Abstract
Conserved eukaryotic signaling elements play an important role in the development of fungal pathogens on their hosts. Chk1, a mitogen-activated protein kinase (MAPK), functions in virulence, mating, and sporulation of the maize leaf pathogen Cochliobolus heterostrophus. Suppression subtractive hybridization was used to identify fungal genes whose expression on the host plant is affected in chk1 deletion mutants. Two of the genes isolated in this screen were predicted to encode cellulolytic enzymes: a cellobiohydrolase, CBH7, and an endoglucanase, EG6. Expression of EG6 and CBH7 was followed by the fusion of their upstream regulatory regions to the coding sequence of the green fluorescent protein. Induction of both genes began at the onset of invasive growth and reached its maximal extent during leaf necrosis. Furthermore, EG6 was induced preferentially within necrotic lesions. Disruption of MAPK CHK1 resulted in a delay in the penetration of hyphae into the leaf and a concomitant delay in the induction of expression of both cellulase genes. In saprophytic culture, the absence of Chk1 resulted in a marked delay in the induction of CBH7 expression by crystalline cellulose. EG6 was expressed at a basal level in culture, and this expression was found to depend strictly on Chk1. Thus, the Chk1 MAPK signaling pathway is involved in the regulation of two cellulase-encoding genes and is necessary for their timely induction by environmental signals.
INTRODUCTION
A successful fungal pathogen must overcome the physical and chemical barriers set up by the host plant to block infection. Conserved eukaryotic signal transduction proteins participate in a number of steps that lead to disease development. Fungal mitogen-activated protein kinase (MAPK) homologs play important roles in regulating differentiation, survival, and pathogenesis (Xu and Hamer, 1996; Csank et al., 1998; Xu et al., 1998; Lev et al., 1999; Wang and Heitman, 1999; Takano et al., 2000; Xu, 2000; Zheng et al., 2000; Di Pietro et al., 2001; Hou et al., 2002; Mey et al., 2002; Zhang et al., 2002). Nevertheless, little is known about the target genes regulated by fungal MAPKs. The gene expression profiles of mutants blocked in a particular signal transduction pathway, compared with the wild-type profiles, provide a powerful tool to identify targets of the pathway. Knowledge of these targets might provide a clue to the mechanism by which the pathway affects development.
In the rice blast fungus, suppression subtractive hybridization (SSH) was used to isolate genes underexpressed in a mutant lacking the MAPK pmk1 at the appressorium formation stage (Hou et al., 2002). Genome-wide transcriptional profiling was applied to determine how the Kss1 and Fus3 MAPK signaling pathways control pheromone response and dimorphic development in Saccharomyces cerevisiae (Madhani et al., 1999; Roberts et al., 2000). One of the targets of Kss1 is PGU1, a gene that encodes a secreted endopolygalacturonase that may aid invasion of the natural plant substrate of S. cerevisiae. A mutant of the vascular wilt fungus Fusarium oxysporum lacking the MAPK Fmk1 is nonpathogenic on tomato roots. PL1, which encodes a pectate lyase, is underexpressed in this mutant (Di Pietro et al., 2001). Loss of any particular hydrolytic enzyme, however, often has little or no effect on pathogenicity. Degrading enzymes are present as families of multiple enzymes with related activity. Because of such redundancy, the disruption of one or even several genes that encode hydrolytic enzymes can decrease the corresponding activity with no loss of virulence (Scott-Craig et al., 1990, 1998; Schäfer, 1993; Sposato et al., 1995; Apel-Birkhold and Walton, 1996; Ahn et al., 2001). Regulatory genes affect a broad set of targets, and their disruption results in a more severe loss of function. The transcriptional regulator SNF1 of Cochliobolus carbonum is required for the expression of several cell wall–degrading enzymes and for full virulence on maize (Tonukari et al., 2000).
Deletion of the MAPK gene CHK1 in the southern maize leaf blight pathogen Cochliobolus heterostrophus results in mutants that do not form the small appressoria characteristic of this species, are impaired in their mating ability, and lack asexual sporulation (conidiation) (Lev et al., 1999). In maize infected with MAPK deletion mutants, the development of disease symptoms is decreased strikingly, although the mutant eventually penetrates into the leaf. We constructed a SSH library to identify genes expressed in the wild type but not in the mutant during plant infection. Two of the genes isolated were homologous with cellulolytic enzymes. In this study, we characterized the expression of these genes in saprophytic culture and on the host, establishing the contribution of the MAPK Chk1 to their regulation.
RESULTS
Construction and Screening of Differential Libraries
Disruption of MAPK CHK1 leads to the inability to produce conidia and to a dramatic decrease in fungal virulence. Mutant mycelium is viable on the plant epidermis, developing a dense network, but penetration into the leaf and establishment of invasive growth are delayed significantly. The appearance of symptoms upon inoculation with fungal mycelium shows that conidial germination and appressorium formation are not essential for disease, which can be initiated by alternative modes of penetration (Lev et al., 1999). This observation allowed us to establish conditions to compare the gene expression of mutant and wild-type fungi on the host (Figure 1). Inoculation of maize leaves with homogenized wild-type mycelia led to complete necrotic destruction of plant tissue within 24 h. After 24 h of incubation, the Δchk1 mutant did not cause any visible disease symptoms; at 48 h after inoculation, the leaves became yellow in a spotted pattern and scattered necrotic lesions appeared. Even after prolonged incubation, complete necrosis usually was not reached in infections by the mutant, except for the first expanded leaf, which proved to be more sensitive to infection (Figure 1).
Figure 1.
Disease Symptoms Caused by Δchk1 Mutant and Wild-Type Strains.
Leaves were photographed after incubation periods of 24 and 48 h. WT, wild type.
Leaf and fungal material from the advanced pathogenic development stage (after 24 h in the wild type and after 48 h in the mutant) was collected for the SSH library preparation. The glyceraldehyde 3-phosphate dehydrogenase gene GPD was used as a probe to quantify the proportion of fungal RNA in total RNA preparations from infected plant tissue. In preparations of wild-type-infected leaves, fungal RNA was ∼47%, whereas in the RNA of mutant-infected plants, it was ∼23%. We used RNA of uninfected maize leaves to bring the fraction of fungal RNA to 23% in both pools. SSH was performed using a mixture of RNA from wild-type-infected and uninfected leaves as the tester population and RNA from mutant-infected leaves as the driver. A total of 192 SSH-derived clones were screened, and 77 of them were classified initially as differential between the wild type and the mutant in planta. Of these 77 candidates, 52 clones represented different parts of an unknown gene. Of the remaining 25 clones, candidates that appeared more than once were tested by gel blot analysis of RNA from the mutant and the wild type in saprophytic culture and on the plant. Three SSH library clones showed homology with plant genes: Rac1 GTPase, ferredoxin, and a triose/phosphate translocator. Of these, a chickpea Rac-type GTP binding protein is upregulated in the defense response against the ascomycete Ascochyta rabiei (Ichinose et al., 1999). Isolation of relatively few maize genes supports the validity of the method used to balance plant and fungal RNA abundance.
Two SSH Library Clones Are Homologous with Fungal Cellulases
The clone designated EG6 showed homology with endoglucanase IV of Trichoderma reesei. The coding sequence and translation initiation site of EG6 were determined using cDNA library clones and homology. The predicted Eg6 protein consists of 319 amino acids, with a cleavable signal peptide 17 amino acids long at the N terminus (identified by SignalP; Nielsen et al., 1997). Two conserved domains were identified in Eg6: a glycosyl hydrolase family 61 domain and a C-terminal fungal cellulose binding domain. The clone designated CBH7 was found to be highly similar to cellobiohydrolase II of Acremonium cellulolyticus Y-94. Isolation of the entire transcript revealed that CBH7 encodes a 389–amino acid protein with a cleavable signal peptide of 18 amino acids. Cbh7 has a glycosyl hydrolase family 6 conserved domain, but it lacks a cellulose binding domain.
Expression of EG6 and CBH7 in Planta
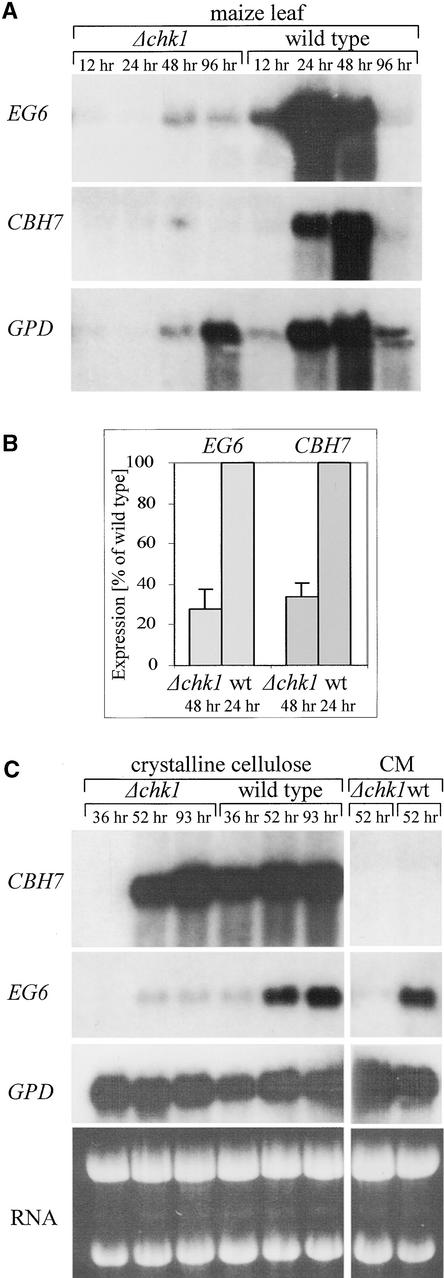
Significant expression of EG6 by the wild-type fungus was detected starting with the early stages of plant infection (12 h after inoculation). At this early stage, injury to the plant tissue was barely visible, whereas in the subsequent 12 h of incubation, the tissue became completely necrotic. At the necrotic stage, EG6 expression was maximal; then it decreased during the next 24 h. After 96 h of incubation, EG6 transcript levels were almost undetectable. Expression of EG6 was detected in the mutant strain after 48 and 96 h of incubation, but it did not reach wild-type levels (Figure 2A). The results of five independent inoculation experiments are summarized in Figure 2B. The CBH7 transcript was detected in the wild type after 24 h of incubation. The transcript level remained high after 48 h and decreased by 96 h after inoculation. In leaves infected with the chk1 deletion mutant, the CBH7 transcript was detected at 48 h of incubation in three of five experiments, and its abundance was consistently lower than in the wild type (Figures 2A and 2B).
Figure 2.
RNA Gel Blot Analysis of Cellulase Gene Expression.
(A) Lanes 1 to 4, RNA of leaves infected with the Δchk1 mutant after 12, 24, 48, and 96 h of incubation, respectively; lanes 5 to 8, RNA of leaves infected with the wild-type strain after 12, 24, 48, and 96 h of incubation, respectively.
(B) Expression of EG6 and CBH7 cellulase genes in the Δchk1 mutant versus the wild type during plant infection. RNA gel blot analyses were performed using Δchk1 mutant-infected plants after 48 h of incubation and wild-type-infected plants after 24 h of incubation. To correct for differences in fungal biomass, the expression of cellulase genes in the mutant and the wild type was normalized to the expression of “housekeeping” genes (GPD or tubulin). EG6 and CBH7 expression in the mutant then was estimated as a percentage of that in the wild type. Error bars indicate standard errors of five independent experiments. wt, wild type.
(C) Cellulase gene expression in culture. Lanes 1 to 6 show fungal cultures grown in the presence of crystalline cellulose. Lanes 1 to 3, RNA of the chk1 deletion mutant grown for 36, 52, and 93 h, respectively; lanes 4 to 6, wild-type fungus grown for 36, 52, and 93 h, respectively. Lanes 7 and 8, chk1 deletion mutant and wild-type strains, respectively, grown in CM for 52 h. Probes were as follows: EG6, coding for endoglucanase; CBH7, coding for cellobiohydrolase; RNA, ethidium bromide staining.
Expression of EG6 and CBH7 in Culture
The development of maize leaf blight caused by the chk1 mutant differs in both rate and severity of symptoms from those caused by the wild-type strain. Delayed penetration of mutant mycelium into the plant tissue, and its inability to cause extensive necrotic lesions, might restrict the availability of inductive signals. To evaluate the direct contribution of Chk1 to cellulase gene regulation, we asked whether CHK1 deletion affects the induction of EG6 and CBH7 in the presence of their substrates in vitro. We used microcrystalline cellulose (Sigmacell) to examine the induction in wild-type and Δchk1 strains. The cellulose-containing medium also included a small amount of maltose, which was used by the fungus during its initial development. After depletion of the maltose, growing mycelium gradually formed aggregates with crystalline cellulose, resulting in complete clearing of the medium. Maltose was depleted at a similar rate by the wild-type and chk1 deletion strains (data not shown). A strong induction of CBH7 expression occurred in the wild type after 36 h of incubation, and the transcript level did not decrease during the course of the experiment (Figure 2C). Induction in the mutant strain was delayed markedly: no CBH7 transcript was detected after 36 h of incubation, but after 52 h, a high level of CBH7 transcript was detected in the mutant. The induced levels of CBH7 at the later culture times were similar in the mutant and the wild type (Figure 2C). Thus, rapid induction of CBH7 on cellulose is strictly Chk1 dependent (Figure 2C, 36 h). At later culture times, additional signals apparently overcome the absence of Chk1 input (Figure 2C, 52 and 93 h).
The expression of EG6 increased between 36 and 52 h of growth in cellulose-containing medium, but the level of expression was similar to that in complete medium (CM). This finding indicates that the expression of EG6 is not restricted to cellulose but rather occurs in the presence of various carbon sources. A further indication comes from experiments in which EG6 expression was tested on minimal medium with different carbon sources: 21 of 23 sources tested did not affect EG6 expression (data not shown). In the chk1 deletion strain grown in cellulose-containing medium, there was a slight upregulation of EG6 that was undetectable when the fungus was grown in CM. Compared with that in the wild type, the expression of EG6 was much lower in Δchk1 in both cellulose-containing medium and CM (Figure 2C). During growth in culture, EG6 expression appeared to reach a certain basal level without the addition of any obvious inductive factor. Similar behavior has been reported for three cellulase genes of T. reesei (Ilmen et al., 1997). This basal EG6 expression was strictly Chk1 dependent (Figure 2C). Strong upregulation by crystalline cellulose of CBH7, but not of EG6, implies that different combinations of signals are required for the induction of these two cellulase genes.
Monitoring of EG6 Expression in Planta
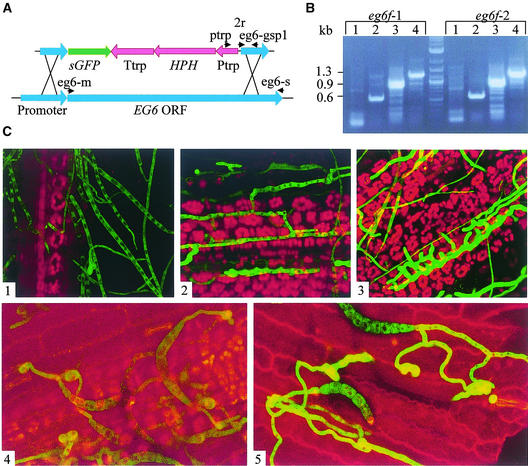
The dramatic increase in the expression of cellulases in planta allows them to be used as reporter genes for pathogenic development. Microscopic examination of the infection process in correlation with changes in reporter gene expression is very helpful in understanding host–pathogen interactions. To preserve the original regulation of EG6 for the green fluorescent protein (GFP) reporter gene, we designed a reporter/disruption construct according to the strategy of Basse et al. (2000). The EG6 coding region was substituted by the GFP coding sequence. The construct included the hygromycin B resistance gene as a selection marker (Figure 3A). EG6 was disrupted by homologous integration of the hygromycin resistance marker by double crossover. More than 20 transformants were obtained, and all of them were indistinguishable from the wild type with regard to growth and sporulation. eg6f transformants also were fully pathogenic, apparently as a result of redundancy. Two independently isolated mutants were chosen for microscopic examination in planta. Maize plants were inoculated evenly with homogenized mycelium of eg6f strains and examined by confocal microscopy. After 4 h of incubation, the mycelium was visible on the leaf surface but was only weakly fluorescent (Figure 3C1). After ∼12 h, fungal hyphae were observed in mesophyll tissue, but the fluorescence of these hyphae was increased only slightly (Figure 3C2). After 27 h of incubation, regions containing very intensely fluorescent mycelia (Figure 3C3) were detected. The appearance of these regions coincided with leaf necrosis. This observation suggests that the induction of EG6 occurs during invasive growth and not during growth on the surface.
Figure 3.
Expression of the EG6 Cellulase Gene.
(A) EG6 disruption/reporter construct. ptrp, 2r, eg6-m, eg6-s, and eg6-gsp1 indicate primers used for transformant verification. HPH, hygromycin phosphotransferase–encoding gene; ORF, open reading frame; Ptrp, constitutive promoter; sGFP, GFP-encoding gene; Ttrp, terminator.
(B) Verification of the EG6 transformants eg6f-1 and eg6f-2. PCR procedures used the following primers: EG6-M and EG6-S (lane 1); Ptrp and EG6-GSP1 (lane 2); 2R and EG6-S (lane 3); and Ptrp and EG6-S (lane 4).
(C) Confocal microscopy of a line expressing GFP under the control of the EG6 promoter. Panels 1 to 3 show micrographs from 4, 12, and 27 h, respectively, after inoculation of leaf with mycelial suspension; panels 4 and 5 show micrographs from 4 and 11 h, respectively, after inoculation of leaf with conidial suspension. Green indicates GFP fluorescence, and red indicates chlorophyll autofluorescence. Parameters for GFP fluorescence detection were as follows: iris, 2.6; gain, 1000.
To compare pathogenic development in the eg6f mutant and the wild type, we used a wild-type strain containing the GFP-encoding sequence under the regulation of the constitutive promoter of GPD. No developmental differences were detected between this control strain and the eg6f line. To follow EG6 promoter activity in germinating conidia, we inoculated maize plants with drops of conidial suspension. After 4 to 6 h of incubation, germ tubes formed appressoria, but no increase in fluorescence was detected (Figure 3C4). Between 11 and 18 h of incubation, regions of highly fluorescent hyphae appeared (Figure 3C5). When conidia were inoculated, the penetration process occurred much faster than when homogenized mycelia were inoculated. Upon spore germination, the fungus did not grow far on the plant surface: the germ tubes rapidly formed appressoria, and the infection peg breached the plant tissue. Faster penetration of hyphae emerging from germinating conidia suggests that the small appressoria of this species, although dispensable for infection, confer an advantage for rapid entry through the plant cuticle and epidermis. At the time of appressorium formation, the two cellulase-encoding genes were not yet upregulated. This finding indicates, again, that the induction of EG6 and CBH7 occurs during invasive growth rather than as an immediate response to leaf surface signals.
Influence of CHK1 Gene Disruption on Expression of the Reporter Gene
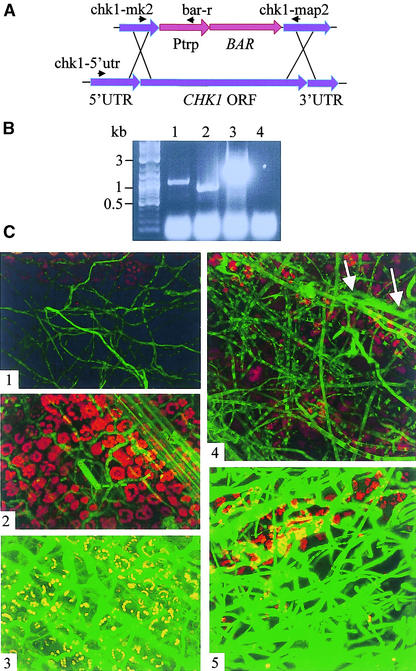
To study the influence of Chk1 on the pattern of EG6 gene expression, we deleted the CHK1 gene from the eg6f disruption/reporter strain. The disruption construct included the bialaphos resistance gene (BAR) as a selection marker under the control of the constitutive promoter Ptrp and two flanking regions homologous with CHK1 genomic sequence (Figure 4A). The resulting mutant was indistinguishable from previously isolated chk1 deletion mutants: the aging central region of colonies grown on solid medium developed an autolytic appearance, and the mutant lacked conidia and showed decreased virulence (Lev et al., 1999). Microscopic examination of eg6f and eg6f/Δchk1 mycelia grown in liquid culture revealed weaker fluorescence of eg6f/Δchk1 compared with that of eg6f. Maize plants were infected with homogenized mycelia of either wild-type or eg6f/Δchk1 reporter lines. After 11 h of incubation, an extensive hyphal network covered the leaf surface, and the growth patterns of the two strains were indistinguishable. Individual highly fluorescent hyphae were observed inside the plant tissue at 11 h after inoculation by eg6f but not by eg6f/Δchk1 (Figures 4C1 and 4C4). After 23 h of incubation of the eg6f-infected plants, the leaf tissue in the lesions was completely necrotic and interspersed with highly fluorescent hyphae (Figure 4C5), whereas eg6f/Δchk1-infected plants showed no disease symptoms. Hyphae of eg6f/Δchk1 were found inside the plant tissue at 23 h, and their fluorescence was increased slightly compared with the low level observed at 11 h (Figure 4C5). After 46 h of incubation, eg6f produced abundant highly fluorescent conidiophores and conidia on the leaf surface, whereas the fluorescence of the internal mycelial network decreased. Scattered necrotic lesions appeared on yellowing eg6f/Δchk1-infected leaves by 46 h, and the fluorescence of mycelia reached a level similar to that of eg6f at the 24-h stage (Figure 4C3).
Figure 4.
Expression of EG6 in the MAPK chk1 Deletion Strain.
(A) CHK1 disruption construct. chk1-5′utr, chk1-mk2, bar-r, and chk1-map2 indicate primers used for transformant verification. BAR, phosphinothricin acetyltransferase–encoding gene; ORF, open reading frame; Ptrp, constitutive promoter; UTR untranslated region.
(B) Verification of CHK1 disruption. Lanes 1 and 3, transformant DNA was used as a template; lanes 2 and 4, wild-type DNA was used as a template. In lanes 1 and 2, amplification was performed with primers CHK1-MK2 and CHK1-MAP2; in lanes 3 and 4, amplification was performed with primers CHK1-5′UTR and BARr.
(C) Influence of CHK1 disruption on EG6 promoter activity. Panels 1 to 3 show micrographs from the Δchk1 strain at 11, 23, and 46 h, respectively, after inoculation; panels 4 and 5 show micrographs from the parental eg6f strain at 11 and 23 h, respectively, after inoculation. White arrows indicate hyphae found inside the plant tissue. Parameters for GFP fluorescence detection were as follows: iris, 5.6; gain, 1100.
Expression of CBH7 during Plant Infection
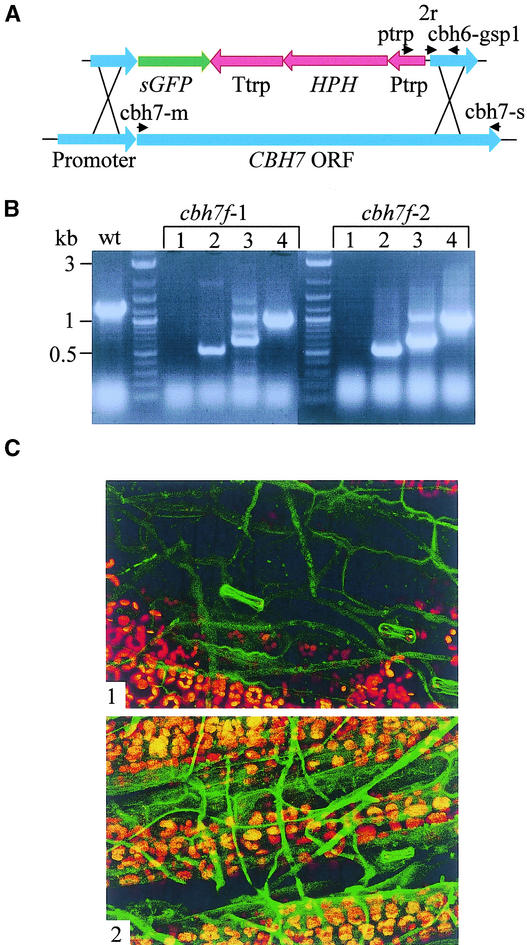
To follow the expression of CBH7 during plant infection, we used a disruption/reporter construct to substitute the CBH7 gene with the GFP coding sequence at the predicted translation initiation site (Figure 5A). The phenotype of cbh7f transformants was indistinguishable from that of the wild-type strain with respect to growth, conidiation, and virulence. Two of six transformants were used further for plant inoculation and confocal microscopy. We followed the expression of GFP driven by the endogenous CBH7 promoter during plant infection. No induction of GFP was detected during the first stages of infection, after inoculation with either conidia or mycelium. After ∼14 h of incubation, hyphae became slightly fluorescent. The fluorescence increased further during the next 10 h (Figures 5C1 and 5C2). Interestingly, no correlation was found between the location of hyphae and the fluorescence level: superficial hyphae covering still-living plant tissue at the late infection stages were fluorescent to the same degree as those found inside necrotic areas.
Figure 5.
Expression of the Cellobiohydrolase Gene CBH7.
(A) CBH7 disruption/reporter construct. ptrp, 2r, cbh7-m, cbh7-s, and cbh7-gsp1 indicate primers used for transformant verification. HPH, hygromycin phosphotransferase–encoding gene; ORF, open reading frame; Ptrp, constitutive promoter; sGFP, GFP-encoding gene; Ttrp, terminator.
(B) Verification of transformants cbh7f-1 and cbh7f-2. PCR procedures used the following primers: CBH7-M and CBH7-S (lane 1); Ptrp and CBH7-GSP1 (lane 2); 2R and CBH7-S (lane 3); and Ptrp and CBH7-S (lane 4). wt, wild type.
(C) Confocal microscopy of the cbh7f line expressing GFP under the control of the CBH7 promoter. Panels 1 and 2 show induction of CBH7 promoter activity at 14 and 24 h, respectively, after plant inoculation with homogenized mycelial suspension. Green indicates GFP fluorescence, and red indicates chlorophyll autofluorescence. Parameters for GFP fluorescence detection were as follows: iris, 4; gain, 1500.
DISCUSSION
Genetic loss-of-function experiments have shown that the MAPK Chk1 is required for sporulation, mating, hyphal integrity, and virulence. Using the SSH method, we isolated two cellulase genes, EG6 and CBH7, that are upregulated strongly in planta during advanced stages of infection. The synchronized induction of both cellulases suggests their synergistic action on the cellulose-based plant cell wall. Both cellulases were underexpressed in Δchk1 mutants in planta, and their average transcript levels reached 27 and 33% of wild-type levels for EG6 and CBH7, respectively.
Fungal mycelium grows on the leaf surface before individual hyphae successfully penetrate and establish invasive growth. As observed by confocal microscopy, this process is much slower in infection by the Δchk1 mutant than by the wild type. Moreover, Chk1-deficient mutants cause necrosis to a lesser extent than the wild type, even when inside the plant tissue (Figure 1). Observation of disease symptoms in concert with microscopic examination of infected plant tissue revealed that both EG6 and CBH7 are expressed mostly during advanced infection stages when necrotic lesions are formed: EG6 was upregulated chiefly in mycelia found inside necrotic areas, whereas CBH7 was induced in mycelia found inside and on the plant tissue, with less direct correlation to necrotic areas. Depending on how tightly cellulase induction is linked to plant cell necrosis, underexpression of both cellulases in the Δchk1 mutant could result from its reduced ability to cause necrotic lesions. However, we found evidence for a more direct correlation between cellulase expression and Chk1 activity: in saprophytic culture, the expression of CBH7 was induced strongly by crystalline cellulose, and this induction was delayed markedly in the Chk1-deficient mutant. During growth of the culture, induction of CBH7 in the mutant overcame the absence of Chk1, and its expression reached the wild-type level (Figure 2C). Expression of EG6 in culture, where there is no obvious inducer, depended strictly on Chk1. We speculate that Chk1 regulates the basal expression of EG6 and that plant signals (coupled to the necrotic stage) gradually overcome Chk1 deficiency, as found for CBH7 expression (Figure 2C). An independent way to prevent the necrotic stage (e.g., an incompatible plant-pathogen pair or additional mutations in the pathogen) would be needed to test the importance of necrotic tissue for the induction of cellulase genes.
MAPK-dependent regulation of EG6 in saprophytic culture (Figure 2C) implies either that there is a signal that activates the MAPK pathway under these conditions or that the pathway is “leaky,” transmitting a basal level of activation in the absence of any specific extracellular signal. Mammalian ERK1/2 MAPKs are able to self-catalyze autophosphorylation in vitro. Moreover, it was shown that another MAPK, p38α, autophosphorylates after interaction with the scaffolding protein TAB1 (Wu et al., 1991; Robbins et al., 1993; Ge et al., 2002; Johnson, 2002). Such an autophosphorylation mechanism, if present in fungi, could explain Chk1-dependent expression when there is no obvious extracellular signal. It also is plausible that signals are generated during saprophytic growth. Such signals could be provided, for example, by relief of catabolite repression, changes in the composition of the medium created by the growth of the culture, oxidative stress, or internal signals related to cell wall deposition.
Many fungal genes expressed specifically in plant–pathogen interactions have been identified, but relatively little is known about the regulation of these genes (Hahn and Mendgen, 1997; Perfect et al., 1999; Kahmann and Basse, 2001; Kim et al., 2001). In this study, the combination of a MAPK disruption mutant with SSH led to the isolation of MAPK-dependent genes expressed in planta. We found several distinct regulatory patterns: first, the Chk1 MAPK pathway was required for the expression of EG6 in culture. Second, the rapid induction of CBH7, and perhaps EG6 as well, was Chk1 dependent. Third, some combinations of signals were able to overcome the lack of Chk1, both on the plant and in culture. Elements to be found in the upstream regulatory regions of these and other fungal genes expressed in planta likely reflect these different regulatory patterns.
METHODS
Fungal Strains and Growth Media
Wild-type C4 and chk1 deletion strains of Cochliobolus heterostrophus, and standard growth conditions, have been described previously (Leach et al., 1982; Wirsel et al., 1996; Lev et al., 1999). For hygromycin B selection, standard complete medium (CM) was prepared without salts. For bialaphos (Duchefa, Haarlem, The Netherlands) resistance selection, CM was prepared without yeast extract and casein hydrolysate, to avoid providing amino acids. The final bialaphos concentration was 100 μg/mL. For induction by crystalline cellulose, fungal strains were grown in shake culture, at 230 rpm and 29 to 30°C in the light, on medium containing salts and trace elements (as in CM) with 1% maltose and 10% Sigmacell cellulose (Sigma) for 36, 52, and 93 h.
Nucleic Acid Isolation
RNA was isolated from mycelia and leaf tissue ground in liquid nitrogen by a modified phenol-SDS method (Ausubel et al., 1987) and purified by lithium acetate precipitation. Crude genomic DNA for PCR analysis was isolated from mycelium ground in liquid nitrogen as described for plant tissue (Edwards et al., 1991).
Plant Inoculation and Normalization of Fungal RNA Quantities
Sweet maize plants (Zea mays var Grand Jubilee) were grown for 15 to 16 days at 22 to 24°C. Wild-type and chk1 disrupted fungal strains were grown in liquid culture in CM with shaking for 3 days. Mycelium was homogenized briefly and, after the addition of 0.05% Tween 80, used for massive inoculation of plants. The plants then were incubated under continuous illumination in moist chambers that provided total containment of the pathogen. Infected leaves were collected for RNA isolation. A GPD gene fragment generated by PCR with primers 5′-CCCTCGCCTGACGCCCCCAT-3′ and 5′-CGAGGACACGGCGGGAGTAA-3′ was used to quantify fungal RNA. Total RNA of mutant and wild-type (with plant RNA added) infected leaves was the starting material for mRNA isolation using the polyATtract kit (Promega).
Construction and Verification of the Subtracted Library
A suppression subtractive library was prepared according to the manufacturer's instructions (PCR-Select cDNA Subtraction Kit; Clontech, Palo Alto, CA). Clones representing transcripts expressed differentially in wild-type and mutant strains were ligated into vector pUC-57 using the InsT/Aclone PCR Product Cloning Kit (MBI Fermentas, Vilnius, Lithuania), transformed to Escherichia coli XL-1 Blue (Stratagene), and amplified directly from bacterial colonies using primers 1 and 2R from the subtraction kit (Clontech). PCR products were denatured by heating at 95°C for 5 min and spotted onto 6× SSC–pretreated (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and dried Nytran SuPerCharge nylon membranes (Schleicher & Schuell). Dot blots were probed with radioactively labeled first-strand cDNA generated by the following method: 0.5 μg of mRNA with 0.5 μg of oligo(dT) primer were incubated at 70°C for 5 min and iced, and 2 μL of reaction buffer, 20 units of RNase inhibitor, 1 μL of 10 mM deoxynucleotide triphosphates, 47 μCi of α-32P-dCTP, and 40 units of reverse transcriptase from Moloney murine leukemia virus were added to a total volume of 20 μL (enzyme and reagents were from Stratagene). After 1 h at 37°C, the reaction was stopped with 5 μL of 0.5 M EDTA and 25 μL of 0.6 N NaOH, and the RNA was hydrolyzed for 30 min at 70°C. The labeled probe was purified on a Sephadex G-100 column in 10 mM Tris·Cl, 1 mM EDTA, pH 8.0. The membranes were hybridized with cDNA pools representing tester populations (wild-type-infected leaves with uninfected leaf mRNA added), driver populations (mutant-infected leaves), wild-type-infected leaves, and wild-type and mutant grown in liquid culture.
Identification of Entire Open Reading Frames and Partial Promoter Sequences of Selected Clones
Clones EG6 and CBH7 derived from the suppression subtractive hybridization were used as probes to screen a cDNA library. Predicted translation initiation sites were identified by PCR screening of multiple positive plaques using sense vector primer and antisense clone-specific primer. PCR genome walking (Universal Genome Walker Kit; Clontech) was used to obtain sequences upstream of the coding regions. The following antisense primers were designed: EG6-GSP1, 5′-CAAGTAGACCAT-CTCGGGACCAGGGTG-3′; EG6-GSP2, 5′-CGGCAGAGGAGAAAGCACCCTGGTTGC-3′; CBH7-GSP1, 5′-ATGGATGGGCAGGAGGAG-ACGCGGAGA-3′; and CBH7-GSP2, 5′-ACGGGGAGAGCCAGCAGCCTTGTAGAC-3′. Genome walking library construction and PCR were performed as recommended by the supplier.
Gene Replacement Constructs
The EG6 disruption/reporter vector was constructed from the following DNA fragments, in order: a 621-bp EG6 promoter fragment, the sGFP coding sequence, the hygromycin B resistance cassette including the constitutive Ptrp promoter, the hygromycin phosphotransferase–encoding gene, and terminator Ttrp, followed by a 482-bp fragment of the EG6 coding sequence. All fragments were assembled in pBluescript SK− (Stratagene) (Figure 3A). The promoter fragment was generated as follows: the genome-walking product (see above) was used as a template to amplify a region of the EG6 promoter ending at the first Met, with the AP2 primer from the Genome Walker Kit and the gene-specific antisense primer EG6-HindIII (5′-gggcaagcTTCTGACTAGCAGTAACGATT-3′), which is homologous (uppercase letters) with the sequence upstream of the start codon, with the addition of a HindIII restriction site. The PCR product was digested with appropriate restriction enzymes and used for ligation.
A CHK1 disruption construct was created by ligation of the 823-bp left flanking region, including part of the CHK1 5′ untranslated region and part of the coding sequence, the phosphinothricin acetyltransferase–encoding gene (BAR) under the regulation of the Ptrp promoter, a 656-bp right flanking region corresponding to part of the CHK1 open reading frame, and part of the 3′ untranslated region, into the pBluescript cloning vector (Figure 4A).
The CBH7 disruption/reporter construct included a 420-bp CBH7 promoter fragment, the sGFP coding sequence, the hygromycin B resistance cassette including the constitutive Ptrp promoter, the hygromycin phosphotransferase–encoding gene, and terminator Ttrp followed by a 456-bp fragment of the CBH7 coding sequence (Figure 5A). The promoter fragment was generated using the same strategy used for the EG6 disruption construct with primer CBH7-HindIII (5′-gggcaagCTTTGAGA-ACCAAGAAGGAA-3′).
DNA for transformation was prepared by PCR amplification of each complete disruption cassette with standard primers T3 and T7 using high-fidelity BIO-X-ACT polymerase (Bioline, Toronto, Canada). Transformation into fungal protoplasts was performed as described previously (Turgeon et al., 1987, 1993).
Verification of Transformants
eg6f transformants were verified by four PCR primer sets: first, EG6-M (5′-CAGAATGAAGTTCTCCGCTGCT-3′) and EG6-S (5′-CCGCTTACT-GGCACTGAGAGT-3′), which is expected to give no product if disruption occurred because the EG6-M sequence should be absent; second, Ptrp (5′-GGTCGTTCACTTACCTTGCTTG-3′) and EG6-GSP1, both sequences present in the disruption vector; third, 2R (Genome Walker Kit primer) and EG6-S (5′-CCGCTTACTGGCACTGAGAGT-3′). The 2R sequence was located in the disruption vector, whereas the EG6-S sequence was found outside of the region used for vector preparation. Primers Ptrp and EG6-S were used in a fourth PCR procedure (Figures 3A and 3B).
To verify the disruption of the CHK1 gene in the eg6f/Δchk1 strain, we performed PCR with primers CHK1-5′UTR (5′-GAGACGCGGGCAGTCCCAGC-3′) and BARr (5′-GGTACCGGCAGGCTGAAGTC-3′) through the junction representing the insertion of the disruption construct by homologous recombination. PCR with primers CHK1-MK2 (5′-CTGCTG-AGCTCCTTCAACCACG-3′) and CHK1-MAP2 (5′-CTGTTCCCGGGTCAGGTTGTCC-3′) was designed to prove the absence of the wild-type CHK1 copy (Figures 4A and 4B).
cbh7f transformants were verified with the following PCR primers: CBH7-M (5′-CAAAGATGCTCTCCAACGTCTT-3′) and CBH7-S (5′-GCTTATTCAAAAGAACCAGACAC-3′); Ptrp and CBH7-GSP1 (5′-ATGGAT-GGGCAGGAGGAGACGCGGAGA-3′); 2R and CBH7-S; and Ptrp and CBH7-S (Figures 5A and 5B).
Confocal Microscopy
Infected leaves were visualized with a MRC-1024 laser confocal scanning microscope (Bio-Rad, Hempstead, UK) with the objective Nikon Plan Fluor ×40/1.30 (Tokyo, Japan). For image acquisition, the “triple-labeling” method was selected. GFP fluorescence was detected with the fluorescein isothiocyanate–optimized channel, and the autofluorescence of the leaf, contributed primarily by chlorophyll, was detected with the Cy5-optimized channel. Image processing was performed with Confocal Assistant 4.02 software (Bio-Rad, Hempstead, UK).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The accession numbers for the sequences mentioned in this article are AY116306 (EG6), AY116307 (CBH7), Y11113 (endoglucanase IV of Trichoderma reesei), BAA74458 (cellobiohydrolase II of Acremonium cellulolyticus Y-94), and AF081380 (GPD).
Acknowledgments
We thank Ira Kolotuev for expert assistance with confocal microscopy and Ofir Degani for valuable advice on cellulolytic enzymes. We are grateful to Amir Sharon for strains, plasmids, and comments on the manuscript, to Eliezer Lifschitz, Hong Ma, and Ramesh Raina for helpful discussions, and to Benjamin Podbilewicz for critical reading of the manuscript. This work was supported by grant ISF 233002 from the Israel Academy of Sciences.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010546.
References
- Ahn, J.H., Sposato, P., Kim, S.I., and Walton, J.D. (2001). Molecular cloning and characterization of cel2 from the fungus Cochliobolus carbonum. Biosci. Biotechnol. Biochem. 65, 1406–1411. [DOI] [PubMed] [Google Scholar]
- Apel-Birkhold, P.C., and Walton, J.D. (1996). Cloning, disruption, and expression of two endo-β1,4-xylanase genes, XYL2 and XYL3, from Cochliobolus carbonum. Appl. Environ. Microbiol. 62, 4129–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kinston, R.E., Moore, D.D., Smith, J.A., Seidman, J.G., and Struhl, K. (1987). Current Protocols in Molecular Biology. (New York: Wiley-Interscience).
- Basse, C.W., Stumpferl, S., and Kahmann, R. (2000). Characterization of a Ustilago maydis gene specifically induced during the biotrophic phase: Evidence for negative as well as positive regulation. Mol. Cell. Biol. 20, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank, C., Schroppel, K., Leberer, E., Harcus, D., Mohamed, O., Meloche, S., Thomas, D.Y., and Whiteway, M. (1998). Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 66, 2713–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro, A., Garcia-MacEira, F.I., Meglecz, E., and Roncero, M.I. (2001). A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39, 1140–1152. [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, B., Gram, H., Di Padova, F., Huang, B., New, L., Ulevitch, R.J., Luo, Y., and Han, J. (2002). MAPKK-independent activation of p38α mediated by TAB1-dependent autophosphorylation of p38α. Science 295, 1291–1294. [DOI] [PubMed] [Google Scholar]
- Hahn, M., and Mendgen, K. (1997). Characterization of in planta-induced rust genes isolated from a haustorium-specific cDNA library. Mol. Plant-Microbe Interact. 10, 427–437. [DOI] [PubMed] [Google Scholar]
- Hou, Z., Xue, C., Peng, Y., Katan, T., Kistler, H.C., and Xu, J.R. (2002). A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant-Microbe Interact. 15, 1119–1127. [DOI] [PubMed] [Google Scholar]
- Ichinose, Y., Toyoda, K., and Barz, W. (1999). cDNA cloning and gene expression of three small GTP-binding proteins in defense response of chickpea. Biochim. Biophys. Acta 1489, 462–466. [DOI] [PubMed] [Google Scholar]
- Ilmen, M., Saloheimo, A., Onnela, M.L., and Penttila, M.E. (1997). Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63, 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, G. (2002). SIGNAL TRANSDUCTION: Scaffolding proteins—More than meets the eye. Science 295, 1249–1250. [DOI] [PubMed] [Google Scholar]
- Kahmann, R., and Basse, C. (2001). Fungal gene expression during pathogenesis-related development and host plant colonization. Curr. Opin. Microbiol. 4, 374–380. [DOI] [PubMed] [Google Scholar]
- Kim, S., Ahn, I.P., and Lee, Y.H. (2001). Analysis of genes expressed during rice-Magnaporthe grisea interactions. Mol. Plant-Microbe Interact. 14, 1340–1346. [DOI] [PubMed] [Google Scholar]
- Leach, J., Lang, B., and Yoder, O. (1982). Methods for selection of mutants and in vitro culture of Cochliobolus heterostrophus. J. Gen. Microbiol. 128, 1719–1729. [Google Scholar]
- Lev, S., Sharon, A., Hadar, R., Ma, H., and Horwitz, B.A. (1999). A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation, and pathogenicity: Diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl. Acad. Sci. USA 96, 13542–13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani, H.D., Galitski, T., Lander, E.S., and Fink, G.R. (1999). Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. USA 96, 12530–12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey, G., Held, K., Scheffer, J., Tenberge, K.B., and Tudzynski, P. (2002). CPMK2, an SLT2-homologous mitogen-activated protein (MAP) kinase, is essential for pathogenesis of Claviceps purpurea on rye: Evidence for a second conserved pathogenesis-related MAP kinase cascade in phytopathogenic fungi. Mol. Microbiol. 46, 305–318. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Perfect, S.E., Hughes, H.B., O'Connell, R.J., and Green, J.R. (1999). Colletotrichum: A model genus for studies on pathology and fungal-plant interactions. Fungal Genet. Biol. 27, 186–198. [DOI] [PubMed] [Google Scholar]
- Robbins, D.J., Zhen, E., Owaki, H., Vanderbilt, C.A., Ebert, D., Geppert, T.D., and Cobb, M.H. (1993). Regulation and properties of extracellular signal-regulated protein kinases 1 and 2 in vitro. J. Biol. Chem. 268, 5097–5106. [PubMed] [Google Scholar]
- Roberts, C.J., et al. (2000). Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287, 873–880. [DOI] [PubMed] [Google Scholar]
- Schäfer, W. (1993). The role of cutinase in fungal pathogenicity. Trends Microbiol. 1, 69–71. [DOI] [PubMed] [Google Scholar]
- Scott-Craig, J.S., Cheng, Y.Q., Cervone, F., De Lorenzo, G., Pitkin, J.W., and Walton, J.D. (1998). Targeted mutants of Cochliobolus carbonum lacking the two major extracellular polygalacturonases. Appl. Environ. Microbiol. 64, 1497–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Craig, J.S., Panaccione, D.G., Cervone, F., and Walton, J.D. (1990). Endopolygalacturonase is not required for pathogenicity of Cochliobolus carbonum on maize. Plant Cell 2, 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sposato, P., Ahn, J.H., and Walton, J.D. (1995). Characterization and disruption of a gene in the maize pathogen Cochliobolus carbonum encoding a cellulase lacking a cellulose binding domain and hinge region. Mol. Plant-Microbe Interact. 8, 602–609. [DOI] [PubMed] [Google Scholar]
- Takano, Y., Kikuchi, T., Kubo, Y., Hamer, J.E., Mise, K., and Furusawa, I. (2000). The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant-Microbe Interact. 13, 374–383. [DOI] [PubMed] [Google Scholar]
- Tonukari, N.J., Scott-Craig, J.S., and Walton, J.D. (2000). The Cochliobolus carbonum SNF1 gene is required for cell wall–degrading enzyme expression and virulence on maize. Plant Cell 12, 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon, B.G., Bohlmann, H., Ciuffetti, L.M., Christiansen, S.K., Yang, G., Schäfer, W., and Yoder, O.C. (1993). Cloning and analysis of the mating type genes from Cochliobolus heterostrophus. Mol. Gen. Genet. 238, 270–284. [DOI] [PubMed] [Google Scholar]
- Turgeon, B.G., Garber, R.C., and Yoder, O.C. (1987). Development of a fungal transformation system based on selection of sequences with promoter activity. Mol. Cell. Biol. 7, 3297–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., and Heitman, J. (1999). Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 2, 358–362. [DOI] [PubMed] [Google Scholar]
- Wirsel, S., Turgeon, B.G., and Yoder, O.C. (1996). Deletion of the Cochliobolus heterostrophus mating-type (MAT) locus promotes the function of MAT transgenes. Curr. Genet. 29, 241–249. [PubMed] [Google Scholar]
- Wu, J., Rossomando, A.J., Her, J.H., Del Vecchio, R., Weber, M.J., and Sturgill, T.W. (1991). Autophosphorylation in vitro of recombinant 42-kilodalton mitogen-activated protein kinase on tyrosine. Proc. Natl. Acad. Sci. USA 88, 9508–9512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J.R. (2000). Map kinases in fungal pathogens. Fungal Genet. Biol. 31, 137–152. [DOI] [PubMed] [Google Scholar]
- Xu, J.R., and Hamer, J.E. (1996). MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10, 2696–2706. [DOI] [PubMed] [Google Scholar]
- Xu, J.R., Staiger, C.J., and Hamer, J.E. (1998). Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. USA 95, 12713–12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Lamm, R., Pillonel, C., Lam, S., and Xu, J.R. (2002). Osmoregulation and fungicide resistance: The Neurospora crassa os-2 gene encodes a HOG1 mitogen-activated protein kinase homologue. Appl. Environ. Microbiol. 68, 532–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L., Campbell, M., Murphy, J., Lam, S., and Xu, J.R. (2000). The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol. Plant-Microbe Interact. 13, 724–732. [DOI] [PubMed] [Google Scholar]