Abstract
To elucidate the relative significance of Lys synthesis and catabolism in determining Lys level in plant seeds, we expressed a bacterial feedback-insensitive dihydrodipicolinate synthase (DHPS) in a seed-specific manner in wild-type Arabidopsis as well as in an Arabidopsis knockout mutant in the Lys catabolism pathway. Transgenic plants expressing the bacterial DHPS, or the knockout mutant, contained ∼12-fold or ∼5-fold higher levels, respectively, of seed free Lys than wild-type plants. However, the combination of these two traits caused a synergistic ∼80-fold increase in seed free Lys level. The dramatic increase in free Lys in the knockout mutant expressing the bacterial DHPS was associated with a significant reduction in the levels of Glu and Asp but also with an unexpected increase in the levels of Gln and Asn. This finding suggested a special regulatory interaction between Lys metabolism and amide amino acid metabolism in seeds. Notably, the level of free Met, which competes with Lys for Asp and Glu as precursors, was increased unexpectedly by up to ∼38-fold in the various transgenic and knockout plants. Together, our results show that Lys catabolism plays a major regulatory role in Lys accumulation in Arabidopsis seeds and reveal novel regulatory networks of seed amino acid metabolism.
INTRODUCTION
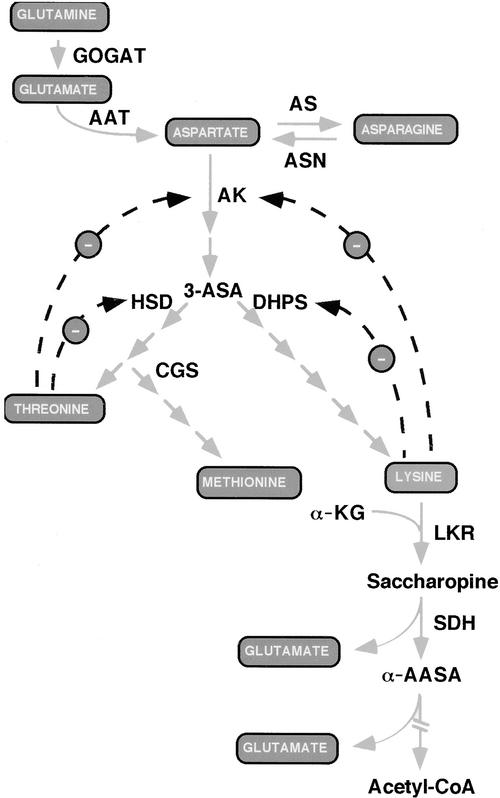
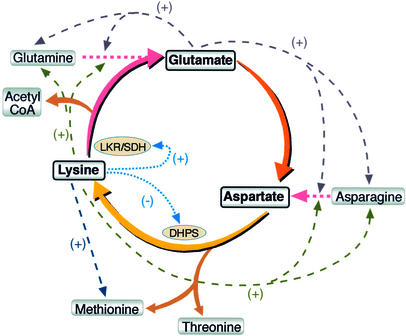
Lys, Met, and Thr are three essential amino acids that are present in limiting amounts in seeds of many crop plants; therefore, they are considered of great nutritional importance in animal feeds and human foods (Galili et al., 1994). These three essential amino acids are synthesized in plants from Asp via several different branches of the Asp family pathway (Figure 1). Asp is synthesized either from Gln via Glu or from Asn (Lam et al., 1995; Galili, 2002), thus linking the Asp family pathway to amide amino acid metabolism (Figure 1).
Figure 1.
Scheme of Amide Amino Acid Metabolism (Top), the Asp Family Pathway (Center), and the Pathway of Lys Catabolism (Bottom).
Dashed arrows with minus signs represent end-product feedback inhibition loops. α-AASA, α-amino adipic semialdehyde; AAT, Asp aminotransferase; AS, Asn synthase; 3-ASA, 3-aspartic semialdehyde; ASN, asparaginase; AK, Asp kinase; CGS; cystathionine γ-synthase; DHPS, dihydrodipicolinate synthase; GOGAT, Glu synthase; HSD, homoserine dehydrogenase; α-KG, α-ketoglutarate; LKR, Lys-ketoglutarate reductase; SDH, saccharopine dehydrogenase. The broken arrow at bottom indicates several enzymatic steps leading to the production of acetyl-CoA.
The Asp family pathway in plants is regulated by several feedback inhibition loops. Asp kinase, the first enzyme in this pathway, is feedback inhibited by Lys and Thr. In addition, Lys also feedback inhibits the enzyme dihydrodipicolinate synthase (DHPS), whereas Thr also feedback inhibits the enzyme homoserine dehydrogenase (Figure 1) (Galili, 1995). Genetic and molecular studies demonstrated that the sensitivity of DHPS to Lys inhibition represents the major rate-limiting step of Lys biosynthesis in vivo (Negrutiu et al., 1984; Galili, 1995). Accordingly, seed-specific expression of feedback-insensitive bacterial DHPS enzymes in various plant species resulted in significant seed Lys overproduction (Karchi et al., 1994; Falco et al., 1995; Mazur et al., 1999). However, this enhanced Lys production was associated with increased activity of the Lys catabolic enzyme Lys-ketoglutarate reductase (LKR) and with enhanced levels of Lys catabolic products (Figure 1) (Karchi et al., 1994, 1995; Falco et al., 1995; Mazur et al., 1999).
To evaluate the regulatory significance of Lys catabolism for seed Lys accumulation, we produced transgenic Arabidopsis plants expressing a bacterial DHPS in a seed-specific manner but lacking Lys catabolism as a result of a T-DNA knockout mutation in the AtLKR/SDH gene (Zhu et al., 2001). This gene encodes the first two enzymes in the Lys catabolism pathway, LKR and saccharopine dehydrogenase (SDH), which are linked on a single bifunctional polypeptide (Zhu et al., 2001). Here, we show that increased Lys biosynthesis coupled with knockout of its catabolism synergistically increases Lys content in mature seeds at the expense of its precursor amino acids Glu and Asp. Moreover, our data also suggest that changes in the flux of Lys metabolism transregulate other pathways of seed amino acid metabolism.
RESULTS
Generation of Transgenic LKR/SDH Knockout Arabidopsis Mutants Expressing a Bacterial DHPS in a Seed-Specific Manner
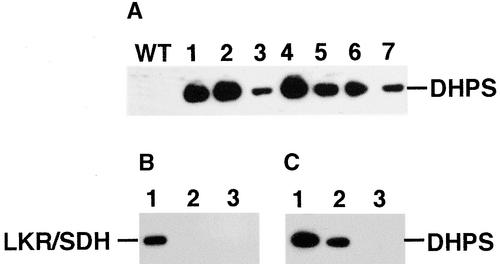
The strategy chosen to generate a transgenic Arabidopsis LKR/SDH knockout mutant expressing a bacterial DHPS in a seed-specific manner is described in detail in Methods. In short, this procedure included the following steps: (1) transformation of wild-type Arabidopsis with a previously described chimeric gene encoding an Escherichia coli DHPS fused to a promoter derived from the bean phaseolin storage protein gene (Karchi et al., 1994); (2) selection for transgenic plants expressing the bacterial DHPS in a seed-specific manner using protein gel blot analysis with anti-bacterial DHPS antibodies (Figure 2A); (3) characterization of independently transformed homozygous lines to ensure that the position of insertion of the transgene in the genome exerts no negative pleiotropic effects besides altering the Lys biosynthesis pathway (data not shown); (4) crossing a transgenic line expressing a high level of the bacterial DHPS (Figure 2A, lane 2) with the LKR/SDH knockout mutant; and (5) screening the progeny containing both the phaseolin-DHPS gene and a homozygous LKR/SDH knockout using PCR and protein gel blot analysis. Figures 2B and 2C depict the desired genotype (hereafter termed LKR/SDH knockout × phaseolin-DHPS) and its two parents using protein gel blots with anti-Arabidopsis LKR and anti-bacterial DHPS antibodies. The parental line expressing the bacterial DHPS contains both an LKR/SDH polypeptide and a bacterial DHPS protein (lanes 1 in Figures 2B and 2C, respectively). The parental LKR/SDH knockout mutant lacks both the LKR/SDH and bacterial DHPS polypeptides (lanes 3 in Figures 2B and 2C, respectively). The selected progeny lack the LKR/SDH polypeptide but contain a bacterial DHPS protein (lanes 2 in Figures 2B and 2C, respectively).
Figure 2.
Generation of Transgenic Plants Expressing a Bacterial DHPS in a Homozygous LKR/SDH Knockout Background.
(A) Arabidopsis plants were transformed with the phaseolin-DHPS construct, and expression of the bacterial DHPS in seeds was tested by protein gel blot analysis using antibodies raised against the bacterial DHPS (Shaul and Galili, 1992). Lanes 1 to 7 represent independently transformed lines. WT, wild type.
(B) and (C) Confirmation of the LKR/SDH knockout × phaseolin-DHPS plant was performed by protein gel blot analysis with anti-LKR (B) and anti-bacterial DHPS (C) antibodies. Lanes 1, 2, and 3 represent proteins extracted from a transgenic plant expressing the bacterial DHPS alone, the LKR/SDH knockout × phaseolin-DHPS candidate, and a homozygous LKR/SDH knockout mutant, respectively.
Increased Lys Synthesis and Reduced Lys Catabolism Synergistically Increase the Lys Content of Mature Seeds
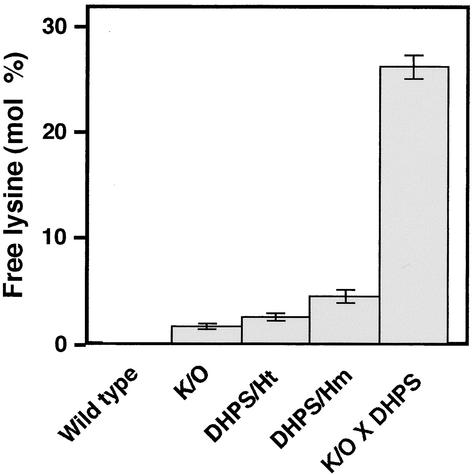
Next, we tested the relative contributions of Lys synthesis and catabolism to the level of free Lys accumulation in seeds. To this end, the LKR/SDH knockout × phaseolin-DHPS line as well as its parents containing either the LKR/SDH knockout mutation or the bacterial DHPS alone (5 to 20 individual plants per genotype) were grown to maturity and seeds were analyzed for their levels of amino acids. The total free amino acid levels per gram of mature seeds were comparable among all genotypes (data not shown), suggesting no significant effects of the bacterial DHPS or of the LKR/SDH knockout on total seed free amino acid levels. As shown in Figure 3 and Table 1, the relative free Lys levels in seeds of the LKR/SDH knockout plants and in transgenic plants heterozygous and homozygous for the phaseolin-DHPS were higher by ∼5-fold, ∼8-fold, and ∼14-fold, respectively, than those of wild-type plants. Notably, the relative free Lys level in seeds of the LKR/SDH knockout × phaseolin-DHPS plants synergistically increased ∼80-fold com-pared with that in wild-type plants (Figure 3), making Lys the most abundant free amino acid in the mature seeds (Table 1).
Figure 3.
Relative Free Lys Levels in Wild-Type and Different Transgenic Arabidopsis Plants.
Relative levels of free Lys are given as mol % of total amino acids. DHPS/Ht and DHPS/Hm, transgenic plants expressing the phaseolin-DHPS construct in heterozygous and homozygous states, respectively; K/O, LKR/SDH knockout mutants; K/O × DHPS, LKR/SDH knockout × phaseolin-DHPS plants. Error bars represent standard errors.
Table 1.
Relative Levels (mol %) of Free Amino Acids in Mature Seeds of Different Arabidopsis Genotypes
| Amino Acid | Wassilewskija | K/O | DHPS/Ht | DHPS/Hm | K/O × DHPS |
|---|---|---|---|---|---|
| Asp | 7.82 ± 0.45 | 6.59 ± 0.35 | 7.74 ± 0.46 | 6.33 ± 0.62 | 1.31 ± 0.04 |
| Glu | 39.75 ± 1.15 | 33.74 ± 0.58 | 35.00 ± 1.95 | 34.04 ± 1.14 | 10.65 ± 0.32 |
| Asn | 14.77 ± 1.04 | 19.61 ± 0.40 | 16.04 ± 1.04 | 14.69 ± 0.34 | 21.10 ± 0.49 |
| Ser | 9.36 ± 0.10 | 9.78 ± 0.41 | 7.56 ± 0.55 | 6.16 ± 0.27 | 4.52 ± 0.08 |
| Gln | 1.26 ± 0.09 | 1.46 ± 0.13 | 2.69 ± 0.43 | 2.75 ± 0.33 | 4.91 ± 0.60 |
| Gly | 2.07 ± 0.11 | 2.03 ± 0.05 | 2.04 ± 0.12 | 1.95 ± 0.16 | 2.02 ± 0.13 |
| His | 0.39 ± 0.04 | 0.36 ± 0.01 | 0.38 ± 0.10 | 0.37 ± 0.07 | 0.88 ± 0.07 |
| Arg | 0.58 ± 0.10 | 0.73 ± 0.08 | 1.24 ± 0.41 | 0.59 ± 0.06 | 0.37 ± 0.06 |
| Thr | 3.51 ± 0.16 | 3.29 ± 0.08 | 2.65 ± 0.12 | 2.34 ± 0.20 | 0.73 ± 0.02 |
| Ala | 9.53 ± 0.26 | 10.08 ± 0.33 | 7.38 ± 0.63 | 6.67 ± 0.23 | 3.91 ± 0.20 |
| Pro | 2.92 ± 0.13 | 2.93 ± 0.16 | 3.73 ± 0.85 | 3.05 ± 0.24 | 3.00 ± 0.44 |
| Tyr | 0.46 ± 0.08 | 0.50 ± 0.03 | 0.36 ± 0.07 | 0.31 ± 0.04 | 0.18 ± 0.02 |
| Val | 4.99 ± 0.28 | 5.34 ± 0.24 | 4.47 ± 0.24 | 4.24 ± 0.13 | 3.08 ± 0.14 |
| Met | 0.40 ± 0.02 | 0.51 ± 0.03 | 4.44 ± 0.75 | 12.80 ± 2.04 | 15.72 ± 0.68 |
| Ile | 1.24 ± 0.07 | 1.32 ± 0.18 | 1.17 ± 0.06 | 1.11 ± 0.05 | 0.97 ± 0.05 |
| Leu | 0.61 ± 0.05 | 0.77 ± 0.04 | 0.58 ± 0.04 | 0.52 ± 0.05 | 0.51 ± 0.04 |
| Lys | 0.33 ± 0.02 | 1.63 ± 0.20 | 2.55 ± 0.44 | 4.48 ± 0.82 | 26.38 ± 0.81 |
The genotypes used are as follows: Wassilewskija, control nontransformed wild type; K/O, LKR/SDH knockout; DHPS/Ht and DHPS/Hm, transgenic plants expressing the phaseolin-DHPS construct in the heterozygous and homozygous states, respectively; K/O × DHPS, LKR/SDH knockout × phaseolin-DHPS. Values represent means of at least five repeats plus or minus standard errors.
We also determined whether the increased free Lys may have contributed to the total seed Lys content. Because most of the Lys in plant seeds is associated with water-soluble albumin proteins (Habben and Larkins, 1994), we measured the relative Lys content (after hydrolysis) in Tris-HCl extracts, which contain both the albumins and free amino acids. The relative Lys content in this extract was ∼4.3-fold higher in the LKR/SDH knockout × phaseolin-DHPS plants than in wild-type plants (data not shown).
The Dramatic Increase in Seed Free Lys in LKR/SDH Knockout × Phaseolin-DHPS Plants Is Associated with Altered Amide Amino Acid Metabolism
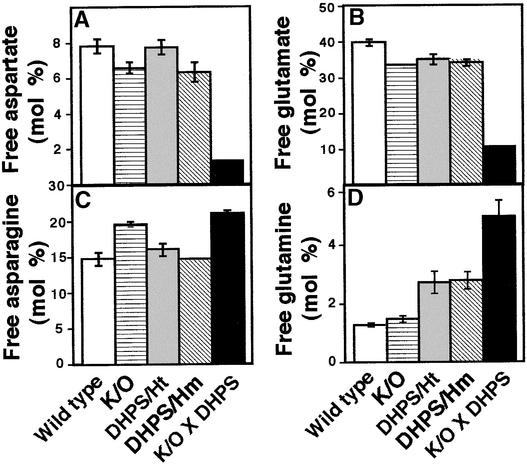
Lys is synthesized from Asp via one of the branches of the Asp family pathway (Figure 1) (Galili, 1995). Asp may be produced in plant seeds either from Glu (which is produced largely from Gln) via Asp aminotransferase or from Asn via asparaginase (Figure 1) (Lam et al., 1995). However, Glu also is a major product of the Lys catabolism pathway (Figure 1) (Galili et al., 2001). Therefore, we sought to determine how the specific changes in Lys metabolism in the various transgenic plants affected the relative levels of Glu, Asp, Gln, and Asn. As shown in Figures 4A and 4B, the relative levels of free Asp and Glu were comparable in the LKR/SDH knockout mutant, the transgenic plants expressing only the bacterial DHPS, and the wild-type plants. However, in the LKR/SDH knockout × phaseolin-DHPS plants, the relative levels of these two free amino acids were reduced substantially, revealing the metabolic significance of the concerted regulation of Lys synthesis and catabolism. The relative level of free Asn was significantly higher in seeds of the LKR/SDH knockout and the LKR/SDH knockout × phaseolin-DHPS plants than in wild-type plants and plants expressing the bacterial DHPS alone (P < 0.01) (Figure 4C). The relative level of free Gln was lowest in the wild-type and LKR/SDH knockout plants, was increased in transgenic plants expressing the bacterial DHPS alone, and was substantially greater in LKR/SDH knockout × phaseolin-DHPS plants (Figure 4D).
Figure 4.
Relative Levels of Free Asp, Glu, Asn, and Gln in Wild-Type and Different Transgenic Arabidopsis Plants.
Relative levels of free Asp (A), Glu (B), Asn (C), and Gln (D) are given as mol % of total amino acids. DHPS/Ht and DHPS/Hm, transgenic plants expressing the phaseolin-DHPS construct in heterozygous and homozygous states, respectively; K/O, LKR/SDH knockout mutants; K/O × DHPS, LKR/SDH knockout × phaseolin-DHPS plants. Error bars represent standard errors.
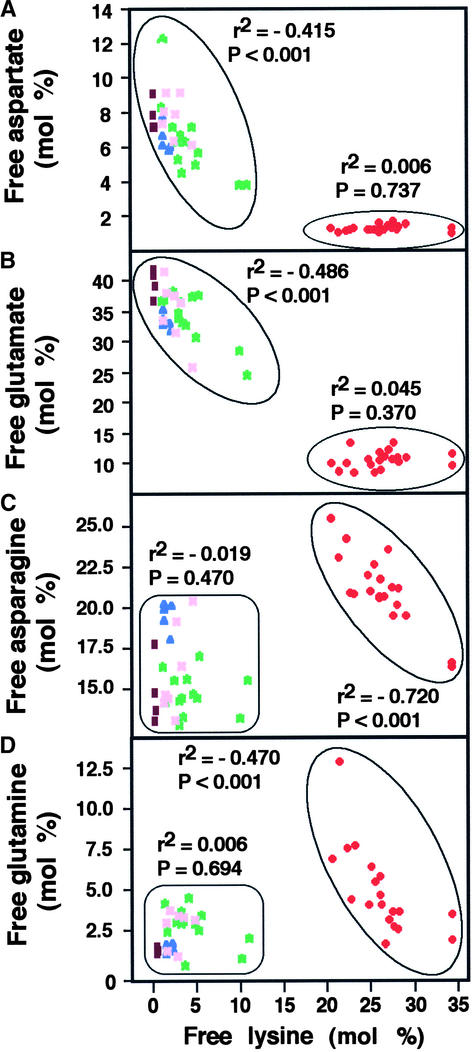
Next, we further dissected the metabolic consequences of the major changes in free Lys as well as Asp, Glu, Asn, and Gln in the LKR/SDH knockout × phaseolin-DHPS plants relative to their parents and the wild-type plants. To this end, we analyzed the correlation between Lys and each of the aforementioned amino acids in each plant individually. We first compared the means of the five genotypes for free Lys (Figure 3) using one-way analysis of variance followed by Fisher's LSD at α = 0.01. The results showed that not all means were equal (P < 0.001), and multiple comparison analysis demonstrated that the mean value of LKR/SDH knockout × phaseolin-DHPS plants was significantly higher than that of the other four genotypes, which, in turn, did not vary significantly among themselves. Thus, we divided LKR/SDH knockout × phaseolin-DHPS plants and the other four genotypes into two separate clusters, hereafter termed group 1 for the combined four genotypes and group 2 for the LKR/SDH knockout × phaseolin-DHPS plants. Free Asp and Glu levels were negatively correlated with free Lys level in group 1, whereas no correlation between these amino acids and Lys was evident in group 2, in which free Asp and Glu levels were the lowest (Figures 5A and 5B). The negative correlation between Glu or Asp and Lys in group 1 was not surprising because Glu and Asp are precursors of Lys. The lack of correlation in group 2 could be attributable to the extensive conversion of Glu and Asp to Lys in the LKR/SDH knockout × phaseolin-DHPS plants. This process significantly reduced the steady state concentrations of Glu and Asp, rendering them rate-limiting precursors for Lys production.
Figure 5.
Correlation between Free Lys and Free Asp, Glu, Asn, or Gln in Wild-Type and Different Transgenic Arabidopsis Plants.
Pearson's correlations between free Lys and each of the other four free amino acids indicated above are given in (A) to (D), respectively. The two distinct populations, designated group 1 and group 2, are circled at left and right, respectively. Numerals near each population represent r2 values of linear regressions followed by significance levels (P). Colored spots represent the following genotypes: brown, wild type; blue, LKR/SDH knockout; pink and green, transgenic plants expressing the phaseolin-DHPS construct in heterozygous and homozygous states, respectively; red, LKR/SDH knockout × phaseolin-DHPS.
The correlation between the levels of free Lys and either free Asn or Gln was clearly distinct from that between free Lys and either free Asp or Glu. In group 1, there was no correlation between free Asn or Gln and free Lys (Figures 5C and 5D). Apparently, the levels of Glu and Asp in group 1 were sufficiently high (Figures 5A and 5B) that no massive conversion of Asn to Asp or Gln to Glu occurred. However, in group 2, the levels of both free Asn and Gln were higher than in group 1 and correlated negatively with the level of free Lys. This could result from the extensive reduction in Glu and Asp levels upon their conversion to Lys (Figures 5A and 5B), which triggered more massive enzymatic conversion of Asn and Gln to these amino acids.
The Dramatic Increase in Seed Free Lys in the LKR/SDH Knockout × Phaseolin-DHPS Line Is Associated with an Altered Profile of Free Thr and Met
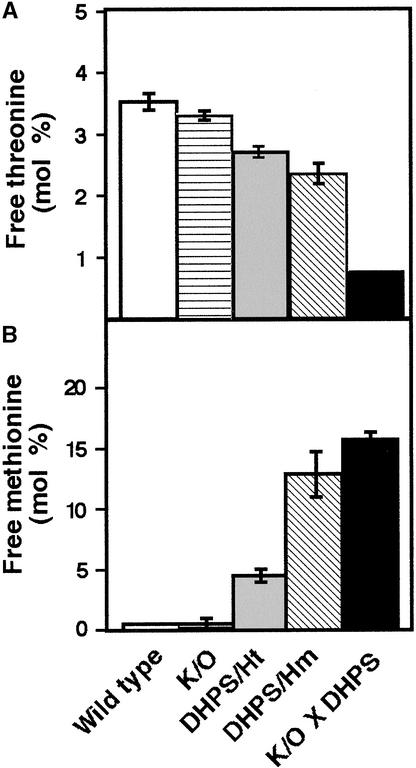
Thr and Met are synthesized by a branch of the Asp family pathway that competes with the Lys branch (Figure 1) (Galili, 1995). Therefore, we tested for changes in the relative levels of these two amino acids in the LKR/SDH knockout × phaseolin-DHPS line compared with its parents and wild-type plants. As shown in Figure 6A, the proportion of free Thr decreased progressively in the LKR/SDH knockout plants, the heterozygous and homozygous transgenic plants expressing the bacterial DHPS, and the LKR/SDH knockout × phaseolin-DHPS plants, reaching an approximately fivefold decline in the latter genotype compared with the level in wild-type plants. In contrast to Thr, the relative level of free Met was increased progressively in the different genotypes, reaching an ∼38-fold increase in the LKR/SDH knockout × phaseolin-DHPS plants compared with the level in wild-type plants (Figure 6B).
Figure 6.
Relative Levels of Free Thr and Met in Wild-Type and Different Transgenic Arabidopsis Plants.
Relative levels of free Thr (A) and Met (B) are given as mol % of total amino acids. DHPS/Ht and DHPS/Hm, transgenic plants expressing the phaseolin-DHPS construct in heterozygous and homozygous states, respectively; K/O, LKR/SDH knockout mutants; K/O × DHPS, LKR/SDH knockout × phaseolin-DHPS plants. Error bars represent standard errors.
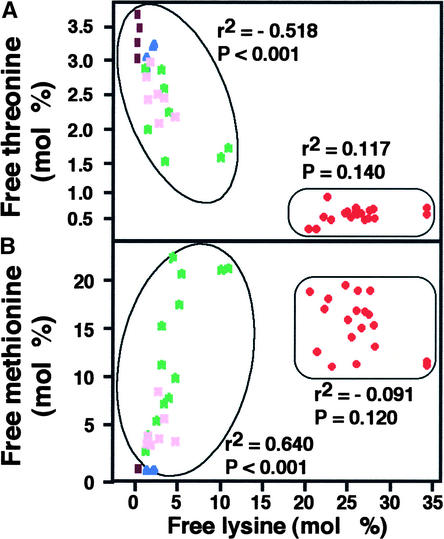
We also analyzed the specific correlation between Lys and Thr or Met in each plant individually. As shown in Figure 7, in all comparisons the individual lines again were clustered into groups 1 and 2, similar to those shown in Figure 5. The relative level of free Thr was negatively correlated with the free Lys level in group 1, whereas no correlation between these two free amino acids was obtained in group 2, in which the lowest levels of free Thr were found (Figure 7A). By contrast, a positive correlation between free Met and free Lys was evident in group 1, but no correlation was obtained between these two free amino acids in group 2, in which relatively high levels of free Met were found (Figure 7B). The lack of correlation between free Lys and free Thr or Met in group 2 may have been caused by the low level of Asp, rendering it a rate-limiting amino acid for the production of Thr and Met (Figure 1).
Figure 7.
Correlation between Free Lys and Free Thr or Met in Wild-Type and Different Transgenic Arabidopsis Plants.
Pearson's correlations between free Lys and free Thr or Met are given in (A) and (B), respectively. The two distinct populations, designated group 1 and group 2, are circled at left and right, respectively. Numerals near each population represent r2 values of linear regressions followed by significance levels (P). Colored spots represent the following genotypes: brown, wild type; blue, LKR/SDH knockout; pink and green, transgenic plants expressing the phaseolin-DHPS construct in heterozygous and homozygous states, respectively; red, LKR/SDH knockout × phaseolin-DHPS.
Effect of Lys Accumulation on the Profiles of Other Seed Amino Acids
Lys overproduction in the different genotypes was associated with a reduction of approximately twofold in the relative levels of Ser, Ala, and Tyr (Table 1). The reduction in Ser may be attributable to the fact that it is a precursor for the synthesis of Met (via Cys), whose level was increased significantly in different genotypes. The reductions in Ala and Tyr may have occurred because their synthesis requires the transamination of Glu, whose level was reduced significantly in the different genotypes.
Effect of Lys Overaccumulation on Seed Germination
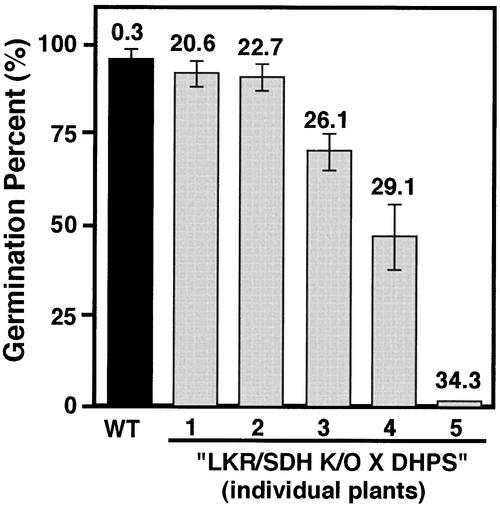
Despite their high Lys content, seeds of most LKR/SDH knockout × phaseolin-DHPS plants germinated with efficiencies similar to those of wild-type plants (Figure 8). However, seeds from some LKR/SDH knockout × phaseolin-DHPS plants had lower germination efficiencies (Figure 8), and the seedlings derived from these plants had some yellowish spots on their cotyledons (data not shown). Nevertheless, these seedlings reached maturity and produced seeds just like wild-type plants, suggesting that the inhibitory effect does not persist beyond the initial stages of seed germination and seedling establishment. Notably, the retarded germination was evident in plants whose average seed Lys level was the highest (Figure 8).
Figure 8.
Association between Average Free Lys Content and Rate of Seed Germination.
Germination percentage of seeds derived from wild-type (WT) plants (black bar) as well as from individual plants (gray bars 1 to 5) of the LKR/SDH knockout × phaseolin-DHPS genotype (LKR/SDH K/O × DHPS). The average relative levels of free Lys (mol %) in mature seeds of each plant are given at top.
DISCUSSION
Lys Catabolism Represents a Major Limiting Factor for Lys Accumulation in Arabidopsis Seeds That Possess Feedback-Insensitive DHPS
Expression of bacterial DHPS enzymes in seeds of various plant species resulted in huge variations in free Lys levels in mature seeds (Karchi et al., 1994; Falco et al., 1995; Mazur et al., 1999), but the relative contribution of Lys synthesis and catabolism in regulating seed Lys levels in these plant species was not studied. In the present report, we used seed-specific expression of a bacterial DHPS in a homozygous LKR/SDH knockout Arabidopsis mutant to show that increasing Lys synthesis and reducing its catabolism synergistically increased Lys content in plant seeds. Although each of these traits, when present alone, increased free Lys levels in seeds by ∼5-fold to ∼14-fold, the synergistic interaction between them yielded a dramatic increase of ∼80-fold in the relative level of this amino acid. This finding implies that under conditions of deregulated Lys synthesis, Lys catabolism becomes the dominant regulator of Lys accumulation in Arabidopsis seeds.
Analysis of the LKR/SDH Knockout × Phaseolin-DHPS Plants Reveals Novel Regulatory Circuits of Amino Acid Metabolism in Seeds
Lys overaccumulation in the LKR/SDH knockout × phaseolin-DHPS line as well as in its parental lines was associated with altered levels of several amino acids that are metabolically related to it. These included Gln, Glu, Asn, and Asp, the precursors of the Asp family pathway leading to Lys, Thr, and Met biosynthesis (Figure 1). Although some of these changes were anticipated, others were unexpected, as discussed below.
There was significant reduction in Asp and Glu in the LKR/SDH knockout × phaseolin-DHPS plants (Figures 4A, 4B, 5A, and 5B) compared with their parents and the wild-type plants. This finding was not surprising because these amino acids are direct precursors of Lys biosynthesis (Figure 1). However, because Gln is the precursor of Glu and Asn is the precursor of Asp, we expected that the levels of free Gln and Asn also would be reduced significantly in these plants, yet the opposite was observed (Figures 4C, 4D, 5C, and 5D). Nevertheless, the increased levels of free Gln and Asn in the LKR/SDH knockout × phaseolin-DHPS plants were negatively correlated with free Lys level (Figures 4C and 4D). Although analysis of steady state levels of amino acids in mature seeds does not precisely reflect metabolic processes during seed development, we hypothesize that Gln and Asn levels were increased in the LKR/SDH knockout × phaseolin-DHPS plants and thus were converted more efficiently via Glu and Asp to Lys.
The progressive reduction in free Thr with the increase in free Lys in the different transgenic plants (Figures 6A and 7A) was anticipated, because these two amino acids compete for Asp as a common precursor (Galili, 1995). However, the positive correlation between free Lys and free Met levels in the different transgenic plants (Figures 6B and 7B) was unexpected, because Met synthesis also should compete with Lys synthesis for Asp (Figure 1) (Galili, 1995).
Considering these results, we hypothesize that Lys metabolism plays a central regulatory role in seed amino acid metabolism, particularly in the metabolism of amide amino acids and Met, as illustrated in Figure 9. Because Glu is both an immediate precursor for the Asp family pathway (Galili, 2002) and a major end product in the Lys catabolism pathway (Galili et al., 2001), we hypothesize that Glu, Asp, and Lys constitute an important metabolic regulatory loop. The flux of this loop is regulated strongly by the sensitivity of DHPS activity to Lys feedback inhibition as well as by a Lys-dependent stimulation of Lys catabolism (Galili, 1995, 2002; Galili et al., 2001). These control mechanisms may function not only to equilibrate Lys levels but also to maintain balanced levels of Glu and Asp, two important regulatory amino acids of plant metabolism whose levels must be regulated tightly (Lam et al., 1995; Stitt, 1999; Stitt et al., 2002). When the fine regulation of the Glu-Asp-Lys loop by DHPS activity and Lys catabolism is impaired, as in the LKR/SDH knockout × phaseolin-DHPS plants, Glu and Asp levels are reduced greatly because of their efficient conversion to Lys. This, in turn, activates another regulatory circuit that stimulates the level of Gln and Asn and their conversion to Glu and Asp, respectively. To date, we do not know what signaling molecule(s) triggers this new regulatory circuit. A favorable candidate is Glu, because this molecule is known as an important signaling molecule, operating at least in part by changing calcium homeostasis (Lam et al., 1998; Dennison and Spalding, 2000; Kim et al., 2001). However, we cannot exclude the possibility that other metabolites, such as Lys or any of its intermediate(s), are involved in this regulation.
Figure 9.
Model Depicting Glu, Asp, and Lys at the Core of an Important Regulatory Loop in Plant Amino Acid Metabolism.
The hypothesized Glu-Asp-Lys metabolic loop is presented in the center, linked by orange, yellow, and pink arrows, respectively. A different branch of the Asp family pathway (brown arrows) synthesizes Thr and Met. The pathway of Lys catabolism produces acetyl-CoA (brown arrow). Two of the most important regulators in this loop are the extreme sensitivity of DHPS activity to feedback inhibition by Lys and the stimulation of LKR/SDH activity by excess Lys levels (light blue arrows) (for discussion, see Galili et al., 2001; Galili, 2002). Impairment of these two important regulatory elements causes extensive conversion of Glu and Asp to Lys, thereby activating several other metabolic regulatory circuits. We believe that the rapid reduction in Glu, an important regulatory amino acid, triggers increased synthesis of Gln and Asn and their conversion to Glu and Asp, respectively (dashed gray arrows). However, we cannot exclude the possibility that the increased Lys level triggers this metabolic change as well (dashed green arrows). In addition, the increased Lys level also triggers another novel regulatory circuit, resulting in a significant increase in Met level (dashed blue arrow). The significance of this novel regulatory circuit is unknown.
This study also revealed another novel and unanticipated transregulatory circuit between Lys and Met metabolism in Arabidopsis seeds. Because Met is regulated extensively by its synthesis and catabolism (Amir et al., 2002), either of these two metabolic fluxes, or both, could be regulated in trans by Lys. At present, we do not know the cellular and metabolic significance of this transregulatory circuit and the mechanism that controls it. Together, our results show that amino acid metabolism in plant seeds is controlled by a complex network of interacting metabolic fluxes and control mechanisms, the identification of which poses an exciting challenge for future research.
Effect of Amino Acid Metabolism during Seed Development on Germination and Seedling Establishment
Seeds of the LKR/SDH knockout × phaseolin-DHPS plants that accumulated superabundant Lys levels exhibited retarded germination and seedling establishment. Similar observations were reported for high-Lys seeds of soybean and rapeseed plants (Falco et al., 1995). Because Lys is known to be toxic to plants (Jacobs et al., 1987), it is possible that the observed germination retardation was the result of excess accumulation of this amino acid. However, we cannot be sure that the retarded germination was caused by relatively higher seed Lys content, because Lys levels represented an average of many seeds per plant and not of individual seeds that were tested for germination frequency. Moreover, because the levels of a number of other amino acids also were altered in the LKR/SDH knockout × phaseolin-DHPS plants (Table 1), such changes or their combinations also may have contributed to the germination retardation effect.
Relevance of This Study to Increasing the Nutritional Quality of Crop Plants by Metabolic Engineering
Although the regulation of Lys metabolism may vary significantly among seeds of different plant species (Galili and Hofgen, 2002), it is likely that the principal regulatory mechanisms are quite conserved. Thus, our results imply that the expression of a feedback-insensitive DHPS coupled with a reduction in Lys catabolism may be the preferred choice for increasing Lys accumulation in seeds of crop plants to remarkably high levels. The unexpected positive correlation between Lys overaccumulation and enhanced seed Met in Arabidopsis also is encouraging, because Met is another important essential amino acid for humans and livestock (Galili and Hofgen, 2002). Future studies should aim to determine whether similar cross-regulation between Lys and Met metabolism also exists in seeds of other plants.
METHODS
Chimeric Genes, Mutants, Primers, and Plant Growth and Transformation
The chimeric phaseolin dihydrodipicolinate synthase (DHPS) and the Arabidopsis thaliana Lys-ketoglutarate reductase/saccharopine dehydrogenase (LKR/SDH) knockout mutant were described previously (Karchi et al., 1994; Zhu et al., 2001). The following specific primers were used in the screening approach. LKR/SDH-specific primers were P5 (5′-GATGAAAATGATCAACGATGCT-3′) and P9 (5′-CTCGGTTAGCTAATCCAA-ATG-3′). DHPS-specific primers were Pf-dapA (5′-TGGCACCACTGG-CGAGTCC-3′) and Pr-dapA (5′-CCACTGTCGGTGATTGGTGTC-3′). Arabidopsis plants were grown in pots under controlled greenhouse conditions (16-h photoperiod at 23 ± 5°C) and transformed as described previously (Clough and Bent, 1998).
Generation of Transgenic LKR/SDH Knockout Arabidopsis Mutants Expressing a Bacterial DHPS in a Seed-Specific Manner
The following strategy was used to generate a transgenic Arabidopsis LKR/SDH knockout mutant expressing a bacterial DHPS in a seed-specific manner (LKR/SDH knockout × phaseolin-DHPS). Wild-type Arabidopsis, ecotype Wassilewskija, was transformed with a previously described chimeric gene encoding a bacterial DHPS fused to a promoter derived from the bean phaseolin storage protein (Karchi et al., 1994). This construct was used previously to express the bacterial DHPS in a seed-specific manner in transgenic tobacco plants (Karchi et al., 1994). Transgenic Arabidopsis plants expressing the bacterial DHPS in seeds were subjected to protein gel blot analysis with anti-bacterial DHPS antibodies, as described previously for tobacco (Karchi et al., 1994), and a line expressing a relatively high DHPS level was selected for crossing to the LKR/SDH knockout mutant. Selection for progeny containing both the phaseolin-DHPS gene and the T-DNA–tagged LKR/SDH gene in heterogous states then was performed by dual PCR analysis, using T-DNA–specific primers (Zhu et al., 2001) and bacterial DHPS–specific primers. This plant was selfed, and progeny containing both a bacterial DHPS and a homozygous LKR/SDH knockout were selected by dual screen. This screen included PCR analysis using LKR/SDH-specific primers that span the ∼7.6-kb T-DNA insertion site from both sides (Zhu et al., 2001). These primers cannot produce an amplification product of the LKR/SDH gene containing the large T-DNA insert, but they can amplify a fragment when combined with primers derived from the T-DNA (Zhu et al., 2001). The candidate lines were confirmed further by protein gel blot analysis with anti-bacterial DHPS antibodies and anti-LKR monoclonal antibody.
Amino Acid Analysis
Free amino acids were extracted from mature Arabidopsis seeds as described previously (Karchi et al., 1993). For extraction of albumins plus free amino acids, mature seeds were extracted in 10 mM Tris-HCl, pH 7.5, containing a protease inhibitor cocktail at 4°C. This extract was subjected to hydrolysis in 6 M HCl for 22 h at 115°C before amino acid analysis. Amino acid analysis was performed by reverse-phase HPLC on a C-18 column (Waters, Milford, MA) using a precolumn phenylisothiocyanate derivatization. Each amino acid analysis was performed with 5 to 20 replications using independent preparations from different plants for each genotype.
Statistical Analysis
The statistical analyses used were one-way analysis of variance followed by multiple comparison (Fisher's LSD) and Pearson's correlation. The JMP-4 computer program (SAS Institute, Cory, NC) was used for this analysis.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Avihai Danon for critical reading of the manuscript and helpful comments and Johann Schaller and Michele Stark for their invaluable help with the amino acid analysis. This work was supported by grants from the FrameWork Program of the Commission of the European Communities, the Israel Academy of Sciences and Humanities, and the National Council for Research and Development, Israel. G.G. is an incumbent of the Bronfman Chair in Plant Sciences.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.009647.
References
- Amir, R., Hacham, Y., and Galili, G. (2002). Cystathionine gamma-synthase and threonine synthase operate in concert to regulate carbon flow towards methionine in plants. Trends Plant Sci. 7 153–156. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Dennison, K.L., and Spalding, E.P. (2000). Glutamate-gated calcium fluxes in Arabidopsis. Plant Physiol. 124 1511–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco, S.C., Guida, T., Locke, M., Mauvais, J., Sandres, C., Ward, R.T., and Webber, P. (1995). Transgenic canola and soybean seeds with increased lysine. Bio/Technology 13 577–582. [DOI] [PubMed] [Google Scholar]
- Galili, G. (1995). Regulation of lysine and threonine synthesis. Plant Cell 7 899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili, G. (2002). New insights into the regulation and functional significance of lysine metabolism in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 53 27–43. [DOI] [PubMed] [Google Scholar]
- Galili, G., and Hofgen, R. (2002). Metabolic engineering of amino acids and storage proteins in plants. Metab. Eng. 4 3–11. [DOI] [PubMed] [Google Scholar]
- Galili, G., Shaul, O., Perl, A., and Karchi, H. (1994). Synthesis and accumulation of the essential amino acids lysine and threonine in seeds. In Seed Development and Germination, H. Kigel and G. Galili, eds (New York: Marcel Dekker), pp. 811–831.
- Galili, G., Tang, G., Zhu, X., and Gakiere, B. (2001). Lysine catabolism: A stress and development super-regulated metabolic pathway. Curr. Opin. Plant Biol. 4 261–266. [DOI] [PubMed] [Google Scholar]
- Habben, J.E., and Larkins, B.A. (1994). Improving protein quality in seeds. In Seed Development and Germination, H. Kigel and G. Galili, eds (New York: Marcel Dekker), pp. 791–810.
- Jacobs, M., Negrutiu, I., Dirks, R., and Cammaerts, D. (1987). Selection programs for isolation and analysis of mutants in plant cell cultures. In Plant Biology, Vol. 3, C.E. Green, D.A. Somers, W.P. Hackett, and D.D. Biesboer, eds (New York: Alan R. Liss), pp. 243–264.
- Karchi, H., Miron, D., Ben-Yaacov, S., and Galili, G. (1995). The lysine-dependent stimulation of lysine catabolism in tobacco seeds requires calcium and protein phosphorylation. Plant Cell 7 1963–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchi, H., Shaul, O., and Galili, G. (1993). Seed specific expression of a bacterial desensitized aspartate kinase increases the production of seed threonine and methionine in transgenic tobacco. Plant J. 3 721–727. [Google Scholar]
- Karchi, H., Shaul, O., and Galili, G. (1994). Lysine synthesis and catabolism are coordinately regulated during tobacco seed development. Proc. Natl. Acad. Sci. USA 91 2577–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.A., Kwak, J.M., Jae, S.K., Wang, M.H., and Nam, H.G. (2001). Overexpression of the AtGluR2 gene encoding an Arabidopsis homolog of mammalian glutamate receptors impairs calcium utilization and sensitivity to ionic stress in transgenic plants. Plant Cell Physiol. 15 74–84. [DOI] [PubMed] [Google Scholar]
- Lam, H.M., Chiu, J., Hsieh, M.H., Meisel, L., Oliveira, I.C., Shin, M., and Coruzzi, G. (1998). Glutamate-receptor genes in plants. Nature 396 125–126. [DOI] [PubMed] [Google Scholar]
- Lam, H.-M., Coschigano, K., Schultz, C., Melo-Oliveira, R., Tjagen, G., Oliveira, I., Ngai, N., Hsieh, M.-H., and Coruzzi, G.M. (1995). Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell 7 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur, B., Krebbers, E., and Tingey, S. (1999). Gene discovery and product development for grain quality traits. Science 285 372–375. [DOI] [PubMed] [Google Scholar]
- Negrutiu, I., Cattoir-Reynearts, A., Verbruggen, I., and Jacobs, M. (1984). Lysine overproducer mutants with an altered dihydrodipicolinate synthase from protoplast culture of Nicotiana sylvestris (Spegazzini and Comes). Theor. Appl. Genet. 68 11–20. [DOI] [PubMed] [Google Scholar]
- Shaul, O., and Galili, G. (1992). Increased lysine synthesis in transgenic tobacco plants expressing a bacterial dihydrodipicolinate synthase in their chloroplasts. Plant J. 2 203–209. [DOI] [PubMed] [Google Scholar]
- Stitt, M. (1999). Nitrate and the regulation of primary metabolism, allocation and growth. Curr. Opin. Plant Sci. 2 178–186. [DOI] [PubMed] [Google Scholar]
- Stitt, M., Muller, C., Matt, P., Gibon, Y., Carillo, P., Morcuende, R., Scheible, W.R., and Krapp, A. (2002). Steps towards an integrated view of nitrogen metabolism. J. Exp. Bot. 53 959–970. [DOI] [PubMed] [Google Scholar]
- Zhu, X., Tang, G., Granier, F., Bouchez, D., and Galili, G. (2001). A T-DNA insertion knockout of the bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase gene elevates lysine levels in Arabidopsis seeds. Plant Physiol. 126 1539–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]