants were transformed with the construct by floral dip (5). Only plants carrying the transgene, as determined by PCR, were used for bacterial inoculation.
Bacterial Growth Assay.
Five-week-old plants were syringe-infiltrated with bacteria at 105 colony-forming units (cfu)/ml, and bacteria numbers in leaves were determined as described (2).
Fungal Inoculation.
Botrytis cinerea (6) was grown on potato dextrose agar for sporulation. Spores were suspended in 0.01% Tween 20 at 5 × 105 spores per ml for inoculation (6).
Overexpression of NHO1 in Plants.
Two NHO1-overexpression constructs were generated. The 7-kb XbaI fragment containing At1g80460 was first inserted into pBluescript SK(−) (Stratagene). The At1g80460 fragment was then excised with BamHI and SacI and inserted in the corresponding sites of pBI121. The resulting construct carried the 7-kb At1g80460 genomic DNA fragment under the control of the Cauliflower Mosaic Virus 35S promoter. The construct was introduced into A. tumefaciens strain GV3101 and subsequently transformed into the nho1 mutant. The second NHO1-overexpression construct directly placed the At1g80460 ORF under the control of the Cauliflower Mosaic Virus 35S promoter. The At1g80460 ORF was PCR-amplified with oligonucleotide primers 5′-TTTCTAGACCTTGAGATAACAACAAAGCATCG-3′ and 5′-TTGAGCTCAAGACAGAGCCCAACTGTGGTAAA-3′, and inserted into the XbaI and SacI sites of pBI121. This construct was used to transform wild-type Col-0 plants. All plants used for bacterial inoculation had been verified by PCR for the presence of transgenes.
Results
NHO1 Is Required for Resistance to B. cinera.
NHO1 is required for nonspecific resistance to multiple Pseudomonas strains including a plant-associated nonpathogenic Pseudomonas fluorescens strain. However, the nho1 mutant does not support the growth of Escherichia coli when infiltrated into the plant (2). To test whether NHO1 is also required for resistance to pathogens other than Pseudomonas bacteria, we inoculated nho1 and Col-0 plants with B. cinerea. The nho1 mutant reproducibly displayed a high level of susceptibility to this fungus. Whereas the wild-type leaves showed only minor disease symptoms, nho1 leaves were completely colonized by fungal mycelia and died within 7 days after inoculation (Fig. 1). However, nho1 was not affected for resistance to Alternaria brassicicola, Peronospora trifoliorum, and Xanthomonas oryzae pv. oryzae (data not shown), suggesting that NHO1 is required for resistance only to certain pathogen groups.
Figure 1.
Botrytis cinerea resistance. At least 20 wild-type (Col-0) and 20 nho1 plants were inoculated with B. cinerea spores and incubated for 7 days before being photographed. All wild-type leaves showed mild symptoms, whereas all nho1 leaves collapsed after 7 days. The experiment was repeated twice with similar results.
Cloning of NHO1.
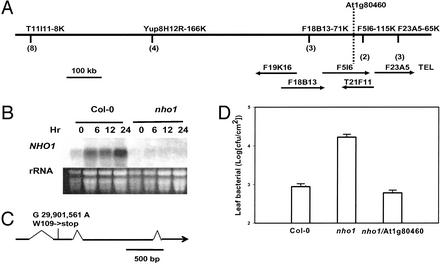
NHO1 was mapped to the bottom of chromosome I on the telomeric side of marker F18B13-71K (2). Further mapping by using ≈2,000 nho1 × Ler F2 plants positioned NHO1 between markers F18B13-71K and F5I6-115K, a 150-kb interval that contained ≈30 genes (Fig. 2A).
Figure 2.
Cloning of NHO1. (A) The chromosomal region containing NHO1. Markers are shown above the chromosome (bar), and bacterial artificial chromosome clones spanning NHO1 are given under the chromosome. The number of recombinants for a marker is given in parentheses. (B) RNA blot analysis of wild-type (Col-0) and nho1 plants for NHO1 expression after inoculation with P. s. pv. phaseolicola at 106 cfu/ml. RNA was isolated at the indicated times. Wild-type plants were also inoculated with 10 mM MgCl2 (mock) as control. Ethidium bromide staining of the RNA gel indicates the amount of RNA loaded. (C) Schematic representation of the coding regions (solid lines), introns (gaps), and the mutation in the nho1 mutant. (D) Complementation of nho1 by At1g80460. Leaves of wild-type (Col-0), nho1, and transgenic nho1 plants carrying the At1g80460 transgene were syringe-infiltrated with P. s. pv. phaseolicola. Bacteria populations were determined 4 days postinoculation from at least four Col-0 or nho1 plants or 32 primary transgenic plants. Error bars are standard errors.
Additional evidence that this interval contained NHO1 came from microarray and RNA blot analyses of nho1 and wild-type plants (Fig. 2B). Selected microarrays (subarrays) containing Arabidopsis ESTs enriched for genes that are either induced or repressed by DC3000 infection (R.T. and S.H., unpublished results) were used to compare transcript levels between nho1 and wild-type (Col-0) plants 18 h after infiltration of P. s. pv. phaseolicola bacteria. Three independent hybridizations repeatedly detected one single transcript species that was underrepresented by ≈10-fold in the nho1 mutant compared with wild-type plants (nho1/Col-0 ratios: 0.13, 0.12, and 0.10). None of other mRNA species was significantly different between nho1 and wild-type plants. The corresponding gene (At1g80460) is carried by bacterial artificial chromosomes F5I6 and T21F11 and maps between F18B13-71K and F5I6-115K (Fig. 2A). RNA gel blot analysis was carried out to verify the microarray results (Fig. 2B). The At1g80460 mRNA in Col-0 plants was strongly induced by the P. s. pv. phaseolicola infection. In contrast, the At1g80460 transcript was nearly undetectable in nho1 plants.
At1g80460 contains four exons and three introns and encodes a protein of 522 aa. Sequencing analysis of the entire At1g80460 gene (from ≈1.5 kb upstream of the start codon to 1 kb downstream of the stop codon) revealed a single nucleotide substitution (G29,901,501A, chromosome I) that introduced an early stop codon in the second exon of the gene (Fig. 2C). The results from genetic mapping, gene expression, and sequencing provided strong evidence that the At1g80460 gene is NHO1.
Complementation tests by gene transformation confirmed that At1g80460 is indeed NHO1. A 7-kb genomic fragment containing the coding region plus a 3.1-kb upstream sequence and a 1.9-kb downstream sequence was transformed into nho1 plants. Thirty-two positive primary (T1) transgenic plants were tested for resistance to P. s. pv. Phaseolicola; all showed resistance similar to that of wild-type plants. Fig. 2D shows bacterial populations in wild-type, nho1, and the transgenic plant leaves. Twelve nho1/At1g80460 T2 transgenic families were inoculated with P. s. pv. phaseolicola, and all showed decreased bacterial populations compared with nontransgenic nho1 plants (data not shown). These results demonstrate that the At1g80460 gene is NHO1.
Induction of NHO1 Transcripts by Nonhost Pseudomonas Bacteria.
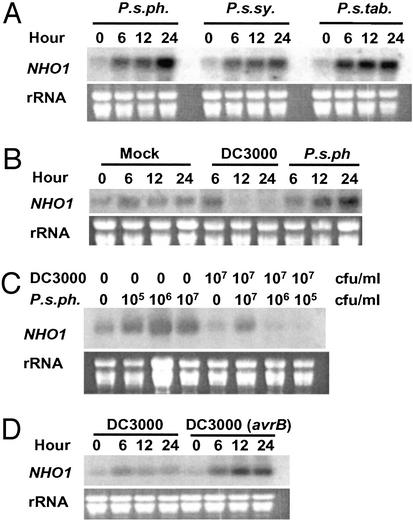
We have previously proposed that NHO1 defines a general resistance mechanism (2). The induction of NHO1 transcripts by P. s. pv. phaseolicola (Fig. 2B) is characteristic of an inducible defense response gene. Similar to numerous other defense-related genes that also respond to wound or mechanic stresses (7, 8), a transient induction of NHO1 was observed on mock-inoculation at the 6-h time point, followed by a decline of expression 12–24 h after the treatment. We examined NHO1 expression in response to two additional nonhost strains, P. s. pv. syringae and P. s. pv. tabaci (Fig. 3A). Similar to P. s. pv. phaseolicola, these strains induced NHO1 transcripts to high levels 12–24 h after inoculation. These results establish NHO1 as an inducible resistance gene in Arabidopsis.
Figure 3.
Expression analysis of NHO1. (A) RNA blot analysis of wild-type Col-0 plants after vacuum infiltration of P. s. pv. phaseolicola (P. s. ph.), P. s. pv. syringae (P. s. sy.), or P. s. pv. tabaci (P. s. tab.) at 106 cfu/ml. (B) Suppression of NHO1 transcript accumulation by DC3000. Wild-type Col-0 plants were vacuum-infiltrated with 107 cfu/ml DC3000 or P. s. pv. phaseolicola (P. s. ph.), or mock-inoculated. (C) Suppression of NHO1 expression in plants coinoculated with DC3000 and P. s. pv. phaseolicola. Wild-type plants were either mock-inoculated (0) or coinfiltrated with DC3000 and P. s. pv. phaseolicola bacteria at the indicated concentrations (cfu/ml) for 24 h before RNA was isolated for gel blot analysis. Shown is expression of NHO1 mRNA in Col-0 plants in response to DC3000 bacteria containing avrB at 106 cfu/ml (D). Leaves infiltrated with 107 cfu/ml DC3000 (avrB) bacteria developed necrosis 24 h after inoculation. All experiments were repeated at least three times with similar results.
Suppression of NHO1 Expression by Virulent Bacteria.
To test the hypothesis that NHO1 is targeted by bacterial virulence mechanisms, we examined NHO1 expression in response to virulent bacteria. Wild-type Arabidopsis plants (Col-0) were vacuum-infiltrated with DC3000 bacteria, and NHO1 transcript levels were examined. In repeated experiments, NHO1 transcripts were nearly abolished 12 and 24 h after inoculation (Fig. 3B), in contrast to the increase of NHO1 transcripts in P. s. pv. phaseolicola-inoculated plants. DC3000 therefore seemed to actively suppress the NHO1 expression. An alternative interpretation is that DC3000 lacks an elicitor for NHO1 induction. However, NHO1 transcript levels in DC3000-inoculated plants (at 12 and 24 h) were lower than that in mock-inoculated plants, arguing for an active suppression by DC3000 bacteria. The suppression seems to be dose-dependent, and the maximum suppression occurs when 107 cfu/ml bacteria were used (data not shown). To further investigate the NHO1 suppression by DC3000, we coinoculated wild-type plants with DC3000 and P. s. pv. phaseolicola bacteria (Fig. 3C). The presence of DC3000 bacteria suppressed NHO1 induction by P. s. pv. phaseolicola in trans, suggesting that this suppression occurred outside of bacterial cells. Together, these results demonstrate that DC3000 actively suppressed NHO1 expression in Arabidopsis.
Gene-for-Gene Resistance Activates NHO1 Expression.
NHO1 is required for gene-for-gene resistance against DC3000 bacteria containing an avirulence gene (2). Col-0 contains the resistance gene RPM1, which mediates resistance to bacteria containing avrB (9). DC3000 bacteria containing avrB were unable to suppress NHO1 expression; instead, they strongly induced NHO1 mRNA in Col-0 plants 12–24 h after inoculation (Fig. 3D). The level and timing of induction seemed to be similar to that induced by P. s. pv. phaseolicola.
Increased Expression of NHO1 Reduces Bacterial Growth in Plants.
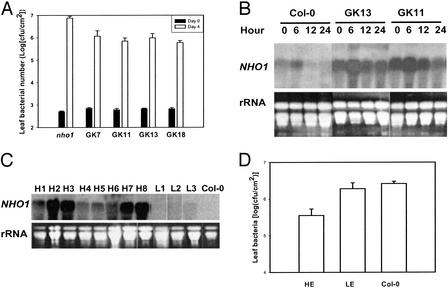
The active suppression of NHO1 expression by DC3000 supports our hypothesis that suppression of general resistance in a host plant represents a critical strategy for a virulent pathogen on this plant species. To test this hypothesis, we asked whether increased expression of NHO1 in plants diminished the virulence of DC3000 bacteria. A 35S∷NHO1 construct was introduced into the nho1 mutant. RNA gel blot analysis identified four transgenic lines (GK7, GK11, GK13, and GK18) constitutively expressing NHO1 at higher levels in the absence of inoculation (data not shown). When tested for resistance to DC3000, they displayed an ≈10-fold reduction of bacterial numbers compared with nontransgenic nho1 plants 4 days after inoculation (Fig. 3A). RNA gel blot analysis on two lines (GK11 and GK13) indicated that DC3000 bacteria were unable to eliminate NHO1 transcripts in the plants (Fig. 4B). Wild-type plants overexpressing NHO1 behaved similarly and showed enhanced resistance to DC3000 bacteria. All primary transgenic plants that expressed NHO1 transcripts at high levels (H1–8; Fig. 4C) showed significant reduction (5- to 16-fold) of bacterial population compared with nontransgenic Col-0 plants (Fig. 4D), whereas the three transgenic plants with wild-type level expression (L1–3) had bacterial numbers similar to those in nontransgenic plants. Therefore, NHO1 overexpression led to increased resistance to virulent bacteria, providing strong evidence that the suppression of NHO1 is important for parasitism of DC3000 in Arabidopsis.
Figure 4.
Resistance of NHO1-overexpression plants to virulent bacteria. (A) DC3000 bacterial populations in nontransgenic nho1 and four transgenic nho1 lines that overexpressed NHO1. Transgenic plants in the second generation were assayed for bacterial numbers 4 days after inoculation. Error bars indicate standard errors. Two independent experiments were performed with similar results. (B) Expression of the NHO1 transcript in nontransgenic Col-0 and two transgenic nho1 lines overexpressing NHO1(nho1/GK11 and nho1/GK13) after P. s. pv. tomato DC3000 inoculation. (C) NHO1 transcript levels in primary Col-0 transgenic plants carrying the 35S∷NHO1 transgene in the absence of bacterial infection. H and L, respectively, denote lines with high and low level transcripts. (D) Leaf bacteria of DC3000 in nontransgenic Col-0 plants or the primary transgenic Col-0 plants expressing NHO1 at a high (HE; an average of eight plants) or low (LE; an average of three plants) level. Two sets of primary Col-0 35S∷NHO1 plants were independently tested with similar results.
COI1 Is Required for NHO1 Suppression by DC3000.
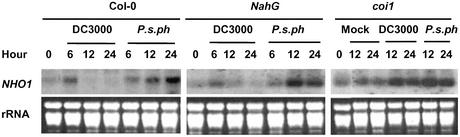
Salicylate, JA, and ethylene are three major defense hormones in plants (10). We therefore tested whether NHO1 expression in response to bacterial infection was affected by transgenic/mutant plants affected in salicylate and JA signaling. Transgenic plants carrying the NahG gene that encodes salicylate hydroxylase (11) and coi1 mutant (12) plants that are abolished in JA signaling were inoculated with P. s. pv. phaseolicola or DC3000 bacteria and examined for NHO1 expression. Fig. 5 shows that NahG and coi1 plants showed similar NHO1 induction in response to the nonhost strain, indicating that the induction of NHO1 did not require either JA or SA signaling. Surprisingly, DC3000 bacteria induced, instead of suppressed, the NHO1 expression in coi1 plants. These results indicated that the JA signaling pathway was specifically required for the NHO1 suppression by DC3000 bacteria and that DC3000 bacteria had the potential to induce NHO1 expression in plants.
Figure 5.
The suppression of NHO1 by DC3000 requires COI1. The wild-type (Col-0), NahG, and coi1 plants were infiltrated with 107 cfu/ml DC3000, P. s. pv. phaseolicola (P.s.ph.), or MgCl2 buffer (mock), and RNA was isolated at indicated times for RNA blot analysis.
NHO1 Encodes a Glycerol Kinase.
A blast search (blastp, version 2.2.3) showed that the predicted protein of At1g80460 shared strong homologies with glycerol kinases from plants, animals, fungi, and bacteria. Glycerol kinases form a distinct subfamily within the FGGY family of carbohydrate kinases (13). Fig. 6 shows a multiple alignment of the predicted NHO1 protein with the glycerol kinases from human and E. coli and a putative tomato glycerol kinase. NHO1 shares 46%, 48%, and 77% amino acid identity with its respective counterparts in E. coli, human, and tomato. NHO1 contains the two signature motives of the FGGY family of carbohydrate kinases and the conserved ATP-binding motif (14). Furthermore, NHO1 contains all four conserved amino acid residues involved in glycerol binding (14).
Figure 6.
Comparison of NHO1 with known glycerol kinases. clustalw (version 1.8) (http://searchlauncher.bcm.tmc.edu/multialign/multialign.html) was used to align the predicted NHO1 amino acid sequence, a putative tomato glycerol kinase (GenBank accession no. TC101532), human glycerol kinase (GenBank accession no. CAB54859), and E. coli glycerol kinase (GenBank accession no. NP_312878). The ATP-binding motif is shaded. The FGGY signature motives are underlined. Residues involved in glycerol binding are indicated by asterisks.
Glycerol kinases convert glycerol to glycerol 3-phosphate and is a rate-limiting enzyme for glycerol metabolism. At1g80460 is the only Arabidopsis gene encoding glycerol kinase. A second gene (At1g15050) mistakenly annotated as a glycerol kinase in the TAIR (The Arabidopsis Information Resource) database actually encodes an AUX_IAA family DNA binding factor (pfam02309; ref. 15).
Discussion
Glycerol 3-phosphate can be directed into three metabolic pathways: glycolysis, gluconeogenesis, and synthesis of phospholipids. In humans, glycerol deficiency is a genetic disease that causes hyperglycerolemia, mental retardation, and developmental disabilities (16). The nho1 mutant also accumulates a high concentration of glycerol (data not shown). The mutant was slightly smaller in size when compared with the wild-type plant, but was otherwise normal under the experimental conditions used. The susceptibility of nho1 might result from either a shortage of G3P that affects one or more of these metabolic pathways or an increased accumulation of glycerol. The alteration of the glycerol metabolism may affect the level of intermediaries such as glycerol, sugars, fatty acids, or lipids. One consequence of this mutation may be an altered apoplast environment more conducive to bacterial growth. Alternatively, some of these intermediaries may play an important role in plant defense. For example, the G3P level affects fatty acid compositions. In plants, chloroplast glycerolipids are synthesized through a prokaryotic pathway and a eukaryotic pathway. A reduction of the G3P pool reduces the flux of fatty acids into the prokaryotic pathway but increases the flux into the eukaryotic pathway, leading to a decrease of hexadecatrienoic acid but an increase of 18 carbon fatty acids (17). SSI2, a negative regulator of PR gene expression, encodes a fatty acid desaturase (18). Mutants suppressing the ssi2-fatty acid deficiency are reduced in hexadecatrienoic acid and show defects in SA-mediated resistance to Pseudomonas bacteria (J. Shah, personal communication). The importance of lipids in disease resistance has also been demonstrated by the putative lipase genes EDS1 and PAD4, which are required for disease resistance to P. syringae and Peronospora parasitica (19, 20). Furthermore, the recently cloned DIR1 gene, which is required for systemic acquired resistance, encodes a lipid transfer protein (21). Although the biochemical basis of glycerol kinase in plant defense remains to be elucidated, the correlation of NHO1 transcript levels with resistance/susceptible reactions and the increased resistance to DC3000 in NHO1 overexpression plants suggest an important role of these biochemical processes in disease resistance or parasitism.
NHO1 is also required for resistance to B. cinerea but not the tested isolates of A. brassicicola, P. trifoliorum, and X. oryzae pv. oryzae. It is not clear what, if any, characteristics shared by B. cinerea and A. brassicicola are related to the NHO1 function. Inoculation of Arabidopsis with Botrytis failed to induce NHO1 transcripts (data not shown).
Several possibilities exist for nonhost resistance. Nonhost plants may possess passive and/or active defense mechanisms that nonspecifically target nonhost phytopathogens. It is also possible that compatibility factors are lacking between nonhost pathogens and nonhost plants. Our results show that these two possibilities are interconnected. We demonstrate that NHO1 encodes an inducible glycerol kinase that is nonspecifically required for resistance to Pseudomonas bacteria. Nonhost Pseudomonas bacteria are unable to overcome this resistance, perhaps because of a lack of adaptation to crucifer plants during evolution. The regulation of NHO1 during bacterial infection suggests a complex interplay between plant defense and pathogen virulence.
Nonspecific recognition of general elicitors produced by nonhost pathogens likely plays an important role in nonhost resistance (2, 22). Pathogen surface molecules, also referred to as Pathogen-Associated Molecular Patterns, which are shared among members of a pathogen group, are known to induce innate immunity in both animals and plants (23, 24). NHO1 is induced nonspecifically by different Pseudomonas bacteria. All three tested nonhost Pseudomonas pathovars induced NHO1. DC3000 also seemed to possess the ability to induce NHO1, at least in coi1 plants. It is possible that a PAMP shared by Pseudomonas bacteria induces the expression of NHO1.
Active suppression of host defenses is a crucial part of parasitism for pathogens. For example, RNA silencing in plants is an active defense against viral infection. Some viruses actively suppress this process, enabling infection and propagation in their host plants (25). Active suppression of host defenses is also well known for animal bacterial pathogens (26). In plant bacterial pathogens, the avrRpt2, avrRpm1, virPphA, and avrPphF genes contribute to virulence, apparently by suppressing host defense (27–30). However, their host targets remain to be determined. The NHO1-mediated resistance is an apparent target of bacterial virulence, although we do not know which DC3000 gene(s) is required for NHO1 suppression. Consistent with this finding, ectopic expression of NHO1 significantly decreased the growth of DC3000 bacteria in plants.
DC3000 bacteria may actively manipulate a signaling pathway in plants to suppress NHO1 expression. Consistent with this hypothesis, we found that an intact JA signaling pathway is required for this suppression. This finding is exciting, because it suggests that the JA pathway is targeted by DC3000 to overcome the resistance mediated by NHO1 and, perhaps, other general defense genes. Bacterial pathogens such as P. s. pv. tomato are known to mimic JA signaling, and that has been thought to play a role in bacterial virulence (12). It is interesting to note that coi1 plants are more resistant to DC3000 bacteria than the wild-type plants (31), although we do not know whether the inability of suppressing NHO1 accounts for this increased resistance. The bacterial mechanisms required for NHO1 suppression are still under investigation.
It is striking that DC3000 bacteria carrying the avrB gene reactivated the NHO1 expression. Therefore, gene-for-gene resistance rendered the suppression of NHO1 by DC3000 inoperative. In fact, NHO1 is required for gene-for-gene resistance (2), suggesting that the activation of NHO1 expression is an integral part of gene-for-gene resistance. Two possibilities exist for the NHO1 induction by incompatible bacteria. The gene-for-gene recognition may directly activate NHO1 transcription independent of Pathogen-Associated Molecular Pattern recognition. Alternatively, gene-for-gene recognition may indirectly activate NHO1 transcription by impeding the NHO1 gene repression caused by DC3000, and the observed NHO1 expression may actually reflect the induction caused by Pathogen-Associated Molecular Pattern. Regardless, NHO1 defines a common defense component activated by both general and gene-for-gene resistance. Thus, the gene-for-gene resistance is superimposed on the preexisting general resistance. Interestingly, recent evidence suggests that NbSGT1, a protein required for multiple gene-for-gene resistance pathways, is also required for the resistance of Nicotiana benthemiana to two bacterial pathogens P. s. pv. maculicola and Xanthomonas axonopodis pv. vesicatoria (32). NbSGT1 is not required for resistance to the Brassicaceae pathogen X. campestris pv. campestris.
Understanding nonhost disease resistance remains a major challenge in plant pathology. Results described here demonstrate that NHO1 is an inducible resistance gene required for nonspecific resistance to nonhost Pseudomonas bacteria. The active induction of NHO1 by nonhost pathogens, suppression of NHO1 by virulent bacteria, and reactivation of NHO1 by gene-for-gene resistance indicate that NHO1 is repeatedly deployed by host for defense and targeted by pathogen for parasitism, highlighting a central role in plant-pathogen coevolution.
Acknowledgments
We thank Drs. Frank White, Barbara Valent, and Greg Martin for critical review of the manuscript, Dr. Donald Stuteville for assistance in P. trifoliorum inoculation, and Doug Baker for excellent technical assistance. We are also grateful to Dr. Jyoti Shah for sharing unpublished results. This paper is a contribution of the Kansas Agriculture Experimental Station (no. 03-97-J). This work is supported by U.S. Department of Agriculture National Research Initiative Grants 2001-35319-09847 and 2001-35319-10895 (to J.-M.Z.).
Abbreviations
- JA
jasmonic acids
- cfu
colony-forming unit
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Heath M C. Can J Plant Pathol. 1987;9:389–397. [Google Scholar]
- 2.Lu M, Tang X, Zhou J-M. Plant Cell. 2001;13:437–447. doi: 10.1105/tpc.13.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell C J, Ecker J R. Genomics. 1994;1:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Penaloza-Vazquez A, Chakrabarty A M, Bender C L. Mol Microbiol. 1999;33:712–7720. doi: 10.1046/j.1365-2958.1999.01516.x. [DOI] [PubMed] [Google Scholar]
- 5.Clough S J, Bent A F. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 6.Thomma B P H J, Eggermont K, Penninckx I A M A, Mauch-Mani B, Vogelsang R, Cammue B P A, Broekaert W F. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durrant W E, Rowland O, Piedras P, Hommand-Kosack K E, Jones J D G. Plant Cell. 2000;12:965–977. doi: 10.1105/tpc.12.6.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheong Y H, Chang H S, Gupta R, Wang X, Zhu T, Luan S. Plant Physiol. 2002;129:661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grant M R, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes R W, Dangl J L. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 10.Reymond P, Farmer E E. Curr Opin Plant Biol. 1998;1:404–411. doi: 10.1016/s1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- 11.Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 12.Xie D X, Feys B F, James S, Nieto-Rostro M, Turner J G. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 13.Reizer A, Deutscher J, Saier M H, Jr, Reizer J. Mol Microbiol. 1991;5:1081–1089. doi: 10.1111/j.1365-2958.1991.tb01880.x. [DOI] [PubMed] [Google Scholar]
- 14.Hurley J H, Faber H R, Worthylake D, Meadow N D, Roseman S, Pettigrew D W, Remington S J. Science. 1993;259:673–677. [PubMed] [Google Scholar]
- 15.Abel S, Nguyen M D, Theologis A. J Mol Biol. 1995;251:533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- 16.Rose C I, Haines D S M. J Clin Invest. 1978;61:163–170. doi: 10.1172/JCI108914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miquel M, Cassagne C, Browse J A. Plant Physiol. 1998;117:923–930. doi: 10.1104/pp.117.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kachroo P, Shanklin J, Shah J, Whittle E J, Klessig D F. Proc Natl Acad Sci USA. 2001;98:9448–9453. doi: 10.1073/pnas.151258398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falk A, Feys B J, Frost L N, Jones J D, Daniels M J, Parker J E. Proc Natl Acad Sci USA. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jirage D, Tootle T L, Reuber T L, Frost L N, Feys B J, Parker J E, Ausubel F M, Glazebrook J. Proc Natl Acad Sci USA. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maldonado A M, Doerner P, Dixon R A, Lamb C J, Cameron R K A. Nature. 2002;419:399–403. doi: 10.1038/nature00962. [DOI] [PubMed] [Google Scholar]
- 22.Felix G, Duran J D, Volko S, Boller T. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 23.Nurnberger T, Brunner F. Curr Opin Plant Biol. 2002;5:318–324. doi: 10.1016/s1369-5266(02)00265-0. [DOI] [PubMed] [Google Scholar]
- 24.Gomez-Gomez L, Boller T. Trends Plant Sci. 2002;7:251–256. doi: 10.1016/s1360-1385(02)02261-6. [DOI] [PubMed] [Google Scholar]
- 25.Mlotshwa S, Voinnet O, Mette M F, Matzke M, Vaucheret H, Ding S W, Pruss G, Vance V B. Plant Cell. 2002;14:S289–S301. doi: 10.1105/tpc.001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galan J E, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 27.Chen Z, Kloek A P, Boch J, Katagiri F, Kunkel B N. Mol Plant Microbe Interact. 2000;13:1312–1321. doi: 10.1094/MPMI.2000.13.12.1312. [DOI] [PubMed] [Google Scholar]
- 28.Ritter C, Dangl J L. Mol Plant Microbe Interact. 1995;8:444–453. doi: 10.1094/mpmi-8-0444. [DOI] [PubMed] [Google Scholar]
- 29.Jackson R W, Athanassopoulos E, Tsiamis G, Mansfield J W, Sesma A, Arnold D L, Gibbon M J, Murillo J, Taylor J D, Vivian A. Proc Natl Acad Sci USA. 1999;96:16875–16880. doi: 10.1073/pnas.96.19.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsiamis G, Mansfield J W, Hockenhull R, Jackson R W, Sesma A, Athanassopoulos E, Bennett M A, Stevens C, Vivian A, Taylor J D, Murillo J. EMBO J. 2000;19:3204–3214. doi: 10.1093/emboj/19.13.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kloek A P, Verbsky M L, Sharma S B, Schoelz J E, Vogel J, Klessig D F, Kunkel B N. Plant J. 2001;26:509–522. doi: 10.1046/j.1365-313x.2001.01050.x. [DOI] [PubMed] [Google Scholar]
- 32.Peart J R, Lu R, Sadanandom A, Malcuit I, Moffett P, Brice D C, Schauser L, Jaggard D A, Xiao S, Coleman M J, et al. Proc Natl Acad Sci USA. 2002;99:10865–10869. doi: 10.1073/pnas.152330599. [DOI] [PMC free article] [PubMed] [Google Scholar]