Abstract
The ABC model, which was accepted for almost a decade as a paradigm for flower development in angiosperms, has been subjected recently to a significant modification with the introduction of the new class of E-function genes. This function is required for the proper action of the B- and C-class homeotic proteins and is provided in Arabidopsis by the SEPALLATA1/2/3 MADS box transcription factors. A triple mutant in these partially redundant genes displays homeotic conversion of petals, stamens, and carpels into sepaloid organs and loss of determinacy in the center of the flower. A similar phenotype was obtained by cosuppression of the MADS box gene FBP2 in petunia. Here, we provide evidence that this phenotype is caused by the downregulation of both FBP2 and the paralog FBP5. Functional complementation of the sepallata mutant by FBP2 and our finding that the FBP2 protein forms multimeric complexes with other floral homeotic MADS box proteins indicate that FBP2 represents the same E function as SEP3 in Arabidopsis.
INTRODUCTION
MADS box genes are transcriptional regulators that act as switches between many different morphological stages; therefore, they have been chosen as subjects for phylogenetic analysis and comparative studies among different taxa in an attempt to elucidate the evolution of morphological characters (Doyle, 1994; Purugganan et al., 1995; Tandre et al., 1995; Theissen et al., 1996; Münster et al., 1997; Winter et al., 1999).
Genetic analysis of floral homeotic mutants in the model species snapdragon and Arabidopsis led to the identification of the first MADS box genes in plants (Schwarz-Sommer et al., 1990; Yanofsky et al., 1990) and provided genetic and molecular evidence for the classic ABC model (Coen and Meyerowitz, 1991). According to this model, floral homeotic genes belonging to these three classes and acting alone and/or in combination determine the identity of the floral organs. More of these genes were cloned subsequently, and most of them appeared to be members of the MADS box gene family (for review, see Riechmann and Meyerowitz, 1997). For its simplicity and what at first seemed its wide applicability, the ABC model has been accepted as a general model for flower organ determination in angiosperms. Nevertheless, the isolation of MADS box genes from numerous distantly related species revealed a more complex regulation of organ identity, making it clear that variations have to be considered to adapt the model to different species. Furthermore, it became apparent that the role of MADS box genes is not confined only to the flower domain.
B and C functions and their respective genes are considered the most conserved among eudicots, and only subtle variations have been noted between the two model species mentioned above (Davies et al., 1999), whereas a slightly different regulation of whorls 2 and 3 has been observed in petunia. Homeotic conversion of the second and third whorls is displayed in petunia plants with mutations in the FLORAL BINDING PROTEIN1 (FBP1) gene (a GLOBOSA homolog), whereas mutations in pMADS1, which shares similar sequence and expression patterns with the snapdragon gene DEFICIENS, cause homeotic conversion in petals only (Angenent et al., 1992; van der Krol et al., 1993; Tsuchimoto et al., 2000). Furthermore, indications for redundancy in the C-function genes have been observed in species such as petunia, cucumber, and gerbera, in which duplication led to more than one AGAMOUS homolog (Tsuchimoto et al., 1993; Kater et al., 1998; Yu et al., 1999). A few years after the ABC model was proposed, studies on ovule development in petunia led to the isolation of two MADS box genes, FBP7 and FBP11, whose expression is turned on just before ovule primordia arise and is maintained in the seed coat after fertilization. Ectopic expression and cosuppression studies indicated that their activity is necessary to determine the identity of ovules, which could be regarded as a separate floral organ, and a new organ identity activity (the D activity, mediated by FBP7 and FBP11) was added to the model (Angenent et al., 1995; Colombo et al., 1995, 1997).
Although these deviations did not greatly affect the robustness of the ABC model and its acceptance as a general paradigm for floral organ determination, the results of more recent studies called for a revision of the whole theory and a reformulation of the model itself, introducing a new class of genes referred to as Identity-mediating or E-function genes (Egea-Cortines and Davies, 2000; Theissen, 2001). However, the first evidence of this new class of MADS box genes dates back to 1994, when the downregulation of the FBP2 and TM5 genes caused very similar phenotypes in petunia and tomato plants, respectively, with homeotic conversion of whorls 2, 3, and 4 simultaneously and loss of determinacy in the center of the flower (Angenent et al., 1994; Pnueli et al., 1994). A further indication that these factors are essential for floral organ fate came from the analysis of Arabidopsis plants with mutations in the SEPALLATA genes SEP1, SEP2, and SEP3, showing the same phenotypic alterations observed in the two Solanaceae species (Pelaz et al., 2000). How these factors act at the molecular level has been suggested in a recent study with snapdragon (Egea-Cortines et al., 1999), in which for the first time the authors raised the possibility that MADS box factors act as multimeric complexes in the regulation of gene expression and evidence was provided about interactions between E proteins and B- and C-class proteins. Very strong support for the recently revised ABC model came from the work of Honma and Goto (2001), who proved that the simultaneous expression of three different MADS box genes is necessary for the specification of flower organ identity and that it is sufficient to cause the homeotic conversion of leaves to floral organs.
The history of MADS box proteins in plants and the research flourishing around these factors teach us that major breakthroughs in this field are the result of information collected from different species and comparison between plants in the search for homologies and differences. In this respect, the detailed analysis of the petunia FBP2 subfamily presented here should provide information needed for a better understanding of flower development in this and other species. The combined results obtained by different approaches indicate that FBP2 is the functional equivalent of the Arabidopsis SEP3 gene in mediating the function of other floral homeotic genes. The protein–protein interaction studies suggest how this mediation occurs at the molecular level: complex formation between FBP2 and class-B protein heterodimers is shown here for the petunia proteins. Furthermore, three-hybrid experiments provided a powerful tool for revealing structural differences among sequence-related proteins, a useful piece of information in defining functional redundancy.
RESULTS
Constitutive Expression of FBP2 in Arabidopsis Resembles That of 35S-SEP3 Plants
The role of FBP2 in mediating the identity of the three inner floral whorls was documented several years ago by cosuppression experiments in petunia (Angenent et al., 1994). A similar function has been attributed to the related SEP genes from Arabidopsis (Pelaz et al., 2000). Although overexpression phenotypes do not provide solid evidence about the functions of the overexpressed genes, the comparison between such phenotypes may give indications about the relatedness of the genes themselves. The constitutive expression of SEP3 in Arabidopsis has been shown to affect flowering time: plants that carry the 35S-SEP3 construct bolt after producing four or five rosette leaves, and the primary inflorescence terminates after only a few flowers have been formed (Pelaz et al., 2001).
To determine whether the petunia FBP2 gene could generate comparable alterations in the heterologous system, a 35S-FBP2 construct was transformed into Arabidopsis. Forty-five transgenic plants were generated, all expressing the petunia gene to various degrees. The plants with the most severe phenotypes (10 transgenic lines), which correlated to the higher overexpressors (data not shown), exhibited a drastic reduction in flowering time. Figure 1B shows the phenotype of a 35S-FBP2 transgenic plant that flowered after producing only two rosette leaves. In extreme cases, transformants flowered directly after the cotyledon stage, often with a single terminal flower. Although this early-flowering phenotype has been observed more often when MADS box genes are overexpressed (Mandel and Yanofsky, 1995; Kyozuka et al., 1997; Sung et al., 1999), it demonstrates that FBP2 and SEP3 have a similar effect on flowering time when expressed ectopically.
Figure 1.
Arabidopsis and Petunia sep-Like Mutants and Complementation of the Arabidopsis Mutant by FBP2 Overexpression.
(A) Wild-type Arabidopsis plant flowering after the production of 9 to 12 rosette leaves (rl).
(B) Severe Arabidopsis 35S-FBP2 plant flowering extremely early after the formation of just one curled rosette leaf (rl) and terminating in a single flower (tfl).
(C) Flower of the triple sep1 sep2 sep3 mutant, with the formation of sepals in whorls 1, 2, and 3 and a new floral bud developing from the center of the flower (whorl 4).
(D) Wild-type petunia (W115) inflorescence. Bracts (b), flowers (f), and inflorescence (i) are indicated.
(E) Inflorescence of a petunia fbp2 cosuppression plant.
(F) Flower of the petunia fbp2 cosuppression plant, with homeotic conversions identical to those found for the Arabidopsis sep1 sep2 sep3 mutant flower (C).
(G) Flower from an Arabidopsis sep1 sep2 sep3 35S-FBP2 plant (No. 270) showing a mild complementation of the sep mutation. Whorl-2 organs are petaloid, and in whorl 4, an aberrant bud appears on a short stem.
(H) Flower from another Arabidopsis sep1 sep2 sep3 35S-FBP2 (No. 23) plant, with petals in whorl 2, staminoid organs in whorl 3, and an ovary-like structure in whorl 4 containing stigmatic tissue (st) on top.
(I) Late flower from the Arabidopsis sep1 sep2 sep3 35S-FBP2 plant No. 23 with pollen-bearing stamens in whorl 3. Part of the sepals and petals are removed.
(J) Close-up of the stamen in (I).
(K) Close-up of an ovary-like structure in whorl 4 from a flower of the Arabidopsis sep1 sep2 sep3 35S-FBP2 plant No. 23. The two carpels are fused almost completely and have stigmatic tissue (st) on top. Inside, sepal-like organs develop (not visible).
Bars in (A) and (B) = 1 cm.
Partial Complementation of the sep Triple Mutant by FBP2
The Arabidopsis sep1 sep2 sep3 triple mutant exhibited homeotic conversion of the three inner floral whorls to sepaloid organs and loss of determinacy in the center of the flower, where a new inflorescence arose (Figure 1C). Such a phenotype also was observed in fbp2 petunia cosuppression plants, suggesting a similar function for the petunia gene (Figures 1E and 1F). To determine whether FBP2 is the functional homolog of SEP3, complementation experiments of the Arabidopsis triple mutant were performed with the petunia gene. For this complementation experiment, seeds from an Arabidopsis plant heterozygous for the sep1 and sep2 mutations and homozygous for sep3 were used. Twenty-eight plants germinated, and the genotypes were checked by PCR, because all of the genes carried insertions, either of the En-1 transposable element (sep2 and sep3) or of T-DNA (sep1) (Pelaz et al., 2000).
A significant bias in the expected segregation of the progeny was observed, and none of the plants was found to be homozygous for the mutation in the SEP2 gene. Eleven of the 28 Arabidopsis plants were suitable for the complementation experiment, because they carried at least one mutant allele for all three genes. These plants were transformed by infiltration with an Agrobacterium tumefaciens strain harboring the FBP2 gene under the control of the constitutive 35S promoter of the Cauliflower mosaic virus. Because of the kanamycin resistance carried by the T-DNA in the sep1 mutant allele, the constitutive expression of Green Fluorescent Protein (GFP) was chosen as a selection marker for the progeny of the 35S-FBP2–transformed plants. All of the seeds obtained after transformation were germinated on normal medium without antibiotics, and the seedlings were selected on the basis of GFP fluorescence. One hundred fourteen seedlings that showed GFP expression were transferred to the soil, and the expression of the petunia FBP2 gene was determined by RNA gel blot analysis (results not shown). Seventy-seven plants appeared to be FBP2 expressors, and their genotypes were assessed subsequently by PCR. Eight plants from the segregating population were found to be homozygous for all three mutant alleles.
The phenotypes of these plants were examined in detail to detect a possible complementation by the petunia FBP2 gene. The majority (six of eight plants) showed a partial to almost complete complementation of the sep mutant phenotype. Flowers of these plants are shown in Figures 1G to 1J. In some plants, the complementation was less pronounced and only the second-whorl sepals were reverted to petaloid organs (Figure 1G), whereas other plants showed complementation in the second and third floral whorls, resulting in the formation of white petals and stamens with pollen-bearing anthers (Figures 1I to 1J). However, the fourth whorl failed to show complete reversion to a wild-type pistil: a stem originating from the center of the flower terminated in an ovary-like structure with stigmatic tissue on top and sepal-like organs inside (Figure 1K). The complete restoration of petals and stamens and the partial restoration of the pistil, observed in most of the flowers, demonstrate that the petunia FBP2 gene represents the SEP function.
FBP2 Subfamily Members: Isolation and Phylogenetic Analysis
The SEP function in Arabidopsis is represented by three redundant genes, SEP1, SEP2, and SEP3 (Pelaz et al., 2000). Because this functional redundancy also may appear in petunia, we isolated putative FBP2-like genes by screening at low stringency a young inflorescence and a corolla cDNA library with the MADS box regions of FBP1 and FBP2 petunia genes (Angenent et al., 1992, 1994). Five clones were identified that belonged to the FBP2 clade: full-length cDNAs of FBP4 and FBP23 (Immink et al., 2003) were isolated from young inflorescences, whereas FBP5, FBP9 (Immink et al., 2002), and pMADS12 were obtained from a corolla library.
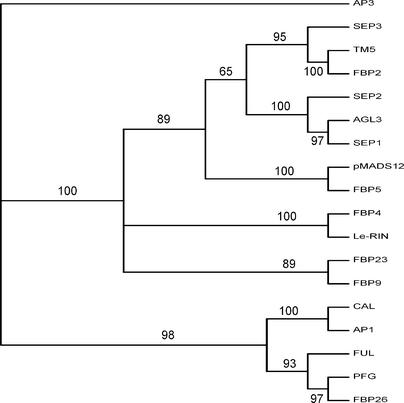
To establish the phylogenetic relationships between the petunia sequences and MADS box proteins from Arabidopsis, a phylogenetic tree was constructed using the MADS box, the I and K regions of 12 different sequences belonging to the SEP3 (AGL9) clade, including two tomato proteins, TM5 (Pnueli et al., 1994) and LeMADS-RIN (Vrebalov et al., 2002), and 5 sequences from the AP1 clade (Purugganan et al., 1995; Immink et al., 1999; Alvarez-Buylla et al., 2000). The AP3 protein from Arabidopsis was used as an outgroup. This phylogenetic analysis, the results of which are shown in Figure 2, reveals that FBP2 is the only protein that closely matches SEP3, with 83% identical amino acids, whereas pMADS12 and FBP5, although belonging to the same subgroup, appear to be less related to any of the Arabidopsis proteins. Two different subgroups include FBP9, FBP23, FBP4, and the tomato protein LeMADS-RIN.
Figure 2.
Phylogenetic Tree of Selected Members of the AP1 and FBP2/SEP MADS Box Protein Family.
The FBP, pMADS, and PFG proteins are from petunia, Le-RIN and TM5 are from tomato, and all others are from Arabidopsis. The phylogenetic analysis was performed with the MADS, I, and K regions of the proteins. Bootstrap values are indicated above each branch. The Arabidopsis AP3 protein (class B) was used as an outgroup.
Expression Analysis in Wild-Type Petunia and FBP2 Cosuppression Plants
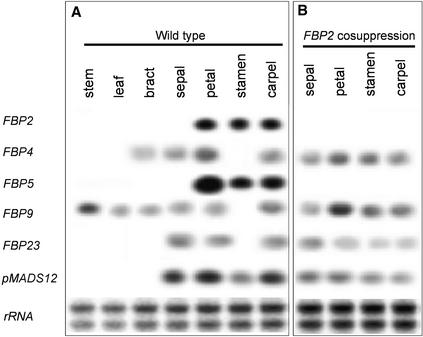
A first impression of the expression pattern in wild-type petunia tissues was obtained by RNA gel blot hybridization. Total RNA was extracted from stems, leaves, bracts, sepals, petals, stamens, and carpels and hybridized with a probe spanning the C-terminal region of the petunia genes. According to the results of the RNA gel blot analysis, shown in Figure 3A, the genes can be divided into two groups: those that are expressed exclusively in floral organs and those whose transcript also is detected in vegetative tissues. FBP2, FBP5, and pMADS12 belong to the first group, and their transcripts were detected in all three inner floral whorls. Furthermore, pMADS12 expression expanded into the first whorl. A similar pattern of expression has been reported for the SEP genes. SEP2 and SEP3 RNAs were detected in Arabidopsis petals, stamens, and carpels, including the ovules, whereas SEP1 was expressed ubiquitously throughout the whole flower (Flanagan and Ma, 1994; Savidge et al., 1995; Mandel and Yanofsky, 1998).
Figure 3.
Expression Patterns of Petunia FBP2 Family Members in Petunia Wild-Type and FBP2 Cosuppression Plants Determined by RNA Gel Blot Analyses.
(A) Expression of FBP2 family members in various tissues of wild-type petunia plants.
(B) Expression of FBP2 family members in floral organs of FBP2 cosuppression plants. Both FBP2 and FBP5 are strongly downregulated in these plants.
In each lane, 10 μg of total RNA was loaded. Equal loading of RNA was demonstrated by staining a representative gel with radiant red before blotting (rRNA).
On the other hand, the expression of FBP4, FBP9, and FBP23 was excluded from the stamens but present in the rest of the flower. Outside the floral domains, FBP4 transcript accumulated in bracts, and FBP9 accumulated in all green tissues of the plant, whereas no transcript of FBP23 was detected in vegetative tissues. Notably, FBP4 and FBP23 were expressed in seed pods (data not shown), which correspond to the fleshy fruits of the tomato plant, where the expression of the sequence-related LeMADS-RIN has been demonstrated to induce ripening (Vrebalov et al., 2002).
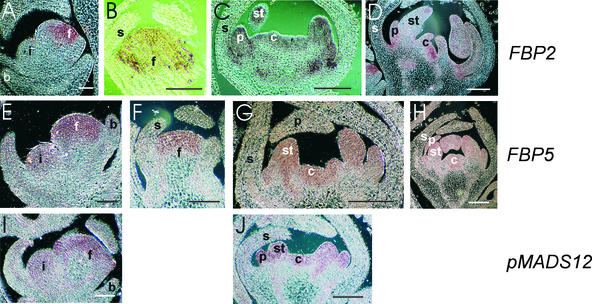
Because the petunia genes FBP2, FBP5, and pMADS12 are the most related to the Arabidopsis SEP genes, their spatial and temporal expression was monitored by in situ hybridization, as shown in Figure 4. The earliest stage at which FBP2 transcript was detected was in the central domain of the floral meristem, after it has been bifurcated from the inflorescence meristem (Figure 4A). During flower development, the expression remained localized in petal, stamen, and carpel primordia, where it persisted until full maturity of these organs (Figures 4B to 4D). Hybridizing signal also was observed at the adaxial side of the sepals (Figure 4C). A slightly earlier expression was found for FBP5 (Figure 4E) and pMADS12 (Figure 4I), whose transcripts appeared first in the inflorescence meristem. In contrast to FBP2, the expression of both FBP5 and pMADS12 was detectable throughout the floral meristem, whereas at later stages when sepal primordia emerge, it was confined to the central domain, like FBP2 (Figures 4F to 4H and 4J). The pattern of expression during later stages was in agreement with the results obtained by RNA gel blot hybridization.
Figure 4.
In Situ Localization of FBP2, FBP5, and pMADS12 Transcripts in Wild-Type Petunia.
Longitudinal sections were hybridized to digoxigenin-labeled antisense RNA fragments of FBP2 ([A] to [D]), FBP5 ([E] to [H]), and pMADS12 ([I] and [J]).
(A) FBP2 expression in a very young floral meristem. No signal is obtained in the inflorescence meristem and bracts.
(B) FBP2 expression in the central part of an early-stage floral bud. No signal is present in the developing sepals.
(C) Young floral bud with developing sepal, petal, and stamen primordia. The carpel primordia just arise. FBP2 mRNA can be detected in the three inner floral whorls.
(D) FBP2 expression in a slightly later stage floral bud than that shown in (C).
(E) Same stage meristems as in (A) hybridized to an antisense FBP5 probe. In contrast to FBP2, FBP5 also is expressed in the inflorescence meristem.
(F) FBP5 expression in an early floral bud showing a pattern similar to that obtained for FBP2 (B).
(G) and (H) FBP5 expression in the inner three floral whorls. Stages are comparable to those shown in (C) and (D), respectively.
(I) Expression of pMADS12 in the inflorescence and floral meristem. The expression pattern resembles the expression of FBP5 at this stage (E).
(J) pMADS12 expression in a same stage floral bud as that shown in (C) and (G).
b, bract; c, carpel; f, floral meristem; i, inflorescence meristem; p, petal; s, sepal; st, stamen. Bars in (A), (B), (E), (F), and (I) = 200 μm; bars in (C), (D), (G), (H), and (J) = 500 μm.
The functional role of FBP2 in specifying the identity of the central domain of the floral meristem was revealed several years ago (Angenent et al., 1994) through cosuppression studies. However, as a result of the strategy adopted, it was possible that other related sequences not yet identified were downregulated together with FBP2. Therefore, the expression of FBP2 subfamily members was tested in the mutated floral organs of FBP2 cosuppression plants. These plants, as well as the Arabidopsis sep1 sep2 sep3 triple mutant, exhibited homeotic conversion of the three inner floral whorls to sepaloid organs and loss of determinacy in the center of the flower, where a new inflorescence arose (Figures 1E and 1F). RNA gel blot analyses, as illustrated in Figure 3B, revealed that both FBP2 and FBP5 transcripts were absent in the cosuppression plants, whereas in wild-type plants, they were present at relatively high levels. All other MADS box genes tested showed little or no variation in their patterns of expression, although small differences occurred between wild-type and mutant plants, which most likely were attributable to changes in the identity of the organs in the cosuppression plants. Therefore, we conclude that the FBP2 cosuppression mutant is a double mutant of FBP2 and FBP5. Nevertheless, we cannot exclude the notion that other MADS box genes, which have not been identified, are downregulated together with FBP2 and FBP5 in the cosuppression mutants.
Protein–Protein Interactions of the FBP2 Subfamily Members
Interactions among MADS box proteins are the molecular basis for the ABC(DE) model, and in most cases, the specificity of such interactions has been conserved throughout angiosperm evolution. Hence, the elucidation of protein complexes provides additional clues for the identification of paralogs and functional homologs within and among species, respectively. Therefore, protein interaction studies were performed between FBP2-like MADS box proteins from petunia and the Arabidopsis SEP3 protein. All of the FBP2 subfamily members, except for pMADS12, gave autoactivation of the yeast reporter gene in the GAL4 system; therefore, the yeast Cytotrap two-hybrid system (Aronheim et al., 1997) was used to test the interactions between the FBP2 subfamily members and a collection of known petunia MADS box proteins. The results of the screening are summarized in Table 1. The petunia FBP2 and FBP5 proteins, and the Arabidopsis SEP3 protein, showed nearly matching patterns of interaction: they all interacted with the class-C proteins FBP6 and pMADS3, with the class-D proteins FBP7 and FBP11, with UNS, FBP21, and FBP22, whose transcripts are present in the vegetative tissues and in the first and second floral whorls, and with FBP29, another MADS box protein present mainly in vegetative tissues (Immink et al., 2003). The only exception was FBP23, which did not form a heterodimer with SEP3, whereas FBP2 and FBP5 interacted with FBP23, although these interactions were detected in only one direction. Despite the fact that pMADS12 and FBP5 are very similar in sequence, they showed significant differences in their dimerization partners: pMADS12 was able to interact with only one of the two C- and D-type proteins and did not dimerize with FBP22 and FBP28. Even more discrepancy occurred in the subgroup that includes FBP4, FBP9, and FBP23, in which FBP4 interacted with FBP21 only, whereas FBP9 was more similar to FBP2 with respect to their interaction patterns.
Table 1.
MADS Box Protein Interactions Determined by the Yeast Cytotrap Two-Hybrid System
| FBP23 | FBP6 | pMADS3 | FBP7 | FBP11 | UNS | FBP21 | FBP22 | FBP28 | FBP29 | |
|---|---|---|---|---|---|---|---|---|---|---|
| SEP3 | + | + | + | + | + | ± | + | + | + | |
| FBP2 | ± | + | + | + | + | + | ± | + | + | ± |
| FBP5 | ± | + | + | + | + | + | ± | + | + | + |
| pMADS12 | + | ± | + | ± | ||||||
| FBP9 | ± | + | + | + | ± | + | + | |||
| FBP23 | ± | ± | ± | + | + | + | ||||
| FBP4 | ± |
An interaction observed in both directions (in pSOS and pMYR) is indicated with +, and an interaction in only one direction is indicated with ±.
Higher Order Complex Formation
The ability of MADS box proteins to form multimeric complexes has been known since 1999, when a modified two-hybrid system was used to show that the snapdragon proteins DEF and GLO together interact with SQUAMOSA and that the complex DEF/GLO/SQUA/SQUA binds to DNA with greater affinity than the dimer DEF/GLO (Egea-Cortines et al., 1999). Similar complexes between dimers comprising the B-type proteins AP3/PI and AP1/SEP3 also may exist in Arabidopsis. In triple overexpressors of AP3, PI, and SEP3, conversion of leaves to petals occurred, whereas this conversion was even more pronounced when AP1 also was expressed ectopically, suggesting that a multimeric complex between AP1/SEP3/PI/AP3 proteins controls petal identity (Honma and Goto, 2001).
Given these findings, we analyzed the multimeric complex formations among the petunia MADS box proteins. The petunia class-B MADS box proteins pMADS1 and FBP1 were tested against the FBP2 subfamily members by means of the yeast three-hybrid system. The results, reported in Table 2, show that only FBP2, FBP4, and to a lesser extent FBP5 were able to interact with the heterodimer pMADS1/FBP1, whereas the other members of the subfamily could not. Slightly different results were obtained with the heterodimer pMADS1/pMADS2, in which pMADS2 is the second B-type protein in the PISTILLATA lineage of petunia (Angenent et al., 1993; Kush et al., 1993). No active complex was formed when FBP5 was fused to the GAL4 activation domain. Noteworthy is the fact that all of the positive interactions were obtained on −LTA (Leu-The-adenine) and −LTH (Leu-Thr-His) with 10 mM 3-amino-1,2,4-triazole medium, which are selective for strong interaction, and that all of the screenings were performed at 30°C, the normal yeast growth condition. By contrast, experiments with AP3/PI and SEP3 were performed at room temperature, because screenings at a higher temperature yielded no positive colonies (Honma and Goto, 2001). None of the FBP2-like proteins appeared able to dimerize with the petunia B-type proteins pMADS1, pMADS2, and FBP1 (data not shown) (Immink et al., 2003).
Table 2.
Higher Order Complex Formation as Determined by a Yeast GAL4 Three-Hybrid Assay
| Ternary Complex | −LTA, 30°C | −LTH + 10 mM 3AT, 30°C |
|---|---|---|
| BD-pMADS1 + pRED-FBP1 + AD-FBP2 | ++ | ++ |
| BD-pMADS1 + pRED-FBP1 + AD-FBP4 | ++ | ++ |
| BD-pMADS1 + pRED-FBP1 + AD-FBP5 | + | + |
| BD-pMADS1 + pRED-FBP1 + AD-FBP9 | − | − |
| BD-pMADS1 + pRED-FBP1 + AD-pMADS12 | − | − |
| BD-pMADS1 + pRED-FBP1 + AD-FBP23 | − | − |
| BD-pMADS1 + pRED-pMADS2 + AD-FBP2 | ++ | ++ |
| BD-pMADS1 + pRED-pMADS2 + AD-FBP4 | ++ | ++ |
| BD-pMADS1 + pRED-pMADS2 + AD-FBP5 | − | − |
| BD-pMADS1 + pRED-pMADS2 + AD-FBP9 | − | − |
| BD-pMADS1 + pRED-pMADS2 + AD-pMADS12 | − | − |
| BD-pMADS1 + pRED-pMADS2 + AD-FBP23 | − | − |
The class-B proteins pMADS1, FBP1, and pMADS2 are in BD and pRED vectors, whereas the FBP2-like genes are inserted in the AD vector. ++ indicates strong interaction, + indicates weak interaction, and − indicates no growth of the yeast cells on the selective medium. A, adenine; 3AT, 3-amino- 1,2,4-triazole; H, His; L, Leu; T, Thr.
Because MADS box proteins are known to bind DNA as dimers and the binding affinity was shown to be enhanced in ternary complexes (Egea-Cortines et al., 1999), the possible formation of a complex between B types, C types, and FBP2 was tested with the GAL4 system in yeast. Yeast two-hybrid experiments performed in the past have shown that no direct interactions occur between petunia B- and C-class proteins (Immink et al., 2003). Like most of the proteins that belong to the AP1/SEP3 superclade, the C terminus of FBP2 possesses transcriptional activity in yeast; therefore, part of this domain (59 amino acids) was removed from the FBP2 protein. The resulting truncated protein (FBP2ΔC) possesses no transcriptional activation activity and still is able to dimerize specifically with other petunia MADS box proteins (Immink et al., 2003). Nevertheless, partial deletion of the C terminus might affect higher order complex formation, because Egea-Cortines et al. (1999) and Honma and Goto (2001) clearly demonstrated that the C terminus is involved in these interactions. To determine the effect of the FBP2 truncation, the interaction between FBP2ΔC and the FBP1/pMADS1 heterodimer was tested. This experiment revealed that FBP2ΔC acts as the full-length FBP2 protein (Table 2) and still is able to form a complex with the B-type proteins (Table 3). Subsequently, pMADS1 (B2) together with pMADS2 (B3) or FBP1 (B1) and either FBP6 (C1) or pMADS3 (C2) were coexpressed with FBP2ΔC, and yeast colonies were selected on the appropriate selective medium. Despite the high similarity between B1 and B3 proteins and between C1 and C2 proteins, the formation of the complex was very specific, because only B1B2C2E and B2B3C1E combinations were positive in the yeast screening (Table 3).
Table 3.
Higher Order Complex Formation as Determined by a Yeast GAL4 Four-Hybrid Assay
The heterodimers FBP2ΔC(E)/FBP6(C1) and FBP2ΔC(E)/pMADS3(C2) are used as “bait,” and the heterodimers pMADS1(B2)/FBP1(B1) and pMADS1(B2)/pMADS2(B3) are used as “prey.” Selection for complex formation was performed at room temperature (RT). ++ indicates strong interaction, + indicates weak interaction and − indicates no growth of the yeast cells on the selective medium. The hypothesized complexes are indicated. A, adenine; 3AT, 3-amino-1,2,4-triazole; H, His; L, Leu; T, Thr.
DISCUSSION
The classic ABC model (Coen and Meyerowitz, 1991) has been accepted for almost a decade to describe the development of floral organs through the action of homeotic genes. Because most of the corresponding proteins are part of the MADS box family of transcription factors, which are known to bind DNA as dimers, pairs of MADS box proteins belonging to class A, B, or C were considered to be sufficient to determine the identity of every single floral organ within the flower (Riechmann and Meyerowitz, 1997). A recent breakthrough in the study of flower development, however, was made possible by the isolation of the Arabidopsis sep1 sep2 sep3 triple mutant: the expression of these genes with partially redundant function is required for the proper action of B and C homeotic genes, adding a new class to the previous ABC model (Pelaz et al., 2000). Homeotic activity as a result of direct interaction between B- or C-class proteins and SEP proteins was supported by the evidence that MADS box proteins can form larger complexes in which at least three members are involved (Egea-Cortines et al., 1999; Honma and Goto, 2001).
The presence of genes from diverse flowering plant species in the SEP-like (previously AGL2-like) subfamily suggests the importance of this clade in flower development and its conserved function during evolution (Theissen et al., 1996; Münster et al., 1997). However, only a few of these genes have been characterized thoroughly through mutant studies, and slight differences were found between some of them. The downregulation of the gerbera SEP-like gene GRCD1 (GERBERA REGULATOR OF CAPITULUM DEVELOPMENT1) caused homeotic conversion in one whorl only. The sterile staminodes of the ray florets were transformed into petals, indicating that gerbera might have whorl-specific SEP-like genes (Kotilainen et al., 2000). On the other hand, missense mutations in OsMADS1 (Oryza sativa MADS-box1) caused phenotypic alterations in lodicules, which were converted to palea- and lemma-like organs that resembled the phenotype of B-type mutants in grasses (Ambrose et al., 2000; Jeon et al., 2000). The only mutant phenotypes that featured all of the alterations displayed by the recent Arabidopsis sep mutant were described in 1994 in the Solanaceae species petunia and tomato (Angenent et al., 1994; Pnueli et al., 1994): cosuppression of the petunia FBP2 gene caused homeotic conversion of the three inner floral whorls to sepaloid organs as well as loss of the determinacy in the center of the flower. A similar phenotype was generated by the downregulation of the tomato TM5 gene in antisense transgenic plants.
FBP2 Is the Functional Equivalent of SEP3
In this study, we provide evidence that the petunia gene FBP2 represents the same function as the SEP3 gene in Arabidopsis. Clues came from expression pattern studies, phenotypic alterations produced in wild-type and mutant Arabidopsis, and the similar pattern of interactions when protein complexes are formed.
Overexpression of FBP2 in Arabidopsis caused early flowering, a single terminal flower, and curled rosette and cauline leaves, all characteristics that were described for 35S-SEP3 transgenic Arabidopsis plants (Pelaz et al., 2001). However, early flowering in Arabidopsis, which often is associated with curled leaves and a terminated inflorescence, is a phenotype that appears in many transgenic plants when a member of the AP1 or AGL2 clade, or another meristem identity gene such as LFY or FUL, is expressed constitutively (Mandel and Yanofsky, 1995; Kyozuka et al., 1997; Sung et al., 1999; Berbel et al., 2001). Gain-of-function phenotypes can be the result of the interference of the transgene with unrelated processes, especially when the gene is expressed outside of its physiological environment and at extremely high levels. Therefore, overexpression analysis, although providing useful indications, is insufficient to establish genetic functions and relatedness among different genes.
The most convincing evidence of functional homology between FBP2 and the SEP genes came from the complementation experiment in which SEP function was restored almost completely in the sep1 sep2 sep3 mutant by the introduction of the petunia gene. Whether FBP2 supplies the function of one or more SEP genes is difficult to assess and also not particularly relevant, because the Arabidopsis genes are functionally redundant. Nonetheless, a few circumstantial pieces of evidence suggest that SEP3 is the ortholog of FBP2. SEP3 is the closest to FBP2 in the phylogenetic analysis, and their expression patterns are very similar. Furthermore, FBP2 and SEP3 are expressed in the same floral domains and during similar developmental stages in petunia and Arabidopsis. Both transcripts first appear in the central portion of the flower meristem and later are restricted to the three inner whorls, although their expression also was detected at low levels on the adaxial side of the sepals (Mandel and Yanofsky, 1998). SEP1, on the other hand, is ubiquitously and highly expressed during early and intermediate stages of flower development without being restricted to particular floral domains (Flanagan and Ma, 1994). Moreover, SEP3 interacts with the same petunia MADS box proteins that FBP2 does, indicating that these proteins also share similar structural features and specificity to form dimers. A ternary complex between AP3/PI and SEP3 has been reported by Honma and Goto (2001), and a complex with a similar constitution of petunia proteins, including FBP2, has been confirmed in this study. These data strongly indicate that FBP2 and SEP3 are functionally equivalent and form higher order complexes with other class-B and class-C MADS box proteins that specify the identity of the inner three whorls.
Two Redundant Genes in Petunia
Whether the same partial redundancy observed among the SEP genes in Arabidopsis occurs in petunia as well could not be excluded by the cosuppression experiments that led to the mutant phenotype in petunia. Because of the type of experiment, it still was possible that more genes were downregulated in the transgenic plants. Therefore, we searched extensively for new FBP2-like genes, and the expression of these newly isolated FBP genes was examined in the FBP2 cosuppression plant. Among the five new petunia genes, FBP5 was the only one from which expression was abolished almost completely. This finding indicates that the SEP phenotype observed in the petunia cosuppression mutant is most likely the result of the downregulation of the two redundant genes FBP2 and FBP5. Besides the high degree of sequence similarity between FBP2 and FBP5 and the double fbp2 fbp5 cosuppression mutant, other functional tests support the notion that these genes are functionally redundant. They have identical dimerization partners among the 23 available petunia proteins, and both form ternary complexes with the class-B proteins pMADS1 and FBP1. Strikingly, the only observed difference between the two petunia proteins is the inability of FBP5 to form a ternary complex with the class-B dimer pMADS1/pMADS2, whereas three-hybrid studies with FBP2 revealed that these complexes are formed efficiently in yeast. pMADS2 is another putative class-B gene, which is similar to FBP1 and the B genes GLOBOSA and PISTILLATA from snapdragon and Arabidopsis, respectively (Kush et al., 1993; Angenent et al., 1994). Although it is reasonable to suggest that FBP1 and pMADS2 are functionally redundant, evidence for this is missing. Considered together, these data suggest that FBP5 might be diverged from FBP2 with respect to ternary complex formation with the dimer pMADS1/pMADS2, but it is doubtful whether this will have biological consequences because of the redundancy among the B proteins.
Multimeric Complex Formation
The finding that a ternary complex could be formed in yeast between the snapdragon DEF-GLO heterodimer and the SQUA protein (Egea-Cortines et al., 1999) and the more recent discovery that higher order complexes of MADS box gene products direct floral organ identity in Arabidopsis (Honma and Goto, 2001) provided a molecular explanation for some of the incongruities of the ABC model. The SEP genes have been shown to be necessary for the development of petals, stamens, and carpels in Arabidopsis (Pelaz et al., 2000); therefore, they represent a new class of floral organ identity genes, termed E-function genes (Theissen, 2001).
The SEP3 protein interacts in yeast with AP1 and the dimer of the B-function proteins PI-AP3, suggesting that the complex B1B2AE might direct petal formation. Similarly, SEP3 forms quaternary complexes in yeast with the class-C protein AG and the PI-AP3 dimer, indicating that the complex B1B2CE might be responsible for stamen identity. The formation of such protein complexes, together with the observation that in plants that overexpress the corresponding genes floral organs are formed outside the flower, led to the proposition of a new model of floral organ identity, the quartet model (Theissen, 2001), according to which four different combinations of four different floral homeotic proteins specify the identity of the different floral organs. Evidence is provided here that the same model might hold for petunia.
The results from the four-hybrid experiments clearly show that a complex in which four different MADS box proteins are involved is formed successfully in yeast. The notion that such a quaternary complex is just a tetramer or a complex of higher order cannot be excluded; however, it involves at least two B-types, one C-type, and one E-type protein in Arabidopsis.
On the other hand, unlike in Arabidopsis, evolution in petunia most likely has led to a duplication in the ancestral B- and C-type genes, resulting in FBP1 and pMADS2 (Angenent et al., 1992; Kush et al., 1993), both of which are related closely to PI, and FBP6 and pMADS3 (Angenent et al., 1993; Tsuchimoto et al., 1993), which are related to AG. Despite the high sequence similarity among the duplicated B and C proteins, there seems to be a high specificity in the pattern of interactions, because the C-type pMADS3 takes part in the protein complex only if the B-type FBP1 also is present, whereas FBP6 requires the presence of pMADS2, leading to the formation of the complexes B1B2C2E and B2B3C1E, respectively. Given these data, it is not possible to establish the biological function of such complexes in the plant. However, we speculate that these two complexes have been recruited for slightly different functions within the flower identity program.
It is apparent that research on floral identity genes has entered into the field of protein complexes, and although small differences in the redundancy of the proteins involved exist between the model species Arabidopsis, snapdragon, and petunia, the formation of MADS box complexes appears to be very conserved.
METHODS
Plant Material
Petunia hybrida lines W115 and W138, Arabidopsis thaliana ecotypes Wassilewskija and Columbia, and transgenic plants were grown under normal greenhouse conditions (22°C, 14 h of light/10 h of dark).
Screening of a cDNA Library and DNA Sequence Analysis
The FBP4, FBP20, FBP21, FBP22, FBP23, FBP26, FBP28, and FBP29 cDNA clones were isolated from a young inflorescence petunia cDNA library (Immink et al., 2003). The FBP5, FBP9 (Immink et al., 2002), and pMADS12 clones were obtained from a pistil cDNA library in λZAPII vector (Stratagene). Approximately 100,000 plaques from each library were screened with a mixed MADS box probe containing the 5′ terminal sequence of FBP1 (362 bp) and FBP2 (364 bp) (Angenent et al., 1992). Hybridization and washing of the Hybond N+ membranes (Amersham) were performed under low-stringency conditions (55°C hybridization and washing with 2× SSC [1× SSC is 0.15 M NaCl and 0.015 M sodium citrate] at 55°C).
The positive clones were isolated and purified, the pBluescript SK− and pAD-GAL4 phagemids were excised in vivo according to the Stratagene protocol, and the inserts were sequenced (BigDye Sequencing Kit; Applied Biosystems, Foster City, CA).
Nucleotide and amino acid sequence comparisons were performed using the CLUSTAL W multiple sequence alignment program (Thompson et al., 1994). Amino acid alignments, including the MADS box, the I region, and the K box, were used to obtain the phylogenetic tree with the Protdist (using the Dayhoff point accepted mutations matrix as a distance matrix) and neighbor-joining programs of the PHYLIP 3.5c package (provided by J. Felsenstein, Department of Genetics, University of Washington, Seattle). Bootstrap analysis was performed with Seqboot on 100 data sets, and branches with values of <50 were collapsed. Also included in the tree were sequences from the Arabidopsis and tomato MADS box gene families.
Construction of the Binary Vector and Plant Transformation
The full-length open reading frame of FBP2 was generated by PCR using a 5′ primer on the ATG containing a HindIII site and a reverse primer designed downstream of the stop codon with a KpnI site. The PCR fragment was placed downstream of the double 35S promoter of Cauliflower mosaic virus in the pGD120 plasmid (Immink et al., 2002), and the expression cassette was cloned subsequently into the pBIN19 binary vector (Bevan, 1984).
Transformation of petunia plants was performed as described previously (Angenent et al., 1993). Transformation of Arabidopsis wild-type and mutant plants was performed using the floral-dip method as described by Clough and Bent (1998); C58C1 was the Agrobacterium tumefaciens strain used for the transformation.
Identification of sep1 sep2 sep3 Mutant Arabidopsis Plants
A population of 28 offspring plants from a plant with the genotype sep1/SEP1; sep2/SEP2; sep3/sep3 was screened for insertions in the SEP1, SEP2, and SEP3 genes. The insertion alleles were identified by PCR as described by Pelaz et al. (2000). The genotypic distribution was as follows: SEP1/SEP1; SEP2/SEP2; sep3/sep3, 3 of 28 plants; SEP1/sep1; SEP2/SEP2; sep3/sep3, 2 plants; sep1/sep1; SEP2/SEP2; sep3/sep3, 4 plants; SEP1/SEP1; SEP2/sep2; sep3/sep3, 3 plants; sep1/sep1; SEP2/sep2; sep3/sep3, 5 plants; SEP1/sep1; SEP2/sep2; sep3/sep3, 11 plants. None of the plants was homozygous for the sep2 mutant allele. The plants belonging to the latter class were used for complementation by a direct transformation with the FBP2 overexpression construct. The FBP2 overexpression construct contained the Green Fluorescent Protein reporter driven by the 35S promoter to enable the selection of transformed seedlings.
RNA Gel Blot Analysis and in Situ Hybridization
Total RNA was isolated from stems, leaves, bracts, sepals, petals, stamens, and carpels of wild-type petunia plants and from the various floral whorls of FBP2 cosuppression plants, according to Verwoerd et al. (1989). Ten micrograms of each RNA sample, denatured with 1.5 M glyoxal, was fractionated on a 1.4% agarose gel and blotted onto a Hybond N+ membrane. Gene-specific fragments from all of the genes tested were used as probes for hybridization. The probes were labeled by random oligonucleotide priming (Feinberg and Vogelstein, 1984), and blots were hybridized as described by Angenent et al. (1992).
In situ hybridizations were performed as described by Cañas et al. (1994). Digoxigenin-labeled RNA probes were synthesized by T7 polymerase–driven in vitro transcription from the PCR fragment containing 3′ ends of FBP2, FBP5, and pMADS12, according to the instructions from Boehringer Mannheim. The amplification products were obtained with the 5′ primer 5′-CCCAAGCTTGGGTTTGAAGAAGATGGTGAGAGG-3′ and the 3′ primer 5′-TAATACGACTCACTATAGGGATAGGTAGTCACC-AATTAATTC-3′ containing the T7 polymerase promoter site. RNA transcripts were hydrolyzed partially for 45 min by incubation at 60°C in 0.1 M Na2CO3/NaHCO3 buffer, pH 10.2.
Two-, Three-, and Four-Hybrid Analysis of Petunia Protein–Protein Interactions
Two-hybrid analyses were performed in the Cytotrap yeast two-hybrid system (Stratagene), as described by Immink et al. (2003). Ternary and quaternary complex formation was studied with a modified yeast two-hybrid GAL4 system. Full-length cDNA copies of FBP2, FBP4, FBP5, FBP6, FBP9, FBP23, pMADS3, and pMADS12 were cloned into the pAD-GAL4 vector, and the coding sequences of pMADS1, pMADS3, FBP2 lacking the coding region for the C-terminal 59 amino acid residues (Immink et al., 2003), and FBP6 were inserted into the pBDcam-GAL4 vector (Stratagene). The third protein was expressed from the pRED-NLSa vector, which contained the full-length ADH1 promoter and the URA3 selectable marker gene. The complete FBP1 and pMADS2 coding sequences and FBP2 lacking the 3′ end were cloned into this vector in frame with the coding region of the SV40 nuclear localization signal.
To test for quaternary complex formation, the full-length pMADS1 coding sequence was cloned into the pTFT1 vector (Egea-Cortines et al., 1999). For all three- and four-hybrid experiments, yeast strain Pj69-4a (James et al., 1996) was used. Selection for interaction was performed on selective medium lacking His and supplemented with 10 mM 3-amino-1,2,4-triazole (Sigma). For the three-hybrid experiments, growth of yeast and hence protein–protein interaction was scored after 5 days of incubation at 30°C, and complex formation was confirmed on selective medium lacking adenine. For the four-hybrid experiments, the latter marker was not used because of the presence of the ADE2 selection marker gene on the pTFT1 vector. Therefore, suspensions of the yeast colonies were spotted onto selective medium lacking His (plus 10 mM 3-amino-1,2,4-triazole) only, and complex formation was scored after 7 days of incubation at room temperature.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Jos Schippers for technical assistance and Marty Yanofsky for the gift of the sep mutant seeds. We also are indebted to Hans Sommer and Annemarie Meijer, who kindly provided the yeast vectors pTFT1 and pRED-NLSa, respectively. This work was supported in part by the Dutch Ministry of Agriculture, Nature Management, and Fisheries (Grant DWK 281-392) and the European Union project BIO4-CT972217.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010280.
References
- Alvarez-Buylla, E.R., Pelaz, S., Liljegren, S.J., Gold, S.E., Burgeff, C., Ditta, G.S., Ribas de Populana, L., Martinez-Castilla, L., and Yanofsky, M.F. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97, 5328–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose, B.A., Lerner, D.R., Ciceri, P., Padilla, C.M., Yanofsky, M.F., and Schmidt, R.J. (2000). Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5, 569–579. [DOI] [PubMed] [Google Scholar]
- Angenent, G.C., Busscher, M., Franken, J., Mol, J.N.M., and Van Tunen, A.J. (1992). Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell 4, 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent, G.C., Franken, J., Busscher, M., Colombo, L., and Van Tunen, A.J. (1993). Petal and stamen formation in petunia is regulated by the homeotic gene Fbp1. Plant J. 4, 101–112. [DOI] [PubMed] [Google Scholar]
- Angenent, G.C., Franken, J., Busscher, M., van Dijken, A., van Went, J.L., Dons, H.J.M., and Van Tunen, A.J. (1995). A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 7, 1569–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent, G.C., Franken, J., Busscher, M., Weiss, D., and Van Tunen, A.J. (1994). Co-suppression of the petunia homeotic gene fbp2 affects the identity of the generative meristem. Plant J. 5, 33–44. [DOI] [PubMed] [Google Scholar]
- Aronheim, A., Zandi, E., Hennemann, H., Elledge, S.J., and Karin, M. (1997). Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17, 3094–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbel, A., Navarro, C., Ferrándiz, C., Cañas, L.A., Madueño, F., and Beltrán, J.-P. (2001). Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J. 25, 441–451. [DOI] [PubMed] [Google Scholar]
- Bevan, M.W. (1984). Binary agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañas, L.A., Busscher, M., Angenent, G.C., Beltran, J.P., and Van Tunen, A.J. (1994). Nuclear localization of the petunia MADS box protein FBP1. Plant J. 6, 597–604. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Colombo, L., Franken, J., Koetje, E., van Went, J.L., Dons, H.J.M., Angenent, G.C., and Van Tunen, A.J. (1995). The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, L., Van Tunen, A.J., Dons, H.J.M., and Angenent, G.C. (1997). Molecular control of flower development in Petunia hybrida. Adv. Bot. Res. 26, 229–250. [Google Scholar]
- Davies, B., Motte, P., Keck, E., Saedler, H., Sommer, H., and Schwarz-Sommer, Z. (1999). PLENA and FARINELLI: Redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 18, 4023–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, J.A. (1994). Origin of the angiosperm flower: A phylogenetic perspective. Plant Syst. Evol. 8, 7–29. [Google Scholar]
- Egea-Cortines, M., and Davies, B. (2000). Beyond the ABCs: Ternary complex formation in the control of floral organ identity. Trends Plant Sci. 5, 471–476. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines, M., Saedler, H., and Sommer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18, 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1984). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137, 266–267. [DOI] [PubMed] [Google Scholar]
- Flanagan, C.A., and Ma, H. (1994). Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant Arabidopsis flowers. Plant Mol. Biol. 26, 581–595. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. [DOI] [PubMed] [Google Scholar]
- Immink, R.G.H., Ferrario, S., Busscher-Lange, J., Kooiker, M., Busscher, M., and Angenent, G.C. (2003). Analysis of the petunia MADS box transcription factor family. Mol. Genet. Genomics 268, 598–606. [DOI] [PubMed] [Google Scholar]
- Immink, R.G.H., Gadella, T.W.J., Ferrario, S., Busscher, M., and Angenent, G.C. (2002). Analysis of MADS box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. USA 99, 2416–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink, R.G.H., Hannapel, D.J., Ferrario, S., Busscher, M., Franken, J., Campagne, M.M.L., and Angenent, G.C. (1999). A petunia MADS box gene involved in the transition from vegetative to reproductive development. Development 126, 5117–5126. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, J.S., Jang, S., Lee, S., Nam, J., Kim, C., Lee, S.H., Chung, Y.Y., Kim, S.R., Lee, Y.H., Cho, Y.G., and An, G. (2000). leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12, 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater, M.M., Colombo, L., Franken, J., Busscher, M., Masiero, S., Campagne, M.M.V., and Angenent, G.C. (1998). Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell 10, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotilainen, M., Elomaa, P., Uimari, A., Albert, V.A., Yu, D., and Teeri, T.H. (2000). GRCD1, an AGL2-like MADS-box gene, participates in the C function during stamen development in Gerbera hybrida. Plant Cell 12, 1893–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kush, A., Brunelle, A., Shevell, D., and Chua, N.-H. (1993). The cDNA sequence of two MADS box proteins in petunia. Plant Physiol. 102, 1051–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka, J., Harcourt, R., Peacock, W.J., and Dennis, E.S. (1997). Eucalyptus has functional equivalents of the Arabidopsis AP1 gene. Plant Mol. Biol. 35, 573–584. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1995). A gene triggering flower formation in Arabidopsis. Nature 377, 522–524. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1998). The Arabidopsis AGL9 MADS box gene is expressed in young flower primordia. Sex. Plant Reprod. 11, 22–28. [Google Scholar]
- Münster, T., Pahnke, J., Di Rosa, A., Kim, J.T., Martin, W., Saedler, H., and Theissen, G. (1997). Floral homeotic genes were recruited from homologous MADS-box genes preexisting in the common ancestor of ferns and seed plants. Proc. Natl. Acad. Sci. USA 94, 2415–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Gustafson-Brown, C., Kohlami, S.E., Crosby, W.L., and Yanofsky, M.F. (2001). APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 26, 385–394. [DOI] [PubMed] [Google Scholar]
- Pnueli, L., Hareven, D., Broday, L., Hurwitz, C., and Lifschitz, E. (1994). The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 6, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan, M.D., Rounsley, S.D., Schmidt, R.J., and Yanofsky, M.F. (1995). Molecular evolution of flower development: Diversification of the plant MADS-box regulatory gene family. Genetics 140, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1997). MADS domain proteins in plant development. Biol. Chem. 378, 1079–1101. [PubMed] [Google Scholar]
- Savidge, B., Rounsley, S.D., and Yanofsky, M. (1995). Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity. Plant Cell 7, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer, Z., Huijser, P., Nacken, W., Saedler, H., and Sommer, H. (1990). Genetic control of flower development in Antirrhinum majus. Science 250, 931–936. [DOI] [PubMed] [Google Scholar]
- Sung, S.K., Yu, G.H., and An, G. (1999). Characterization of MdMADS2, a member of the SQUAMOSA subfamily in apple. Plant Physiol. 120, 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandre, K., Albert, V.A., Sundas, A., and Engström, P. (1995). Conifer homologues to genes that control floral development in angiosperms. Plant Mol. Biol. 27, 69–78. [DOI] [PubMed] [Google Scholar]
- Theissen, G. (2001). Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. [DOI] [PubMed] [Google Scholar]
- Theissen, G., Kim, J.T., and Saedler, H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43, 484–516. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto, S., Mayama, T., Ohtsubo, E., and van der Krol, A. (2000). The whorl-specific action of a petunia class B floral homeotic gene. Genes Cells 5, 89–99. [DOI] [PubMed] [Google Scholar]
- Tsuchimoto, S., van der Krol, A., and Chua, N.-H. (1993). Ectopic expression of pMADS3 in transgenic petunia phenocopies the petunia blind mutant. Plant Cell 5, 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krol, A.R., Brunelle, A., Tsuchimoto, S., and Chua, N.-H. (1993). Functional analysis of petunia floral homeotic MADS box gene pMADS1. Genes Dev. 7, 1214–1228. [DOI] [PubMed] [Google Scholar]
- Verwoerd, T.C., Dekker, B.M.M., and Hoekema, A. (1989). A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., Schuch, W., and Giovannoni, J. (2002). A MADS-box gene necessary for fruit ripening at the tomato Ripening-inhibitor (Rin) locus. Science 296, 343–346. [DOI] [PubMed] [Google Scholar]
- Winter, K.-U., Becker, A., Münster, T., Kim, J.T., Saedler, H., and Theissen, G. (1999). MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proc. Natl. Acad. Sci. USA 96, 7342–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldman, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene Agamous resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]
- Yu, D.Y., Kotilainen, M., Pollanen, E., Mehto, M., Elomaa, P., Helariutta, Y., Albert, V.A., and Teeri, T.H. (1999). Organ identity genes and modified patterns of flower development in Gerbera hybrida (Asteraceae). Plant J. 17, 51–62. [DOI] [PubMed] [Google Scholar]