Abstract
Alternative splicing is a major contributor to genome complexity, playing a significant role in various cellular functions, including signal transduction, immunity, and development. The spliceosomal machinery is responsible for the processing of nuclear RNA. Several splicing factors associated with this complex are phosphorylated by kinases that possess a conserved LAMMER motif. We demonstrate in BY-2 tobacco cells a novel role for the LAMMER motif in the maintenance of proper subnuclear localization. Furthermore, high expression of the LAMMER kinase in Arabidopsis plants modulated the alternative splicing of specific endogenous genes and resulted in abnormal plant development and a novel transcriptome profile. A prominent feature was the upregulation of genes that play a role in protein turnover, suggesting a moderating function for these gene products in the control of alternative splicing events. Together, these results demonstrate alternative splicing modulation as a result of phosphorylation activity, providing an opportunity to study its global effect on the plasticity of plant development and gene expression at the organism level.
INTRODUCTION
Alternative splicing is a process that selects different splice sites in nuclear pre-mRNA and results in the production of distinct transcripts that will translate into diverse polypeptides. The recent realization that the human genome contains only one-fifth of the initial estimated number of genes has focused attention on alternative splicing as a critical means of increasing genome complexity (Graveley, 2001; Modrek and Lee, 2002). Thus, metazoan genes are spliced differentially under diverse conditions, including developmental stage, organ identity, sex determination, hormone stimuli, and stress responses (Lopez, 1998; Reddy, 2001).
RNA splicing reactions are mediated by the spliceosome complex, which is composed of small nuclear ribonucleoprotein particles (snRNPs) and a large number of accessory proteins (Kramer, 1996). Among the key non-snRNP components are the Ser/Arg (SR) splicing factors, which are characterized by the presence of an RNA recognition motif at their N termini, and the Arg/Ser (RS) domain at their C termini (Zahler et al., 1992; Fu, 1995; Graveley, 2000). Protein–protein interactions mediated by the RS motif as well as RNA recognition of cis-regulatory RNA elements are phosphorylation dependent (Xiao and Manley, 1997). Moreover, changes in SR phosphorylation lead to their redistribution within the nucleus (Colwill et al., 1996b; Kuroyanagi et al., 1998). Distinct kinases have been shown to phosphorylate SR proteins in vitro (Gui et al., 1994; Rossi et al., 1996; Kuroyanagi et al., 1998; Okamoto et al., 1998), among them kinases of the LAMMER family (Colwill et al., 1996b; Du et al., 1998; Golovkin and Reddy, 1999; Savaldi-Goldstein et al., 2000).
The ubiquitous LAMMER kinases share a similar overall structure in their kinase domains and a nearly identical EHLAMMERILG signature at their catalytic subdomain X, giving rise to the name. This motif was shown to be important for kinase activity (Savaldi-Goldstein et al., 2000). LAMMER members have dual specificity autophosphorylation on both Ser/Thr and Tyr residues (Ben David et al., 1991; Howel et al., 1991; Lee et al., 1996; Sessa et al., 1996); however, exogenous substrates analyzed to date are phosphorylated on Ser residues (Colwill et al., 1996a; Lee et al., 1996). The involvement of LAMMER kinases in alternative splicing was demonstrated for the mammalian and Drosophila members Clk/Sty and DOA, respectively. In mammalian cells, overexpressed Clk/Sty leads to a switch in the alternative splice site selection of miniature gene constructs (Duncan et al., 1997). In Drosophila, the loss of function of DOA affects sexual differentiation through specific gene splicing alteration and SR protein hypophosphorylation, providing in vivo support for the association of DOA with SR protein phosphorylation and splicing (Du et al., 1998). Thus, the phosphorylation of SR splicing factors in animal cells can influence the occurrence and specificity of splice site choice.
Plant and animal RNA processing systems share commonalties but have distinct differences. Plant splice cis elements have unique features that distinguish them from those of other eukaryotes, such as the UA-rich or U-rich compositional bias in their introns. Indeed, plant systems generally fail to process animal nuclear pre-mRNAs (Lorkovic et al., 2000b). The Arabidopsis genome maintains a relatively complex set of RNA recognition motif–containing genes. For example, 18 SR proteins are found in Arabidopsis compared with 10 in humans; among them are four mammalian ASF/SF2 homologs (atSRp30, atSRp34/SR1, atSRp34a, and atSRp34b) and SR proteins that contain unusual zinc-knuckle motifs (Lazar et al., 1995; Lorkovic and Barta, 2002). Plant SR proteins have been shown biochemically to play a role in splicing in animal in vitro systems (Lazar et al., 1995; Lopato et al., 1996). In addition, atSRp30 was shown to affect plant development and to change the alternative splicing pattern of specific endogenous genes when overexpressed in Arabidopsis (Lopato et al., 1999). The plant LAMMER kinases differ in structure from their animal counterparts in that they lack the RS domain thought to mediate the kinase–splicing factor interaction. However, PK12 and its Arabidopsis homolog, AFC2, were shown to phosphorylate and bind SR proteins in vitro (Golovkin and Reddy, 1999; Savaldi-Goldstein et al., 2000).
As sessile organisms, plants require special adaptations to cope with environmental changes. This is reflected in the coupling of gene expression during plant development and signal transduction pathways to surrounding stimuli. Thus, the plasticity of plant development offers an opportunity to follow the contribution of alternative splicing modulation to this flexibility. The present work uses two heterologous expression systems, tobacco suspension cells and Arabidopsis plants, to follow in vivo interactions between PK12 LAMMER kinase and atSRp34/SR1 and to examine their biological functions. We show that these components colocalize depending on the presence of the LAMMER motif. Transgenic Arabidopsis lines that express constitutively active PK12 LAMMER kinase exhibited modulation of the alternative splicing pattern of endogenous genes. These changes were associated with developmental defects, including overall size reduction and prolonged life cycle. The transcriptome of plants with high expression levels of PK12 reflects the alteration of gene expression profiles for a wide spectrum of cellular functions, including genes that are involved in stress and protein turnover.
RESULTS
Colocalization of the PK12 LAMMER Kinase with atSRp34/SR1 Splicing Factor Depends on the LAMMER Motif
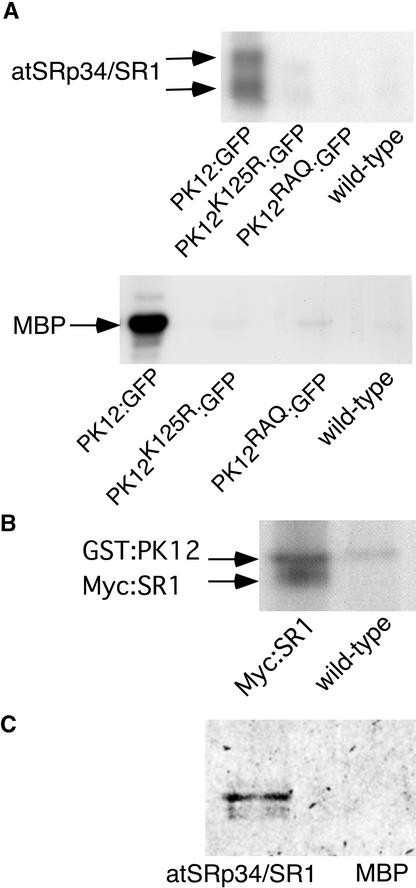
Plant LAMMER kinases have been shown to phosphorylate and bind splicing factors in vitro (Golovkin and Reddy, 1999; Savaldi-Goldstein et al., 2000). In vivo interactions between the kinase and SR splicing factors were followed using the wild-type PK12 and two mutant versions of the kinase, PK12RAQ and PK12K125R (Savaldi-Goldstein et al., 2000). PK12RAQ contains a mutation in the conserved LAMMER motif, a region that was shown to be important for kinase activity but not for binding to the SR protein. PK12K125R is a nonactive kinase, the result of a mutation in the invariant Lys present in subdomain II. For imaging purposes, green fluorescent protein (GFP) and a red variant, dsRED, were fused to the C termini of PK12 and its mutants (PK12:GFP, PK12RAQ:GFP, and PK12K125R:GFP) and to the N terminus of atSRp34/SR1 (RED:SR1), respectively. The fusion constructs were inserted behind the constitutively expressed 35S promoter of Cauliflower mosaic virus and used for stable transformation of tobacco BY-2 suspension cells. The functional fidelity of the fused product was examined in transformed cell extracts by immunoprecipitation with GFP-specific antibodies followed by an in vitro kinase assay using atSRp34/SR1 and myelin basic protein (MBP) as substrates (Figure 1A, top and bottom gel, respectively).
Figure 1.
Kinase and Binding Activities of PK12 Fusion Proteins.
(A) In vitro kinase assay of PK12 fusion proteins. PK12:GFP and its mutant forms were immunoprecipitated from transgenic BY-2 cells using antibodies raised against GFP. Nontransformed cells (wild type) were used as a control. The immunoprecipitate was subjected to an in vitro kinase assay using atSRp34/SR1 (top) or MBP (bottom) as a substrate.
(B) In vitro kinase assay of recombinant PK12 protein using atSRp34/SR1 as a substrate. Myc:SR1 was immunoprecipitated from control and transgenic BY-2 cells and subjected to an in vitro kinase assay in the presence or absence of recombinant GST:PK12. Residual autophosphorylation activity of the latter is shown.
(C) Overlay assay. atSRp34/SR1 and MBP proteins (1 μg) were fractionated on an SDS–polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane then was probed with in vitro Met 35S- labeled PK12:GFP.
The PK12:GFP fusion protein retained phosphorylating activity, as shown by two bands corresponding to different phosphorylation states of atSRp34/SR1 and a single band of phosphorylated MBP. The activities of the mutant derivatives were only slightly above background. For convenience, the MBP substrate was used to monitor the kinase activity subsequently. Myc N-terminally tagged atSRp34/SR1 (Myc:SR1) expressed in BY-2 cells was tested for its ability to undergo phosphorylation by PK12. Myc:SR1 was immunoprecipitated from BY-2 cells, and the precipitate bound to the beads was subjected to kinase assay in the presence or absence of recombinant PK12 (Figure 1B). As shown, Myc:SR1 is a substrate for PK12. The additional band represents glutathione S-transferase (GST):PK12 autophosphorylation activity, and its abundance in the context of Myc:SR1 immunoprecipitate reflects binding to atSRp34/SR1. The binding capability of the PK12:GFP fusion polypeptide to atSRp34/SR1 was verified further by an overlay assay using in vitro–labeled Met 35S-PK12:GFP. As shown in Figure 1C, PK12:GFP bound atSRp34/SR1 but did not bind to the general substrate MBP. Thus, GFP fusion does not appear to affect the PK12 phosphorylating or binding properties, and N-terminally tagged atSRp34/SR1 expressed in BY-2 cells serves as a substrate for PK12 in vitro.
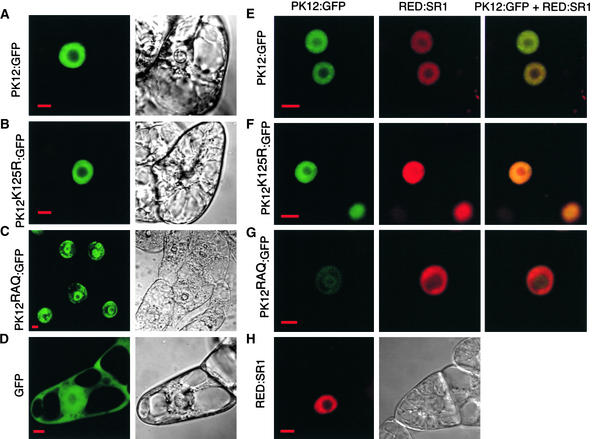
The subcellular distribution of PK12:GFP, PK12K125R:GFP, and PK12RAQ:GFP was examined using a confocal fluorescence microscope. Compared with the GFP control (Figure 2D), PK12:GFP and PK12K125R:GFP appeared as a diffuse pattern limited to the nucleus and were excluded from the nucleoli (Figures 2A and 2B). The latter did not exhibit the prominent speckled pattern reported for the equivalent inactive mammalian LAMMER kinase Clk/Sty (Colwill et al., 1996b; Sacco-Bubulya and Spector, 2002). Unexpectedly, although most of the cells expressing PK12RAQ:GFP showed a normal localization pattern similar to those of PK12:GFP and PK12K125R:GFP, ∼30% of the transformed cells showed aberrant localization of this fusion protein into a “ring structure” (Figure 2C). In these cells, PK12RAQ:GFP was found to accumulate in the periphery of the nucleus in an aggregate manner and on the border of the nucleolus. Cells that contained RED:SR1 exhibited a nucleoplasmic distribution pattern similar to that found for PK12:GFP (Figure 2H), in contrast to the speckled formation that was detected in the mammalian homolog ASF/SF2 (Mintz and Spector, 2000).
Figure 2.
Subcellular Localization of PK12 and SR1 Fusion Proteins.
Confocal optical sections are shown of viable BY-2 cells that overexpress either PK12 or SR1 proteins fused to GFP and dsRED, respectively, or cells that overexpress both PK12:GFP and RED:SR1. The panels at right show color overlays. The fluorescence images displaying either GFP or dsRED signal are shown together with the corresponding Nomarski images. Bars = 10 μm.
(A) PK12:GFP.
(B) PK12K125R:GFP.
(C) PK12RAQ:GFP shown in cells exhibiting nuclear ring structure.
(D) GFP.
(E) PK12:GFP and RED:SR1.
(F) PK12K125R:GFP and RED:SR1.
(G) PK12RAQ:GFP shown in cells displaying nuclear ring structure and RED:SR1.
(H) RED:SR1.
BY-2 cells that overexpress PK12:GFP, PK12K125R:GFP, or PK12RAQ:GFP were transformed with constructs containing the RED:SR1 fusion gene (Figures 2E to 2G). Overlay of the color-coded distribution patterns show that RED:SR1 colocalized with PK12:GFP and PK12K125R:GFP (Figures 2E and 2F). However, in cells that showed aberrant PK12RAQ:GFP localization patterns, RED:SR1 colocalized with PK12RAQ:GFP in the nuclear periphery but was not found in the ring structure or within the nucleolus region (Figure 2G). Thus, PK12RAQ:GFP lost its ability to colocalize with RED:SR1.
Constitutive Kinase Activity of PK12 in Arabidopsis Transgenic Lines
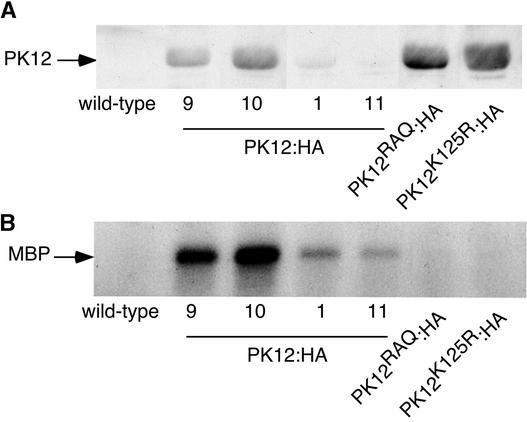
PK12 LAMMER kinase is homologous with the Arabidopsis LAMMER isoform AFC2 (73% identity) and was shown here to bind, phosphorylate, and colocalize with Arabidopsis atSRp34/SR1. To examine its activity in vivo, the PK12 LAMMER kinase was fused to three consecutive repeats of the hemagglutinin (HA) tag at its C-terminal end, cloned behind the constitutive 35S promoter (PK12:HA), and introduced into Arabidopsis. Using the HA tag, independent homozygous lines with low (lines 1 and 11) and high (lines 9 and 10) protein expression levels were selected for further analysis (Figure 3A). Transgenic lines that overexpress PK12RAQ:HA and PK12K125R:HA were generated similarly. The various PK12:HA fusion polypeptides were immunoprecipitated from total plant extracts and subjected to an in vitro kinase assay using MBP as a substrate (Figure 3B). As shown, the PK12:HA fusion had constitutive activity that was correlated with its polypeptide expression level; however, the mutant PK12 derivatives had only residual, near-background activity, as expected.
Figure 3.
Expression and Activity of PK12:HA and Its Derivatives in Transgenic Plant Lines.
(A) Protein gel blot analysis. Equal amounts of total protein were fractionated by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies raised against HA. Four homozygous independent lines with high (lines 9 and 10) and low (lines 1 and 11) wild-type PK12:HA ex-pression levels, and homozygous lines that highly express mutant PK12K125R:HA or PK12RAQ:HA, are shown.
(B) In vitro kinase assay. PK12:HA and its mutant variants were immunoprecipitated from the transgenic lines as shown in (A). The immunoprecipitate then was subjected to an in vitro kinase assay using MBP as a substrate.
Overexpression of PK12:HA Modifies Plant Development
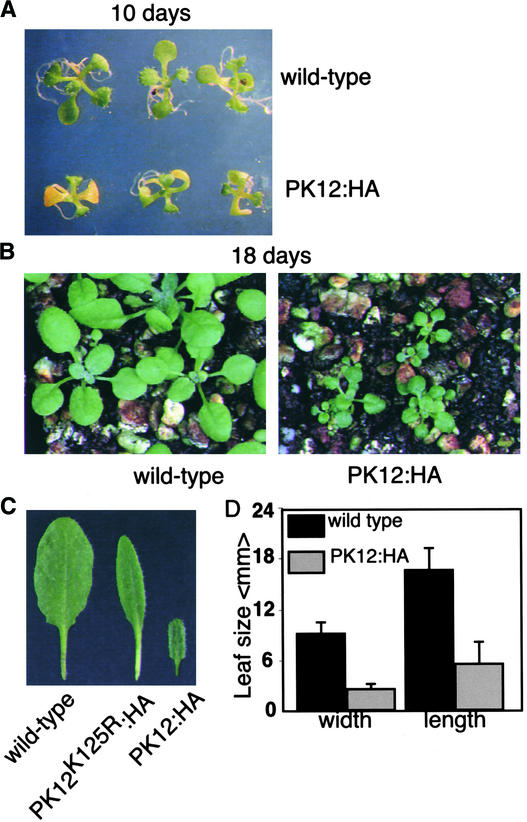
Transgenic lines that showed high levels of PK12 polypeptide expression and kinase activity were diminutive and had prolonged life cycles (Figure 4). When grown under long-day conditions, PK12:HA plants (lines 9 and 10) showed significantly smaller leaf size. Thus, line 10 had an average length of 3.3 mm and an average width of 6.3 mm compared with wild-type plants, with an average length of 20 mm and an average width of 12.1 mm (Figures 4A to 4D). Similar results were obtained for line-9 plants. This phenotype can be attributed to the smaller size of cells, measured as 4.6 ± 1.3 and 7.6 ± 0.5 in 10-mm2 leaf area in wild-type and PK12:HA (line 10) plants, respectively. At 25 days after germination, control plants bolted, compared with 25 to 30 days (line 9) and 35 days (line 10) for PK12:HA plants. Plants that overexpressed high levels of PK12K125R:HA or PK12RAQ:HA or low levels of PK12:HA showed similar flowering time and overall size as wild-type plants (data not shown). Leaves of PK12K125R:HA plants tended to be more narrow (Figure 4C).
Figure 4.
Morphology of PK12:HA Plants.
(A) PK12:HA high-expression plants and wild-type plants were grown for 10 days in agar under long-day conditions.
(B) PK12:HA high-expression plants and wild-type plants were grown for 18 days in soil under long-day conditions.
(C) Leaves from wild-type (left), PK12K125R:HA (middle), and PK12:HA (right) plants.
(D) Average length and width of rosette leaves in wild-type and PK12:HA high-expression lines.
Plants shown are from PK12:HA line 10.
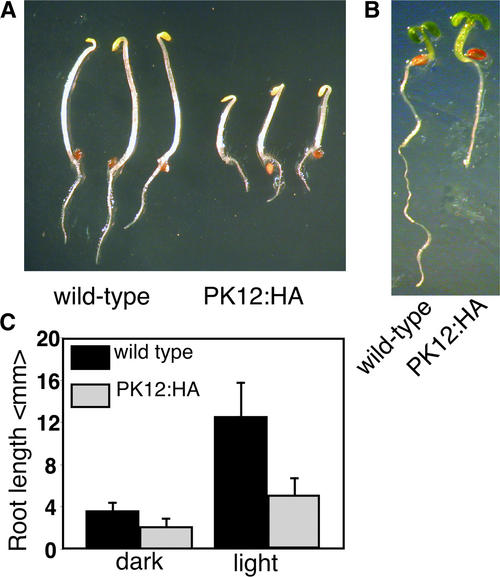
PK12 activity was shown to be induced transiently by ethylene in tobacco and could be involved in generating aspects of the ethylene-dependent morphology (Sessa et al., 1996). There-fore, we examined PK12:HA high-expression lines for additional phenotypic characteristics. In dark-grown wild-type Arabidopsis seedlings, treatment with ethylene induces a triple response (i.e., short roots, short hypocotyls, and exaggerated hook formation) (Guzman and Ecker, 1990). In light-grown seedlings, ethylene can promote the elongation of hypocotyls (Smalle et al., 1997). Plants that contain gain-of-function mutations of the ethylene pathway express these phenotypes constitutively. When PK12:HA seedlings were germinated in the dark in the absence of ethylene, they exhibited constitutively shorter roots and hypocotyls (Figure 5A). Shorter roots were similarly apparent in the light (Figure 5B). The measurement of root length in light-grown and dark-grown seedlings is summarized in Figure 5C. Only minor differences in apical hook curvature, the third characteristic of ethylene treatment, were apparent. Therefore, the PK12 overexpression phenotype exhibits only a subset of the ethylene-dependent morphological changes, which may indicate its involvement in different developmental pathways.
Figure 5.
Constitutive PK12:HA Activity Is Not Involved in the Ethylene Pathway.
(A) Wild-type and high-expression PK12:HA plants germinated for 3 days in the dark.
(B) Wild-type and high-expression PK12:HA plants germinated for 5 days in the light.
(C) Average length of roots in wild-type and PK12:HA plants grown under dark or light conditions.
Plants shown are from PK12:HA line 9.
PK12 Modulates Alternative Splicing in Planta
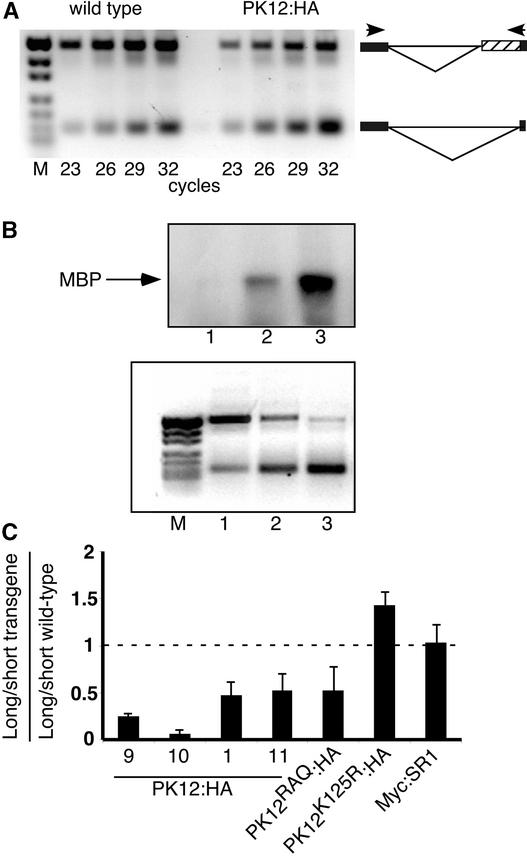
The interaction of PK12 with SR splicing factors suggests that it likely plays a role in modulating splicing. This possibility was examined by following atSRp30, which has been reported previously to appear in alternative transcript forms (Lopato et al., 1999). The transcript patterns of this gene was assayed by a reverse transcriptase–mediated (RT) PCR approach that elucidates alternative splicing (Dinesh-Kumar and Baker, 2000; Auboeuf et al., 2002). In this procedure, specific primer pairs spanning the alternative spliced intron are used for quantitative measurements of splicing activity. As exemplified in Figure 6A for wild-type and PK12:HA (line 1) plants, the PCR cycle first was optimized to determine the linear range of amplification for the transcript sizes. In this case, cycle 26 was selected for analysis. The sequence of the long transcript of atSRp30 revealed that it encodes mRNA3, a result of an alternative 3′ splice site in the 10th intron (Lopato et al., 1999).
Figure 6.
Modulation of Alternative Splicing in the atSRp30 Transcript in PK12:HA Plants.
(A) Quantitative RT-PCR analysis of two spliced variants in the atSRp30 transcript. PCR products were fractionated on an ethidium bromide–stained agarose gel. The reaction was performed on first-strand cDNA generated from total RNA extracted from wild-type and PK12:HA plants (line 1). The PCR cycle was terminated at the cycle number indicated to determine the linear range common to the two alternatively spliced forms. Schemes of the alternatively spliced forms are shown at right. The boxes indicate exons flanking the 10th intron. The hatched box represents the intron fragment that resulted from the use of the 3′ alternative splice site. Primers flanking the alternatively spliced intron are shown as arrowheads. M, molecular mass markers.
(B) In vitro kinase assay and RT-PCR analysis of wild-type (lane 1), low-expressing PK12:HA (lane 2), and high-expressing PK12:HA (lane 3) plants. Top gel, total protein extract was subjected to immunoprecipitation with anti-HA antibodies. The immunoprecipitate then was used in an in vitro kinase assay with MBP as a substrate. Bottom gel, RT-PCR analysis of the atSRp30 transcript was performed on transcripts from the same lines used in the top gel. M, molecular mass markers.
(C) Quantification of alternatively spliced long and short forms of atSRp30 in wild-type and transgenic plants. The PK12:HA lines are as indicated. The ratio ± sd of long to short transcript was calculated in different transgenic plants (for details, see Methods). The resulting value then was divided by the value obtained from the same calculation performed for wild-type plants grown under the same conditions. A ratio of 1 (represented by a dashed line) indicates no change in alternative splicing between wild-type and transgenic plants. The results are representative of at least three independent experiments.
We next compared the relationship between PK12:HA transgene activity and splice variants of atSRp30. To this end, lines that exhibited increasing levels of kinase activity were chosen (Figure 6B, top gel). In this case, a continuous shift of atSRp30 alternative splicing from long to short forms was evident as a function of PK12 kinase activity (Figure 6B, bottom gel). The level of alternative splicing was quantified by calculating the ratio between the long and short forms of atSRp30 transcripts in transgenic lines and comparing it with the equivalent ratio measured in wild-type plants. Lines with low PK12:HA expression showed a moderate decrease in the ratio (Figure 6C, lines 1 and 11), whereas high-expression plants (Figure 6C, lines 9 and 10) had a dramatically reduced ratio compared with wild-type plants. Similar modification of splicing was obtained with high and low PK12:HA transgene expression in the Columbia background as well (data not shown). In addition, measurements taken from transgenic plants that overexpress Myc:SR1 in the same binary vector did not significantly change the splicing pattern observed in these plants (Figure 6C). Thus, the expression of this specific splicing factor, the transformation procedure itself, and the plant background do not influence splicing. We conclude that the active PK12 LAMMER kinase modulates alternative splicing in plants in a dose-dependent manner.
Lines that express high levels of each of the two inactive mutated versions of PK12 showed a more complex effect on splicing. PK12RAQ:HA showed intermediate changes in the splicing pattern, similar to the levels achieved with low expression of PK12:HA. By contrast, the PK12K125R:HA transgene reversed the ratio, rendering more “long” alternative spliced variant than the “short” spliced form compared with the wild type (Figure 6C). In both mutant lines lacking kinase activity, the amount of cross-reactive polypeptide was greater than that detected in the most highly expressed PK12:HA lines (Figure 3). This finding suggests that the presence of inactive PK12 polypeptides can exert a dominant effect on splicing.
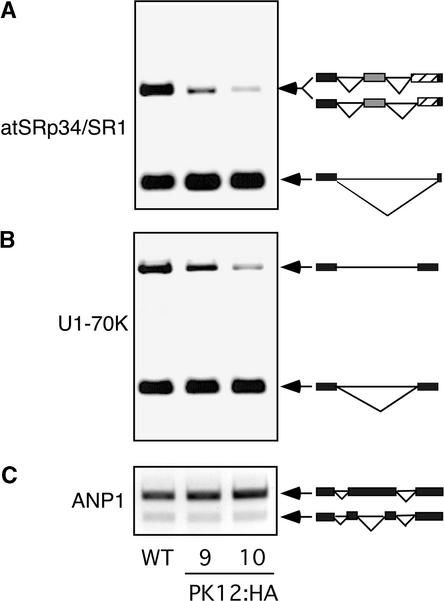
The specificity of the effect of increased PK12 kinase activity was examined in high PK12 expression lines for other genes, including splicing factors atSRp34/SR1 and U1-70K and a mitogen-activated protein kinase kinase kinase, ANP1. In all three genes examined, two main transcripts were detected (Figure 7). The long alternatively spliced form in atSRp34/SR1 is composed of two transcripts that differ from each other by 10 bp. One transcript encodes the previously reported SR1C (Lazar and Goodman, 2000), and the second encodes a newly identified alternative 3′ splice site that is present 10 bp downstream. The longer transcript of U1-70K is a product of intron inclusion (Golovkin and Reddy, 1996). The bands in ANP1 are the result of differential splicing of an intron-like coding frame sequence (Nishihama et al., 1997). As shown in Figures 6B, 7A, and 7B, plants that support the highly expressed PK12:HA transgene consistently displayed a shift in the alternatively spliced mRNAs of splicing factors atSRp30, atSRp34/SR1, and U1-70K. By contrast, the ANP1 transcript showed no variation. The differential effect of PK12 activity on splicing variants suggests a degree of specificity.
Figure 7.
Modulation of Alternative Splicing in High-Expression PK12 Kinase Lines.
RT-PCR was used to amplify the alternatively spliced products in PK12:HA plants (lines 9 and 10) as described for Figure 6A. Schemes describing the corresponding splicing events are shown at right. WT, wild type.
(A) atSRp34/SR1 showing 5′ and 3′ alternative splicing.
(B) U1-70K showing intron retention.
(C) ANP1 showing alternative exon choice.
Transcriptome Activity in PK12:HA Overexpression Plants
RNA processing can have a direct impact on the final gene product or transcript stability; thus, it can exert ripple effects on the profile of gene expression (Proudfoot et al., 2002). To examine the changes in global transcription caused by PK12 LAMMER kinase activity, we determined gene expression in plants at two different stages of growth. Differentially expressed genes common to both stages likely represent the modification of gene expression that results from a direct or indirect response to the kinase activity rather than from occasional environmental conditions or a particular developmental stage. Total RNA was extracted from transgenic PK12:HA (line 10) and wild-type plants at 10 and 21 days and compared. At 10 days, wild-type and transgenic plants both contained three to four apparent true leaves (Figure 4A), whereas at 21 days, both plant types featured rosettes with seven to eight leaves (Figure 4B). For each time point, two biological repetitions were performed, and for each repetition, two hybridizations were performed on duplicate arrays containing either 9000 or 12,000 Arabidopsis ESTs as probes. The results were processed using the GeneSpring microarray analysis package (Table 1) (see Methods). Transcripts with a statistically significant ratio of >1.6-fold change that were common to at least six of the eight hybridizations were compiled and were found to represent 48 different genes. The genes with known functions are listed in Table 1, and those with unknown functions, as well as the complete experimental data, are provided at http://www.ncbi.nlm.nih.gov/geo (accession number GSE274).
Table 1.
Upregulated and Downregulated Transcripts in the PK12 LAMMER Kinase Overexpression Linea
| Upregulated Genes | Downregulated Genes |
|---|---|
| Protein destination | Energy |
| At4g24190: Hsp90-7, 2.4 ± 0.7 (5 ESTs), 2.1 ± 0.2 (2 ESTs) | At1G44575: psbS mRNA, 0.3 ± 0.1, 0.4 ± 0.1 |
| At3g09440: HSP70, 2.2 ± 0.5, 2.0 ± 0.2 (2 ESTs) | At1g29930: PSII cab protein, 0.4 ± 0.1 (12 ESTs), 0.4 ± 0.1 |
| At5g28540: BiP, 2.4 ± 0.4, 2.7 ± 0.8 | At1g31330: PSI reaction center III, 0.4 ± 0.1 (3 ESTs), 0.5 ± 0.1 |
| At5g19180: ECR1, 3.5 ± 0.9, 2.5 ± 1 | At4g12800: PSI chain XI, 0.4 ± 0.1(5 ESTs), 0.6 ± 0.03 |
| At2g26990: CSN2, 3.9 ± 2.7, 3.0 ± 1 | At3g61470: Lhca2, 0.4 ± 0.1 (12 ESTs), 0.6 ± 0.04 |
| At5g10760: cnd41, 29.2 ± 10.2, 4.9 ± 2 | At4g10340: Lhcb5, 0.4 ± 0.1 (7 ESTs), 0.5 ± 0.1 |
| Cell rescue, defense, cell death, and aging | At5g38410: ssRubisco 3b, 0.4 ± 0.1, 0.5 ± 0.1 (10 ESTs) |
| At1g74710: ICS1, 4.2 ± 3.4 (2 ESTs), 2.0 ± 0.4 | At1g32060: PRK precursor, 0.4 ± 0.2 (7 ESTs), 0.5 ± 0.2 |
| At1g72930: RPP5-like, 5.9 ± 2.3, 3.8 ± 0.4 | Metabolism |
| At1g72910: RPP5-like, 2.7 ± 0.7, 3.2 ± 0.8 | At5g46800: carnitine acyl carrier, 0.4 ± 0.1, 0.5 ± 0.02 |
| At1g31580: CxC750, 2.5 ± 0.6, 2.4 ± 0.6 (2 ESTs) | At3g61580: sphingolipid desaturase, 0.3 ± 0.1, 0.5 ± 0.1 |
| At1g02920: GST, 6.6 ± 2.6, 2.2 ± 0.5 | At5g49360: glucosyltransferase, 0.3 ± 0.1, 0.6 ± 0.03 |
| At1g02930: GST, 2.9 ± 1.2 (7 ESTs), 2.0 ± 0.3 | At3g47340: ASN1, 0.4 ± 0.1 (2 ESTs), 0.6 ± 0.03 |
| At1g07600: metallothionein, 4.9 ± 1.6 (3 ESTs), 2 ± 0.5 | At5g49360: xylosidase, 0.4 ± 0.1 (8 ESTs), 0.4 ± 0.1 (2 ESTs) |
| At1g21310: extensin 5 3.8 ± 1.0, 5.7 ± 3.5 | Cell rescue, defense, cell death, and aging |
| Cellular communication/signal transduction | At4g35770: Sen1, 0.3 ± 0.2 (4 ESTs), 0.3 ± 0.1 (2 ESTs) |
| At1g34750: phosphatase type 2C, 1.9 ± 0.15, 1.9 ± 0.3 | Cellular communication/signal transduction |
| Metabolism | At5g21170: AKIN β1, 0.4 ± 0.1, 0.3 ± 0.1 |
| At3g51240: flavanone-3-hydroxylase, 3.0 ± 0.9 (2 ESTs), 1.7 ± 0.3 | Cellular biogenesis |
| At2g45300: EPSP, 10.0 ± 9.9 (3 ESTs), 2.4 ± 0.4 | At5g19770: tubulin α-5 chain, 0.4 ± 0.1, 0.5 ± 0.1 |
| Transcription | |
| At1g36060: RAP2.4, 0.5 ± 0.05, 0.3 ± 0.1 |
Plants analyzed were 10- and 21-day-old plants of PK12:HA line 10. Transcripts were divided into functional categories using definitions provided by MIPS (http://mips.gsf.de/proj/thal/db/tables/tables_func_frame.html). Transcripts (12) without homology with known proteins were not assigned to categories. Transcripts with a ratio of >1.6-fold change in at least three of the four repetitions in both 10- and 21-day-old plants are tabulated, and their average ratios are shown. The numbers of different ESTs that belong to the same gene are indicated in parentheses. PSI and PSII, photosystems I and II, respectively; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.
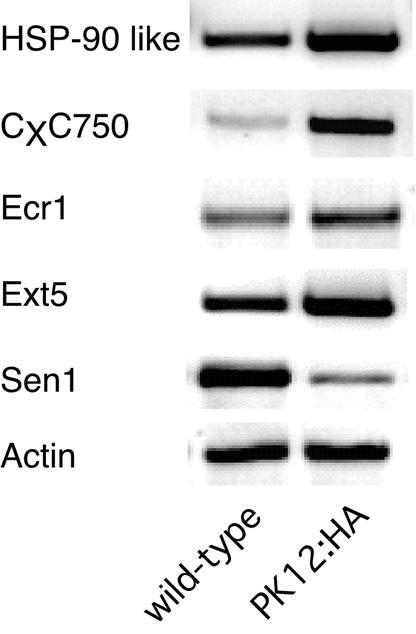
The microarray analysis was confirmed by sampled quantitative RT-PCR of transcripts of Hsp90-7, CxC750, Ecr1, and AtExt5 and the downregulated transcript, Sen1, in which actin transcript was used as a control (Figure 8). The prominent common upregulated genes function in protein destination and turnover. They include the ubiquitous chaperon classes Hsp90, Hsp70, and BiP as well as components of the proteasome system, the ubiquitin-activating enzyme E1 (ECR1) and the proteasome signalosome subunit 2 homolog (CSN2). The other prominent upregulated group includes transcripts that are indicative of cell stress (e.g., GST, metallothionein, pathogen-induced CxC750 mRNA, and isochorismate synthase 1 precursor for salicylic acid biosynthesis [ICS1]). Common downregulated genes are characterized by transcripts that are related chiefly to components of the photosynthetic machinery and metabolism.
Figure 8.
Modulation of Transcriptome in High-Overexpression PK12:HA Lines.
RT-PCR analysis of selected upregulated or downregulated genes in 21-day-old PK12:HA plants.
DISCUSSION
The LAMMER Motif Plays a Role in the Subnuclear Colocalization of PK12 and atSRp34/SR1
PK12 overexpression in transgenic plants has allowed us to gain insight into the overall effect of a LAMMER kinase at the organism level. Three different biological aspects are modulated as a result of the kinase activity of Arabidopsis: alternative splicing, gene expression, and plant phenotype. It is likely that these responses are mediated by the interaction of LAMMER kinases with SR proteins, as shown both by localization studies in living cells (this study) and in vitro (Savaldi-Goldstein et al., 2000). However, we cannot exclude the possibility that other, yet unknown substrates for LAMMER kinases might be involved.
Stable overexpression of the PK12 LAMMER kinase and its mutant derivatives in the presence or absence of atSRp34/SR1 in tobacco BY-2 cells revealed significant similarities and differences with that shown for their animal counterparts. The active mammalian LAMMER member Clk/Sty exhibited a diffuse pattern of distribution in the nucleoplasm, whereas its inactive variant was confined to speckles (Colwill et al., 1996b; Sacco-Bubulya and Spector, 2002). Under certain conditions, a subpopulation of Clk/Sty also was detected in the cytoplasm (Menegay et al., 1999; Yun et al., 2000). In BY-2 cells, both active PK12:GFP and its inactive variant PK12K125R:GFP were found to be dispersed in the nucleoplasm, and no evidence for cytoplasmic localization was detected. It is possible that localization into speckles is a function of cell type and species. For example, the SR protein ASF/SF2 from either human or Drosophila localizes with both speckles and diffuse patterns in HeLa cells, whereas in Drosophila S2 cells, they show only a diffuse pattern (Allemand et al., 2001). In addition, although several spliceosomal snRNPs concentrate in speckles in animal cells, they are distributed in an “interchromatin network” rather than in speckles in BY-2 cells (Beven et al., 1995). Furthermore, distinct plant RNA binding proteins localized in the nucleus in a similar manner (Lambermon et al., 2000; Lorkovic et al., 2000a). Likewise, and in contrast to its animal homolog ASF/SF2, the RED:SR1 splicing factor was found not to appear in nuclear speckles (Mintz and Spector, 2000). atSRp34/SR1 can be immunoprecipitated from BY-2 cells in a phosphorylation-receptive form, and GFP:PK12 binds atSRp34/SR1 in vitro. Thus, it is likely that this phosphorylation step also takes place in vivo. Several splicing factors, most of them of the SR family, are the only known substrates to interact with LAMMER kinases (Colwill et al., 1996b; Du et al., 1998).
Based on our analysis of kinase mutants, the colocalization of the PK12 LAMMER kinase and atSRp34/SR1 was independent of kinase activity. An intriguing result came from the aberrant localization of the LAMMER motif mutant PK12RAQ:GFP. In this case, a significant population of cells adopted a ring-structure pattern. That only a subpopulation conveyed this morphology may be attributable to a specific cell stage or to stochastic critical transgene accumulation in the cells. Interestingly, the mutated p80 coilin polypeptide that is localized normally to coiled bodies in the nucleus of HeLa cells was shown to accumulate around the nucleoli (Bohmann et al., 1995). PK12RAQ:GFP did not colocalize with RED:SR1 in a ring structure. Thus, the LAMMER motif may be required for protein–protein interactions that are important for subnuclear localization. Alternatively, mutations in such disparate molecules as LAMMER kinases and coilin may share in common the ability to lead to transcriptional arrest, a phenomenon that has been shown to induce aberrant accumulation at the nucleolar periphery (Carmofonseca et al., 1992; Smetana et al., 2001). Because splicing and transcription are linked processes (Proudfoot et al., 2002), the overexpression of PK12RAQ:GFP may interfere with normal splicing and slow the transcription processes, resulting in ring structure and an aberrant localization pattern.
Alternative Splicing as a Function of PK12 Activity in Planta
Comparison of Arabidopsis lines that express various levels of active PK12 demonstrated that the observed changes in alternative splicing were correlated with kinase activity. Transcripts atSRp30, atSRp34/SR1, and U1-70K showed an alternative splicing pattern that favored the formation of shorter transcripts in plant lines with high levels of PK12 activity. However, the alternative transcripts of ANP1, a gene unrelated to splicing factors, remained unchanged. Thus, the effect of PK12 shows a degree of specificity, the extent of which remains to be defined. LAMMER kinase activity is unlikely to be exclusive for transcripts of splicing factors, because the Drosophila LAMMER kinase loss-of-function mutant disrupted sex-specific splicing of doublesex pre-mRNA (Du et al., 1998). Plants that overexpress atSRp34/SR1 had no significant effect on alternative splicing of atSRp30. Similarly, atSRp34/SR1 did not affect its own splicing when transfected transiently into Arabidopsis protoplasts (Lazar and Goodman, 2000). However, modification of atSRp34/SR1 splicing has been shown to occur in plants that overexpress atSRp30, albeit at a different position from that shown here (Lopato et al., 1999). Together, these findings suggest that modulation of splicing factor phosphorylation and level results in a distinct alteration of splicing and implies complex control junctions in alternative splice choice. As one would expect from the global deregulation of splicing, both PK12:HA- and atSRp30-overexpressing plants exhibited a pleiotropic phenotype, although of distinct features and in agreement with their differential effect on splicing.
It is intriguing that opposing effects on splicing were obtained in lines that overexpress PK12 and the inactive PK12K125R:HA or PK12RAQ:HA. A possible explanation for this finding would include the formation of dynamic spliceosomal components that would depend on PK12 activity and the subsequent splice site choice. PK12K125R:HA likely confers a hypophosphorylated state on SR proteins compared with the active PK12 kinase, imposing a “mirror image” of splicing patterns compared with active PK12. The LAMMER motif was shown here to be required for the proper localization of the kinase. Its abnormal localization within the nucleus could lead to pleiotropic changes that would influence the selection of the particular transcript splice site.
The Plant LAMMER Kinase Modulates Development and Gene Expression
The role of LAMMER kinases in development was first demonstrated in the Drosophila LAMMER kinase, DOA. Loss-of-function mutations in DOA are lethal, and adult flies that escape death are defective in a variety of structures (Yun et al., 1994). This phenotype is present irrespective of sex type, although DOA is involved in sex determination through alternative splicing (Du et al., 1998). As opposed to a single gene encoding LAMMER in Drosophila, there are three homologs in the Arabidopsis genome. Phylogenetic comparison of sequences (data not shown) indicates that this division is prevalent in other plants and is of ancient origin. Whether the other alleles play redundant roles or add new functions remains to be seen.
PK12 is expected to maintain a low basal activity level under continuous ethylene response (Sessa et al., 1996). This observation is consistent with the normal-type splicing pattern that was observed for atSRp30 and atSRp34/SR1 in ctr1, a mutant in which ethylene response is constitutively active (data not shown). High constitutive PK12 activity is unlikely to play a role in the primary control of ethylene-mediated processes. Indeed, LAMMER kinase overexpression did not induce genes that belong to the ethylene-signaling pathway per se. Instead, genes that correspond to stress and defense response were expressed differentially. The partial triple response observed in PK12:HA plants can result from the modulation of the AP2/EREBP-type DNA binding protein Rap 2.4, because a gain of function of ERF-1 and TINY, both of which are AP2/EREBP-type DNA binding proteins, was shown to cause a partial triple response (Wilson et al., 1996; Solano et al., 1998). Thus, regulation of PK12 could contribute aspects of the triple response through AP2/EREBP gene expression.
Constitutive high PK12 LAMMER kinase activity was shown here to be associated with changes in plant development. The most prominent effects included smaller cells, reduced size of the plants, and prolonged life cycle. The morphological modifications were accompanied by alterations in the differential expression of genes that belong to distinct functional categories. Upregulated genes that were independent of plant age included members of the chaperone and proteasome machinery that are related to increased protein turnover. This category likely is related directly to the effects of the deregulated LAMMER kinase on alternative splicing and the production of variant gene products, suggesting enhanced protein turnover caused by the production of defective polypeptides as a result of aberrant splicing. Upregulated members include a variety of heat-shock proteins, among them Hsp90-7. Interestingly, this heat-shock protein gene product has been implicated as a capacitor for phenotypic variation. Thus, inhibition of its activity by pharmacological agents increased the frequency of exposure to cryptic genetic variation (Queitsch et al., 2002). The upregulation detected here suggests that Hsp chaperones also police variations in the products of alternative splicing.
Among downregulated genes are components of the photosynthetic machinery and the sucrose-sensitive transcripts sen1 and asn1 (Lam et al., 1994; Chung et al., 1997). Photosynthetic capacity is known to be inhibited by the presence of excess free sugars (Oswald et al., 2001), and sen1 and asn1 are downregulated by sugar accumulation (Lam et al., 1994; Chung et al., 1997). Thus, a situation could be imagined in which these seemingly disparate transcripts are reduced as a result of increased sugar levels acting as a unifying signal. Possible changes in sugar metabolism also may explain the modulation of AKIN β1 transcript that is associated with SNF1-related kinases (SnRK1 family) that play a role in sugar signal transduction. The relationship between alternative splicing afforded by plant LAMMER kinase overexpression and sugar signaling remains to be elucidated.
The observed changes in gene expression can be attributed to either direct or indirect effects of altered splicing or splicing components on transcript stability. For example, ASF/SF2 was shown recently to regulate gene expression by binding to a cis element in the 3′ region of the transcript independent of splicing activity (Lemaire et al., 2002). However, the enhanced transcription of components related to protein stability, turnover, and stress and the downregulated transcripts of metabolism and photosynthesis likely represent secondary effects. Measuring global effects of the LAMMER kinase on gene transcripts would require the development of specialized splice-sensitive microarrays. Such microarrays have been used recently in the analysis of yeast constitutive splicing mutants (Clark et al., 2002). Exploitation of these tools will facilitate our understanding of the regulation and specificity of differential splice choice.
METHODS
Plant and Cell Culture Materials and Growth Conditions
Arabidopsis thaliana plants (C-24 ecotype) were grown under white light in a 16-h-light/8-h-dark cycle at 21°C. For RNA and microarray analysis, seeds were washed in 75% ethanol, treated for 10 min in 3% bleach and 0.1% Tween 20, rinsed three times with sterile water, and spread on 0.8% Nitsch plates (Nitsch, 1969). The plates were vernalized for 2 days in darkness at 4°C and then placed in light for 10 days. For seed production, phenotypic analysis, and microarray analysis, seeds were spread onto soil under similar light and temperature conditions. Tobacco (Nicotiana tabacum) BY-2 cells were maintained by weekly dilution in fresh Murashige and Skoog (1962) medium supplemented with 200 mg/L KH2PO4, 1 mg/L thiamine HCl, 100 mg/L myoinositol, and 30 g/L sucrose maintained with shaking at 120 rpm at 25°C.
Constructs and Transgenic Cell Lines and Plants
PK12 cDNA was fused upstream to three repeats of the hemagglutinin (HA) epitope (YPYDVPDYA) in pBluescript SK+ vector. The chimera PK12:HA gene then was subcloned into the polylinker cloning site of the binary vector located between the 35S promoter of Cauliflower mosaic virus and the E9 terminator. PK12K125R and PK12RAQ cDNAs were generated as described previously (Savaldi-Goldstein et al., 2000), fused to HA, and cloned into binary vector in the same mode. Arabidopsis plants were transformed with Agrobacterium tumefaciens strain EHA105 as described by Clough and Bent (1998). Transgenic plants were selected on agar plates supplemented with 100 mg/L kanamycin. T2 plants were tested for segregation on kanamycin and for PK12:HA expression. Six independent lines with different expression levels were selected for further analysis and grown for T3 homozygous plants. For imaging purposes, PK12, PK12K125R, and PK12RAQ cDNAs were fused to the N terminus of GFP (Clontech, Palo Alto, CA). atSRp34/SR1 cDNA was fused to the C terminus of dsRED (Clontech). GFP fusion constructs then were subcloned into binary vector as described above. The RED:SR1 fusion construct was subcloned into a cassette containing the 35S promoter of Cauliflower mosaic virus and the OCS terminator. The cassette was inserted into a binary vector that contains a gene for hygromycin resistance. Binary vectors were introduced into Agrobacterium strain EHA105. For transformation into BY-2 cell lines, 1-week-old cells were diluted 1:1 with fresh medium and grown for 1 day. The cells were diluted 1:12 into fresh medium in a 10-cm Petri dish and incubated with 0.2 mL of confluent-grown Agrobacterium for 4 days at 25°C. The cells then were washed with fresh medium and transferred into selection medium containing 400 mg/L carbenicillin, 200 mg/L cefotaxime, and either 10 mg/L G418 (for kanamycin selection) or 20 mg/L hygromycin or both. Transformed BY-2 cells were maintained by the addition of antibiotic to the medium. Coexpression of the proteins was achieved by transforming stable transgenic cells containing GFP fusion constructs with RED:SR1.
Preparation of Protein Extracts and Protein Gel Blot Analysis
Plant material was homogenized in liquid nitrogen, thawed in NETN buffer (20 mM Tris-HCl, pH 8, 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40) supplemented with freshly added phosphatase inhibitor cocktail (50 mM β-glycerophosphate, 100 μM Na3VO4, 10 mM NaF, and 2 mM EGTA) and protease inhibitor cocktail (10 μg/mL aprotinin, 10 μg/mL leupeptin, 10 μg/mL pepstatin A, and 100 mM phenylmethylsulfonyl fluoride) and centrifuged at 10,000g for 10 min. Protein concentration in the supernatant then was determined using the Bradford method (Bradford, 1976). Proteins were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Immun-Blot Membranes; Bio-Rad). Equal loading of proteins was confirmed by Coomassie Brilliant Blue R 250 staining of the membrane. Blots were probed with monoclonal antibodies raised against the HA tag, and detection was performed by peroxidase-coupled secondary antibodies in conjunction with an enhanced chemiluminescence detection system (Amersham, Uppsala, Sweden). The protein overlay assay was performed as described (Savaldi-Goldstein et al., 2000).
Immunoprecipitation and in Vitro Kinase Assay
Protein A–Sepharose beads (25 μL; Pharmacia) were bound to anti-HA antiserum or to polyclonal antibodies raised in rabbits against GFP (Clontech) in PBS (Gibco BRL) for 1 h at room temperature. The beads coupled to anti-HA or anti-GFP then were recovered by centrifugation and washed three times in PBS and finally in NETN buffer. Beads were incubated with 500 μg of total protein for 2 h at 4°C, and samples were harvested by centrifugation and washed four times in NETN buffer and then in kinase buffer (50 mM Tris-HCl, pH 7.5, and 10 mM MgCl2) supplemented with phosphatase and protease inhibitors as described above but without EGTA. The kinase reaction was started by the addition of a 30-μL solution of 2 μCi of γ-32P-ATP (3000 mCi/mmol; Amersham Pharmacia Biotech), 100 μM ATP, and 200 ng/μL MBP in kinase buffer to the sample. Beads were incubated for 10 min at 30°C, and the reactions were terminated by the addition of protein sample buffer. Proteins were fractionated on a 12% SDS-PAGE gel and exposed for phosphorimager analysis or to film.
Alternative Splicing Assay
Total RNA was extracted with the RNeasy Plant Mini Kit (Qiagen, Valencia, CA). RNA (1.5 μg) was subjected to first-strand synthesis using SuperScript II reverse transcriptase (Gibco BRL) according to the manufacturer's procedure using oligo(dT) as a primer. As a negative control, the same reaction was performed in parallel in the absence of the enzyme. A sample of one-tenth of the reaction then was used for PCR amplification in the linear range (generally 26 cycles). Primers flanking the alternatively spliced intron of U1-70K were as described by Golovkin and Reddy (1996), and primers flanking the alternatively spliced introns of atSRp30 and atSRp34/SR1 were modified from Lopato et al. (1999) with the following sequences: atSRp34/SR1, 5′-AGGAGCAGAAGTCCCAAGGCA-AAG and 3′-AGAAGGTAGAGGAGATCTTGATC; atSRp30, 5′-TGTCACCTGCTAGATCC and 3′-AGATATCACAGGTGAAAC. Primers for ANP1 were as described by Nishihama et al. (1997). PCR conditions were as follows: 2 min at 94°C (first cycle); 30 s at 94°C, 45 s at 53°C, and 45 s at 72°C (23 to 29 cycles); and 5 min at 72°C (last cycle). PCR products were separated on a 1.8% agarose gel containing ethidium bromide (10 μg/mL) and visualized by the Bio-Imaging System (model 202D; DNR-Imaging Systems, Kiryat Anavim, Israel). PCR products that appeared in the captured images were quantified by the Image Gauge V3.41 program (Fuji Film Science, Tokyo, Japan). For sequencing, PCR products were excised from the gel and subcloned into the pGEM-T Easy vector system (Promega).
Fluorescence Microscopy
Transformed BY-2 cells in the logarithmic stage (3 days after subculture) were mounted on slides and visualized by confocal laser microscopy (LSM 510; Zeiss, Oberkochen, Germany). For GFP visualization, excitation with a 488-nm laser line and a 505- to 530-nm band-pass filter for emission was used. For dsRED visualization, excitation with a 543-nm laser line and a 560-nm long-pass filter for emission was used. Image analysis was performed using the standard system operating software provided with the Zeiss 510 microscope (version 2.01).
Microarray Analysis
Total RNA was extracted using the RNeasy Midi Kit (Qiagen) and subjected to reverse transcription reaction. cDNA products then were labeled with Cy3 and Cy5 by the indirect amino-allyl method. Hybridization was performed on slides containing 9216 or 12,000 Arabidopsis ESTs (Keck Biotechnology Resource Laboratory, Yale University, New Haven, CT) (Ma et al., 2001; Zik and Irish, 2003). For each biological repetition, two hybridizations with swapped dye labeling were performed. Separate images were acquired using ScanArray 4000 software (Packard BioScience) for each fluorescence at a resolution of 10 μm per pixel, adjusting the photomultiplier and laser power to achieve an optimal distribution of signals without minimal saturation. Image analysis was performed using QuantArray version 3 software (Perkin-Elmer Life Sciences, Boston, MA). Spots were quantified using the fixed-circle method, measuring the mean of pixels encompassing the spot and subtracting the local background areas. Spots with visible defects were omitted from further analysis by flagging. Data analysis was performed with GeneSpring 4.2 software (Silicon Genetics, Redwood City, CA) applying “one-per-spot” and “one-per-chip” normalization.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We are indebted to Roni Golan, Michal Efrat, and Alon Zaban for technical assistance and to Ruth Levy and Hadas Ner-Gaon for assistance with overlay and PCR techniques. We thank Ron Ophir and Ziv Zeira for assistance in microarray analysis. atSRp34/SR1 cDNA was kindly provided by Howard M. Goodman. This work was supported by an Infrastructure Grant from the Ministry of Science, Technology, and Sports, by Israel Science Foundation Grant 388/02-1, and by the Raymond Burton Fund for Plant Genomic Research.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.011056.
References
- Allemand, E., Gattoni, R., Bourbon, H.M., Stevenin, J., Caceres, J.F., Soret, J., and Tazi, J. (2001). Distinctive features of Drosophila alternative splicing factor RS domain: Implication for specific phosphorylation, shuttling, and splicing activation. Mol. Cell. Biol. 21, 1345–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf, D., Honig, A., Berget, S.M., and O'Malley, B.W. (2002). Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 298, 416–419. [DOI] [PubMed] [Google Scholar]
- Ben David, Y., Letwin, K., Tannock, L., Bernstein, A., and Pawson, T. (1991). A mammalian protein kinase with potential for serine/threonine and tyrosine phosphorylation is related to cell cycle regulators. EMBO J. 10, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beven, A.F., Simpson, G.G., Brown, J.W.S., and Shaw, P.J. (1995). The organization of spliceosomal components in the nuclei of higher plants. J. Cell Sci. 108, 509–518. [DOI] [PubMed] [Google Scholar]
- Bohmann, K., Ferreira, J.A., and Lamond, A.I. (1995). Mutational analysis of P80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J. Cell Biol. 131, 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Carmofonseca, M., Pepperkok, R., Carvalho, M.T., and Lamond, A.I. (1992). Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J. Cell Biol. 117, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, B.C., Lee, S.Y., Oh, S.A., Rhew, T.H., Nam, H.G., and Lee, C.H. (1997). The promoter activity of sen1, a senescence-associated gene of Arabidopsis, is repressed by sugars. J. Plant Physiol. 151, 339–345. [Google Scholar]
- Clark, T.A., Sugnet, C.W., and Ares, M., Jr. (2002). Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296, 907–910. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Colwill, K., Feng, L.L., Yeakley, J.M., Gish, G.D., Caceres, J.F., Pawson, T., and Fu, X.D. (1996. a). SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J. Biol. Chem. 271, 24569–24575. [DOI] [PubMed] [Google Scholar]
- Colwill, K., Pawson, T., Andrews, B., Prasad, J., Manley, J.L., Bell, J.C., and Duncan, P.I. (1996. b). The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 15, 265–275. [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar, S.P., and Baker, B.J. (2000). Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 97, 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, C., McGuffin, M.E., Dauwalder, B., Rabinow, L., and Mattox, W. (1998). Protein phosphorylation plays an essential role in the regulation of alternative splicing and sex determination in Drosophila. Mol. Cell 2, 741–750. [DOI] [PubMed] [Google Scholar]
- Duncan, P.I., Stojdl, D.F., Marius, R.M., and Bell, J.C. (1997). In vivo regulation of alternative pre-mRNA splicing by the Clk1 protein kinase. Mol. Cell. Biol. 17, 5996–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X.D. (1995). The superfamily of arginine/serine-rich splicing factors. RNA 1, 663–680. [PMC free article] [PubMed] [Google Scholar]
- Golovkin, M., and Reddy, A.S.N. (1996). Structure and expression of a plant U1 snRNP 70K gene: Alternative splicing of U1 snRNP 70K pre-mRNAs produces two different transcripts. Plant Cell 8, 1421–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin, M., and Reddy, A.S.N. (1999). An SC35-like protein and a novel serine/arginine-rich protein interact with Arabidopsis U1-70K protein. J. Biol. Chem. 274, 36428–36438. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. (2000). Sorting out the complexity of SR protein functions. RNA 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. (2001). Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 17, 100–107. [DOI] [PubMed] [Google Scholar]
- Gui, J.F., Tronchere, H., Chandler, S.D., and Fu, X.D. (1994). Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc. Natl. Acad. Sci. USA 91, 10824–10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman, P., and Ecker, J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howel, B.W., Afar, D.E.H., Lew, J., Douville, E.M.J., Icely, P.L.E., Gray, D.A., and Bell, J.C. (1991). Tyrosine-phosphorylating enzyme with sequence homology to serine/threonine kinases. Mol. Cell. Biol. 11, 568–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, A. (1996). The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65, 367–409. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi, N., Onogi, H., Wakabayashi, T., and Hagiwara, M. (1998). Novel SR-protein-specific kinase, SRPK2, disassembles nuclear speckles. Biochem. Biophys. Res. Commun. 242, 357–364. [DOI] [PubMed] [Google Scholar]
- Lam, H.M., Peng, S.S.Y., and Coruzzi, G.M. (1994). Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiol. 106, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambermon, M.H.L., Simpson, G.G., Kirk, D.A.W., Hemmings-Mieszczak, M., Klahre, U., and Filipowicz, W. (2000). UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J. 19, 1638–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar, G., and Goodman, H.M. (2000). The Arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol. Biol. 42, 571–581. [DOI] [PubMed] [Google Scholar]
- Lazar, G., Schaal, T., Maniatis, T., and Goodman, H.M. (1995). Identification of a plant serine-arginine-rich protein similar to the mammalian splicing factor Sf2/Asf. Proc. Natl. Acad. Sci. USA 92, 7672–7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., Du, C., Horn, M., and Rabinow, L. (1996). Activity and autophosphorylation of LAMMER protein kinases. J. Biol. Chem. 271, 27299–27303. [DOI] [PubMed] [Google Scholar]
- Lemaire, R., Prasad, J., Kashima, T., Gustafson, J., Manley, J.L., and Lafyatis, R. (2002). Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: A novel function for SR proteins. Genes Dev. 16, 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato, S., Kalyna, M., Dorner, S., Kobayashi, R., Krainer, A.R., and Barta, A. (1999). atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 13, 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato, S., Mayeda, A., Krainer, A.R., and Barta, A. (1996). Pre-mRNA splicing in plants: Characterization of Ser/Arg splicing factors. Proc. Natl. Acad. Sci. USA 93, 3074–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, A.J. (1998). Alternative splicing of pre-mRNA: Developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32, 279–305. [DOI] [PubMed] [Google Scholar]
- Lorkovic, Z.J., and Barta, A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30, 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic, Z.J., Kirk, D.A.W., Klahre, U., Hemmings-Mieszczak, M., and Filipowicz, W. (2000. a). RBP45 and RBP47, two oligouridylate-specific hnRNP-like proteins interacting with poly(A)(+) RNA in nuclei of plant cells. RNA 6, 1610–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic, Z.J., Kirk, D.A.W., Lambermon, M.H.L., and Filipowicz, W. (2000. b). Pre-mRNA splicing in higher plants. Trends Plant Sci. 5, 160–167. [DOI] [PubMed] [Google Scholar]
- Ma, L., Li, J., Qu, L., Hager, J., Chen, Z., Zhao, H., and Deng, X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegay, H., Moeslein, F., and Landreth, G. (1999). The dual specificity protein kinase CLK3 is abundantly expressed in mature mouse spermatozoa. Exp. Cell Res. 253, 463–473. [DOI] [PubMed] [Google Scholar]
- Mintz, P.J., and Spector, D.L. (2000). Compartmentalization of RNA processing factors within nuclear speckles. J. Struct. Biol. 129, 241–251. [DOI] [PubMed] [Google Scholar]
- Modrek, B., and Lee, C. (2002). A genomic view of alternative splicing. Nat. Genet. 30, 13–19. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nishihama, R., Banno, H., Kawahara, E., Irie, K., and Machida, Y. (1997). Possible involvement of differential splicing in regulation of the activity of Arabidopsis ANP1 that is related to mitogen-activated protein kinase kinase kinases (MAPKKKs). Plant J. 12, 39–48. [DOI] [PubMed] [Google Scholar]
- Nitsch, J.P. (1969). Experimental androgenesis in Nicotiana. Phytomorphology 19, 389–404. [Google Scholar]
- Okamoto, Y., Onogi, H., Honda, R., Yasuda, H., Wakabayashi, T., Nimura, Y., and Hagiwara, M. (1998). cdc2 kinase-mediated phosphorylation of splicing factor SF2/ASF. Biochem. Biophys. Res. Commun. 249, 872–878. [DOI] [PubMed] [Google Scholar]
- Oswald, O., Martin, T., Dominy, P.J., and Graham, I.A. (2001). Plastid redox state and sugars: Interactive regulators of nuclear-encoded photosynthetic gene expression. Proc. Natl. Acad. Sci. USA 98, 2047–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. (2002). Integrating mRNA processing with transcription. Cell 108, 510–512. [DOI] [PubMed] [Google Scholar]
- Queitsch, C., Sangster, T.A., and Lindquist, S. (2002). Hsp90 as a capacitor of phenotypic variation. Nature 417, 618–624. [DOI] [PubMed] [Google Scholar]
- Reddy, A.S.N. (2001). Nuclear pre-mRNA splicing in plants. Crit. Rev. Plant Sci. 20, 523–571. [Google Scholar]
- Rossi, F., Labourier, E., Forne, T., Divita, G., Derancourt, J., Riou, J.F., Antoine, E., Cathala, G., Brunel, C., and Tazi, J. (1996). Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature 381, 80–82. [DOI] [PubMed] [Google Scholar]
- Sacco-Bubulya, P., and Spector, D.L. (2002). Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 156, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein, S., Sessa, G., and Fluhr, R. (2000). The ethylene-inducible PK12 kinase mediates the phosphorylation of SR splicing factors. Plant J. 21, 91–96. [DOI] [PubMed] [Google Scholar]
- Sessa, G., Raz, V., Savaldi, S., and Fluhr, R. (1996). PK12, a plant dual-specificity protein kinase of the LAMMER family, is regulated by the hormone ethylene. Plant Cell 8, 2223–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., Haegman, M., Kurepa, J., Van Montagu, M., and Straeten, D.V. (1997). Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc. Natl. Acad. Sci. USA 94, 2756–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smetana, K., Busch, R., Chan, P.K., and Busch, H. (2001). Immunocytochemical localization of nucleophosmin and RH-II/Gu protein in nucleoli of HeLa cells after treatment with actinomycin D. Acta Histochem. 103, 325–333. [DOI] [PubMed] [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q.M., and Ecker, J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, K., Long, D., Swinburne, J., and Coupland, G. (1996). A dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S.H., and Manley, J.L. (1997). Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 11, 334–344. [DOI] [PubMed] [Google Scholar]
- Yun, B., Farkas, R., Lee, K., and Rabinow, L. (1994). The Doa locus encodes a member of a new protein kinase family and is essential for eye and embryonic development in Drosophila melanogaster. Genes Dev. 8, 1160–1173. [DOI] [PubMed] [Google Scholar]
- Yun, B., Lee, K., Farkas, R., Hitte, C., and Rabinow, L. (2000). The LAMMER protein kinase encoded by the Doa locus of Drosophila is required in both somatic and germline cells and is expressed as both nuclear and cytoplasmic isoforms throughout development. Genetics 156, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler, A.M., Lane, W.S., Stolk, J.A., and Roth, M.B. (1992). SR proteins: A conserved family of pre-messenger RNA splicing factors. Genes Dev. 6, 837–847. [DOI] [PubMed] [Google Scholar]
- Zik, M., and Irish, V.F. (2003). Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15, 207–222. [DOI] [PMC free article] [PubMed] [Google Scholar]