Abstract
Cotton is the most important textile crop as a result of its long cellulose-enriched mature fibers. These single-celled hairs initiate at anthesis from the ovule epidermis. To date, genes proven to be critical for fiber development have not been identified. Here, we examined the role of the sucrose synthase gene (Sus) in cotton fiber and seed by transforming cotton with Sus suppression constructs. We focused our analysis on 0 to 3 days after anthesis (DAA) for early fiber development and 25 DAA, when the fiber and seed are maximal in size. Suppression of Sus activity by 70% or more in the ovule epidermis led to a fiberless phenotype. The fiber initials in those ovules were fewer and shrunken or collapsed. The level of Sus suppression correlated strongly with the degree of inhibition of fiber initiation and elongation, probably as a result of the reduction of hexoses. By 25 DAA, a portion of the seeds in the fruit showed Sus suppression only in the seed coat fibers and transfer cells but not in the endosperm and embryo. These transgenic seeds were identical to wild-type seeds except for much reduced fiber growth. However, the remaining seeds in the fruit showed Sus suppression both in the seed coat and in the endosperm and embryo. These seeds were shrunken with loss of the transfer cells and were <5% of wild-type seed weight. These results demonstrate that Sus plays a rate-limiting role in the initiation and elongation of the single-celled fibers. These analyses also show that suppression of Sus only in the maternal seed tissue represses fiber development without affecting embryo development and seed size. Additional suppression in the endosperm and embryo inhibits their own development, which blocks the formation of adjacent seed coat transfer cells and arrests seed development entirely.
INTRODUCTION
The unique feature of the seed of the tetraploid cotton is that ∼30% of the seed coat epidermal cells develop into specialized fibers. Each cotton fiber is a single cell of maternal origin that initiates from the ovule epidermis at or just before anthesis. After a period of rapid elongation for ∼16 to 18 days, the fibers synthesize massive amounts of secondary cell wall cellulose (Basra and Malik, 1984). Thus, cotton fiber represents a model single-cell system to study the control of cell differentiation and elongation (Ruan et al., 2000, 2001), carbon partitioning to cellulose synthesis (Ruan et al., 1997; Haigler et al., 2001), and the interaction between maternal (fiber) and embryonic tissues in seeds.
Research on cotton fiber development during the last decade has been focused largely on gene expression profiles during fiber elongation and secondary cell wall synthesis (Pear et al., 1996; Ruan et al., 2001, and references therein). Several attempts have been made to alter the expression of some unknown fiber-specific genes and genes involved in auxin and cytokinin biosynthesis in the fibers (John, 1999). However, no phenotypic changes were observed in fiber development in the resulting transgenic plants (John, 1999). Indeed, there is no evidence to date that the expression of any particular gene plays an essential role in cotton fiber development.
The developing cotton fiber is a highly active sink cell that uses sucrose for its rapid expansion and cellulose synthesis (Haigler et al., 2001; Ruan et al., 2001). In this cell type, sucrose synthase (Sus; EC 2.4.1.13), not invertase, is the major sucrolytic enzyme (Ruan and Chourey, 1998; Ruan et al., 2000). It catalyzes a reversible reaction but preferentially converts sucrose into fructose and UDP-glucose in planta (Chourey and Nelson, 1979; Geigenberger and Stitt, 1993). A cDNA (SS3) that encodes Sus has been isolated from cotton fiber (Ruan et al., 1997). Previous studies suggest that this Sus gene may play important roles in fiber development. First, the Sus mRNA and its protein are localized specifically in fiber initials, but not in adjacent normal epidermal cells, of cotton ovules on the day of anthesis (Nolte et al., 1995; Ruan and Chourey, 1998). The expression of Sus in the later stage of fiber development may play a role in cell wall cellulose synthesis (Amor et al., 1995; Ruan et al., 1997). On the other hand, the Sus protein also is detected in the endosperm at ∼10 days after anthesis (DAA) (Y.-L. Ruan, unpublished data) and in the embryo from 16 DAA onward (Ruan et al., 1997). It remains unknown whether Sus expression plays a different role in the fiber, a maternal tissue, and in the endosperm or embryo during seed development.
In this study, we examined the role of Sus in cotton fiber and seed development using a reverse genetics approach by transforming cotton plants with Sus suppression gene constructs. We analyzed the cell- and development-specific phenotypes of the transgenic lines with the focus on two issues. First, does Sus play a critical role in fiber cell initiation from the ovule epidermis, and how sensitive is fiber development to changes in Sus expression? Second, can the contribution to fiber and seed development of Sus in the embryonic tissue be differentiated from that in the maternal fibers? The results obtained provide significant new insights into the roles of Sus in the control of plant cell differentiation and expansion and in the interaction between maternal and embryonic tissues of seeds.
RESULTS
Transformation with Sus Suppression Constructs Leads to A Fiberless Phenotype
Previous studies indicated the existence of a small Sus gene family in the tetraploid cotton genome, with one gene (SS3) highly expressed in seeds and fibers (Ruan et al., 1997). To achieve relatively specific suppression of Sus in developing cotton fibers and seeds, we made sense and antisense Sus suppression gene constructs containing only 390 bp of the 3′ end of the SS3 cDNA driven by the constitutive segment-7 promoter (S7) from Subterranean clover stunt virus (see Methods). DNA gel blot analysis using this 390-bp fragment as a probe detected two bands in the tetraploid AD genome of cotton (Gossypium hirsutum), each of which corresponds to only one band detected in a diploid A (G. herbaceum) and D (G. raimondii) genome cotton, respectively (data not shown). This finding suggests that the two bands seen in the G. hirsutum cotton are homeologous genes, representing one Sus gene and originating from the A and D genome progenitors. Because gene suppression by both antisense suppression and cosuppression works through homology-dependent RNA degradation (Waterhouse et al., 2001), the 390-bp Sus sequence used here likely would target only its own Sus gene.
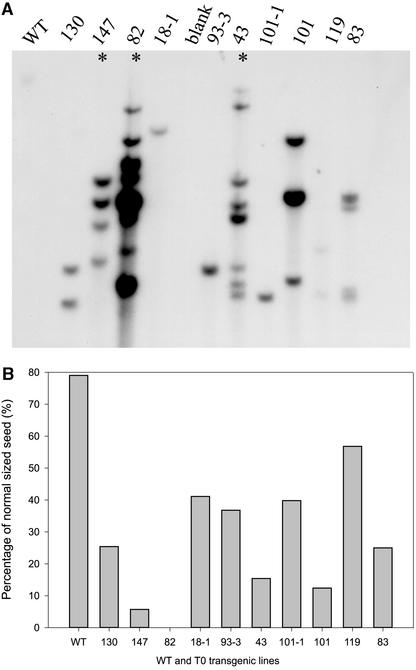
The Sus suppression constructs were introduced into cotton by means of Agrobacterium-mediated transformation. Twenty-two primary (T0) transgenic lines were generated. Among them, seven lines showed a fiberless phenotype early in seed development. The remaining lines showed various degrees of suppression of fiber development. All of the lines were unaffected in vegetative growth and flowering, except for line 82, which was fiberless but also dwarfed. This line aborted all of its fruit prematurely by 10 DAA. The altered fiber phenotypes were observed equally among both sense and antisense transformants. The lines that showed the severe fiberless phenotype all had multiple copies of the transgene (Figure 1A), as is found commonly in transgenic plants that show gene silencing (Waterhouse et al., 2001). These included lines 147, 82, and 43. The high-copy-number T0 lines generally produced a smaller percentage of normal-sized T1 seeds and more inviable shrunken seeds in mature cotton fruit than the lines with lower copy numbers (Figure 1B).
Figure 1.
Transgene Copy Number and Percentage of Normal-Sized Cotton Seeds.
(A) Gel blot analysis of DNA isolated from 10 primary (T0) transformants and wild-type (WT) plants digested with EcoRI and probed with a 1-kb NPTII probe (see Methods). The transformant numbers are shown. Asterisks indicate lines that showed an obvious fiberless seed phenotype at 0 to 6 DAA.
(B) Percentage of normal-sized T1 seeds in mature cotton fruit from the T0 lines shown in (A). The remaining T1 seeds were stunted and inviable. Each value is the mean of ∼350 seeds from 10 mature fruit with se < 10%.
PCR detection of the transgene, coupled with segregation and biochemical analysis, revealed that the normal-sized seeds consisted of all of the null segregants without the transgene in their embryos and a smaller portion of hemizygous seeds with wild-type levels of Sus expression in their embryos. By contrast, the shrunken seeds had no or highly defective embryo development, as a result of strong Sus suppression in the endosperm and embryo. They contained a majority of the predicted hemizygous seeds and all of the homozygous seeds. The simplest example of this segregation may be seen in the single-copy lines 18-1 and 101-1 (Figure 1A). Each of these lines produced ∼40% normal-sized T1 seeds (Figure 1B), including 25% null segregants and 15% hemizygous seeds, based on the following observation. Among the T1 plants grown from 24 normal-sized T1 seeds from each of the two lines, 15 plants did not contain the transgene, as determined by PCR and DNA gel blot analysis, whereas 9 plants contained the transgene but continued to show segregation among the T2 seeds set on these T1 plants. Because 18-1 and 101-1 were single-copy lines, the remaining 60% of stunted inviable T1 seeds would consist of 35% hemizygous and 25% homozygous seeds. All of these seeds showed suppressed embryo development. We refer to the normal-sized seeds as type I and the shrunken seeds as either type II or type III (see Figure 7).
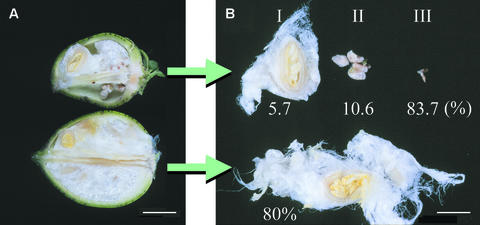
Figure 7.
Phenotypes of Sus-Suppressed Transgenic Cotton Fruit and Seeds at 25 DAA.
(A) Cotton fruit.
(B) Cotton seeds.
The top and bottom samples represent T1 line 147-7 and the wild type, respectively. The numbers in (B) indicate the percentage of a given seed type among the seed population, derived from 10 fruits in each case. Bars = 2.0 cm in (A) and 0.8 cm in (B).
It is noteworthy that in the wild type, Sus protein is expressed only in the fibers, a maternal tissue, and not in the remaining ovule or seed from 0 to 3 DAA (Ruan and Chourey, 1998) (see Figure 3B). Thus, segregation of the transgene in the embryonic tissue would have no impact on fiber and seed development at this early stage. As predicted, we found that the mature normal-sized and shrunken seeds originated from the phenotypically identical seed population in a given fruit from 0 to 3 DAA (i.e., both were fiberless in lines 147, 82, and 43) (Figure 2). To determine whether the fiberless phenotype was caused by the presence of the Sus transgene, 24 normal-sized T1 seeds from each of six selected T0 lines were sown for PCR detection of the transgene in the T1 generation. For each line, six PCR-positive and two PCR-negative segregants were grown for detailed analysis. The fiberless seed phenotype seen in the T0 generation was inherited and present only in T1 individuals that contained the transgene. By contrast, all null segregants showed the normal wild-type phenotype in fiber and seed growth. These results demonstrate that the fiberless phenotype observed is attributable to the presence of the Sus transgene rather than to any pleiotropic effect of cotton transformation and tissue culture.
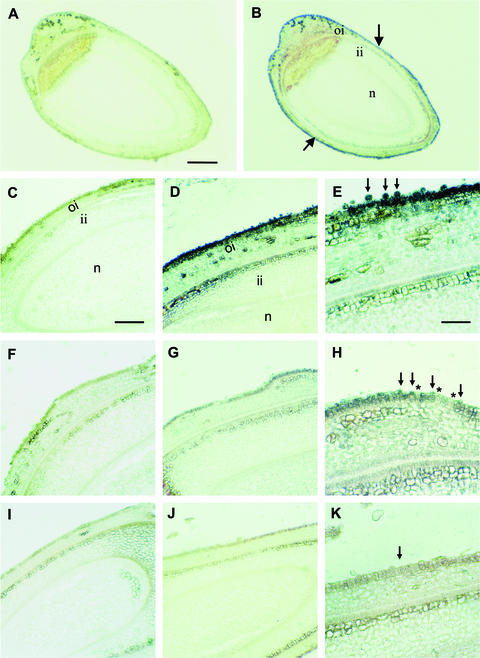
Figure 3.
Immunogold Localization of Sus Protein in Wild-Type and Transgenic Ovules at 0 DAA.
(A) Whole view of a longitudinal section of a wild-type ovule treated with preimmune serum.
(B) A consecutive section of (A) treated with polyclonal antibody against cotton Sus. Note that the Sus protein signal was detected specifically in the ovule epidermis (arrows) but not in the remaining ovule tissues.
(C) A wild-type longitudinal section treated with preimmune serum.
(D) A consecutive section of (C) treated with the Sus antibody showing strong Sus signal in the ovule epidermis and expanding fiber initials.
(E) Magnified view of the epidermis in (D) showing abundant Sus proteins in spherically expanding fiber initials (arrows).
(F) A longitudinal section of ovules from line 147-7 treated with preimmune serum.
(G) A consecutive section of (F) treated with the Sus antibody. Note the very weak Sus signal in the ovule epidermis compared with the wild type in (D).
(H) Magnified view of the epidermis in (G). Note, in comparison with the wild-type ovules (E), that the fiber initials were much smaller and fewer and showed dramatically reduced levels of Sus (arrows). Also note that the detectable Sus signal was restricted to the small fiber initials (arrows) and was not seen in the adjacent epidermal cells (asterisks).
(I) A longitudinal section of ovules from line 82 treated with preimmune serum.
(J) A consecutive section of (I) treated with the Sus antibody. Note that Sus signals were undetectable.
(K) Magnified view of the epidermis in (J) showing no Sus protein signals and/or few fiber cells (arrow).
The black signal represents Sus proteins. ii, inner integument; n, nucellus; oi, outer integument. Bars = 250 μm in (A), 125 μm in (C), and 65 μm in (E). The scale in (B) is the same as that in (A); the scale in (D), (F), (G), (I), and (J) is the same as that in (C); the scale in (H) and (K) is the same as that in (E).
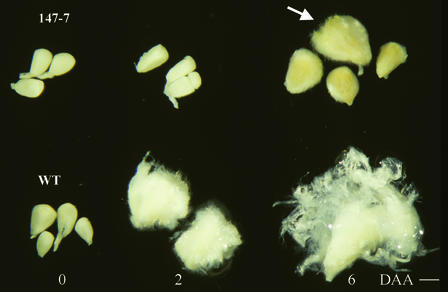
Figure 2.
Fiberless Seed Phenotype of Sus-Suppressed T1 Line 147-7 Compared with Wild-Type Seeds at 0, 2, and 6 DAA.
Note that a small portion of transgenic seeds showed a layer of very short fibers with wild-type (WT) seed size at 6 DAA (arrow). Bar = 0.1 cm.
Suppression of Sus in the Ovule Epidermis Represses Fiber Initiation and Elongation
Figure 2 shows the early fiber and seed phenotype of a T1 segregant, number 7 from line 147 (referred to as 147-7). It represents a typical fiberless phenotype seen in the T0 generation and in T1 individuals, including 43-4 and 101-1-22. At 0 DAA, transgenic ovules were the same as wild-type ovules in size and shape (Figure 2). However, a marked difference emerged 2 days later. Wild-type seeds were well covered with young fibers, whereas transgenic seeds remained fiberless at 2 DAA (Figure 2). This difference became more striking at 6 DAA, although a small portion of the transgenic seeds began to show a layer of short fibers at this stage.
The inhibition of early fiber development at 2 DAA (Figure 2) suggested that the fiber initiation process was disrupted in the transgenic plants. To test this possibility, immunolocalization analysis was conducted on ovules at 0 DAA using a polyclonal antibody raised against Sus protein purified from cotton fiber (Ruan et al., 1997). In wild-type ovules, the Sus protein, as indicated by black immunogold signals, was localized specifically in the ovule epidermis (Figure 3B), where fibers initiate (Ruan and Chourey, 1998), compared with the preimmune control (Figure 3A). For a better comparison, wild-type and transgenic ovules were arranged subsequently on the same slides. In contrast to the strong Sus protein signals seen in wild-type ovule (Figure 3D), much weaker and no Sus expression was detected in the ovule epidermis from lines 147-7 and 82, respectively (Figures 3G and 3J). The preimmune controls showed no background reaction (Figures 3C, 3F, and 3I). Under higher magnification, it is evident that abundant Sus protein was expressed in the spherical fiber initials of the wild-type ovules (Figure 3E). By contrast, the Sus protein was reduced dramatically and the fiber initials were repressed significantly in the ovule epidermis of line 147-7 (Figure 3H). Notably, only those initials that showed low levels of Sus protein had begun to protrude above the ovule surface, whereas the adjacent normal epidermal cells showed little or no signal (Figure 3H). This specific localization of Sus in the fiber initials also was seen in wild-type ovules when the incubation time with Sus antibody was decreased from 30 to 15 min (see Methods). For line 82, Sus protein was undetectable, and virtually no fiber initials were formed in the ovule epidermis (Figure 3K). RNA gel blot analysis revealed that Sus mRNA levels were reduced significantly or undetectable in whole ovules from line 147-7 or line 82, respectively, compared with the wild type (data not shown). This finding shows that the reduction of Sus protein in the ovule epidermis is a result of the decrease in the Sus transcript.
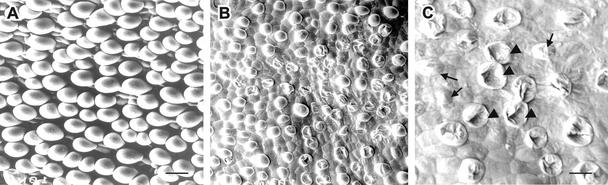
The impact of Sus suppression on the cellular development of fiber initials was visualized further using scanning electron microscopy. Figure 4A shows the evenly arranged spherical fiber cells on the surface of wild-type ovules. By contrast, the fiber initials were much fewer and smaller in line 147-7 (Figure 4B). Higher magnification shows that many of those cells were shrunken and collapsed, whereas some had an abnormal shape, a wrinkled surface, and very weak projection above the ovule surface (Figure 4C). Similar shrunken and collapsed fiber initials also were observed in other fiberless transgenic individuals. Virtually no fiber cells were visible in line 82, in which Sus protein was undetectable (Figure 3K).
Figure 4.
Scanning Electron Microscopy of the Surface of Wild-Type and Transgenic Ovules at 0 DAA.
(A) Wild-type ovule epidermis showing evenly arranged and spherically expanding fiber initials.
(B) Transgenic ovules from 147-7. Note that the fiber cells are much smaller and fewer.
(C) Magnified view of (B) showing the collapsed (arrowheads) and shrunken (arrows) fiber initials.
Bars = 15 μm in (A) and 5 μm in (C). The scale in (B) is the same as that in (A).
Table 1 shows measurements of Sus activity and associated changes in carbohydrate status in transgenic ovules. Sus activity was reduced to 25, 20, and 0% of wild-type levels in the ovules of lines 147-7, 101-1-22, and 82, respectively, which correlated with the reduction of hexoses and starch. Interestingly, the sucrose level remained unchanged. The activities of acid and alkaline invertases were unaffected in the Sus-suppressed ovules.
Table 1.
Sucrose Synthase and Invertase Activities and Carbohydrate Content in Cotton Ovules at 0 DAA
| Line | Enzyme Activity (nmol·mg−1 fresh weight·min−1)
|
Carbohydrate (nmol·mg−1 fresh weight)
|
|||||
|---|---|---|---|---|---|---|---|
| Sus | Acid Invertase | Alkaline Invertase | Sucrose | Glucose | Fructose | Starcha | |
| Wild type | 20.3 ± 2.3b | 4.2 ± 0.3 | 1.5 ± 0.1 | 12.4 ± 0.7 | 6.7 ± 0.2 | 3.5 ± 0.4 | 58.9 ± 0.7 |
| 147-7 | 5.0 ± 0.3 | 3.0 ± 0.3 | 1.5 ± 0.1 | 12.8 ± 2.0 | 4.1 ± 0.1 | 2.7 ± 0.7 | 17.7 ± 1.5 |
| 101-1-22 | 4.1 ± 0.3 | 3.8 ± 0.4 | 1.4 ± 0.2 | 11.6 ± 2.3 | 3.0 ± 0.2 | 2.1 ± 0.2 | 17.0 ± 1.6 |
| 82 | 0.0 ± 0.1 | 4.4 ± 0.2 | 0.9 ± 0.1 | 10.7 ± 0.3 | 2.0 ± 0.1 | 1.8 ± 0.0 | 8.6 ± 1.3 |
Starch content was expressed as nmol glucose equivalent·mg−1 fresh weight.
Each value is the mean ± se of six replicates.
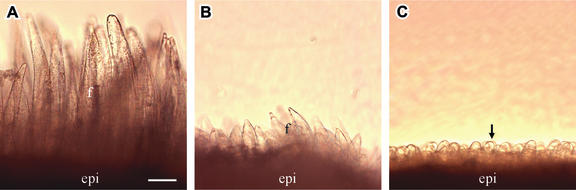
After initiation on 0 DAA, fibers elongated rapidly to ∼250 μm by 3 DAA in wild-type seeds (Figure 5A). This elongation process, however, was inhibited severely in transgenic seeds. Figure 5B shows only ∼20% of the wild-type fiber length in line 147-7. Virtually no elongation occurred in line 82, although some fiber initial–like cells appeared on the seed coat epidermis (Figure 5C).
Figure 5.
Fiber Length of Wild-Type and Transgenic Cotton Seeds at 3 DAA.
(A) Fiber from wild-type seeds.
(B) Fiber from transgenic line 147-7. Note the much reduced fiber length compared with (A).
(C) Fiber from transgenic line 82 showing that fiber elongation was inhibited completely. Some fiber-like epidermal cells are indicated by the arrow.
epi, epidermis; f, fiber. Bar = 55 μm in (A). The scale in (B) and (C) is the same as that in (A).
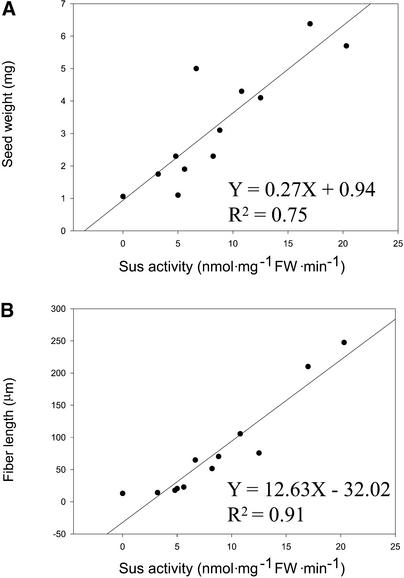
The measurements of Sus activity on ovules at 0 DAA and fiber length and seed weight at 3 DAA were extended further to 10 segregating individuals from three lines plus the asexually propagated line 82 and a wild-type plant. The assay revealed that reduction of Sus activity by 70% or more resulted in a fiberless phenotype early in seed development (Figure 2, Table 1). These include individual in lines 82-1, 147-7, 43-4, and 101-1-22. Regression analysis among the 12 individuals showed a stronger correlation between Sus activity and fiber length than with total seed weight including fibers (cf. Figures 6B and 6A, respectively). This finding is consistent with our observation that fiber is the only tissue that expressed the Sus protein in the seed from 0 to 3 DAA. The linear equation (Figure 6B) predicts that suppression of Sus activity in cotton ovules to 2.54 nmol·mg−1 fresh weight·min−1, which is ∼13% of the wild-type level, will completely inhibit fiber elongation.
Figure 6.
Regression Correlation between the Suppression of Sus Activity in Ovules at 0 DAA and the Reduction of Seed Weight and Fiber Length at 3 DAA among Wild-Type and 11 Sus-Suppressed Transgenic Individuals.
Each value is the mean of at least six replicates. FW, fresh weight.
(A) Seed weight.
(B) Fiber length.
Impact of Sus Suppression on Seed Development
As the cotton seed develops further to 10 DAA onward, Sus protein is expressed not only in the seed coat but also in the endosperm (Y.-L. Ruan, unpublished data) and the embryo (Ruan et al., 1997). To examine the possible interaction between maternal (fiber) and embryonic tissues mediated by Sus, we extended our analysis to the transgenic seed at 25 DAA, when the seed has developed to its maximal size. Figure 7 shows a representative transgenic fruit with seeds at 25 DAA from the four-copy line 147-7 compared with a wild-type fruit. The T2 seeds were derived from self-pollination of the T1 line. The transgenic fruit contained the same number of seeds as the wild-type fruit. However, they had only 5.7% normal-sized seeds with ∼70% reduction of fiber mass (type I). The majority (94.3%) of the seeds were stunted and inviable (types II and III; Figure 7B) and were either hemizygous or homozygous for the transgenes. Because the null segregants constituted only part of the normal-sized seeds (see below), the segregation ratio of null to hemizygous plus homozygous seeds was <6:94 for this line. This finding suggests that the four copies of the transgene in line 147-7 act independently and are not linked as a single locus. The dry weight of the shrunken seeds was <5% of that of wild-type seeds. This heterogeneity in seed development existed in various degrees in all of the other transgenic lines. PCR and segregation analyses revealed that, like the T1 seeds in T0 lines, the normal-sized T2 seeds from the T1 generation consisted of all of the null segregants and a small portion of hemizygous seeds (data not shown) but with unsuppressed Sus expression in their embryos (see below).
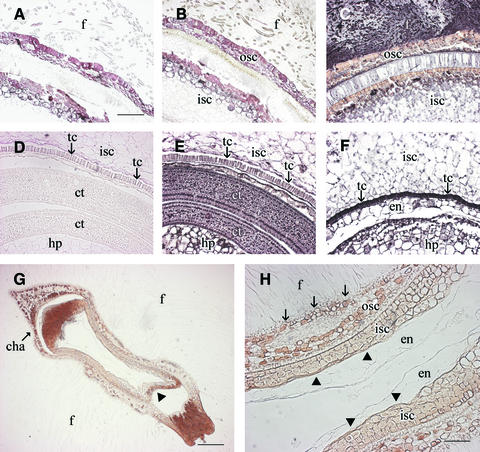
For the developing normal-sized seeds (type I) at 25 DAA, enzyme assays revealed no reduction of Sus activity (8.2 ± 0.4 nmol·mg−1 fresh weight·min−1) in the embryos regardless of the presence or absence of the transgene in this tissue. By contrast, the activity in fibers, a maternal tissue, was reduced by ∼65%. To confirm this finding at the cellular level, immunolocalization was conducted. The immunogold-labeled Sus signals of the type-I seeds were much reduced in the fibers (Figure 8B) and in the transfer cells at the innermost layer of the seed coat (Figure 8E) compared with the strong signals in these cell types of wild-type seeds (Figures 8C and 8F). This finding showed that Sus in the maternal tissue of type-I seeds was repressed, as expected. By contrast, the residual endosperm and the developing embryo, including cotyledons and hypocotyls, showed levels of Sus protein (Figure 8E) similar to those of wild-type seeds (Figure 8F). Although the cotyledons of the wild-type seed are not visible in Figure 8F as a result of a different sectioning position, the Sus signals in this tissue were similar to that shown in Figure 8E. These results confirmed that, in type-I seeds, Sus was suppressed in the seed coat but not in embryonic tissues. Similar results were found for type-I seeds from other transgenic individuals, including 147-5, 43-4, and 101-1-22.
Figure 8.
Immunogold Localization of Sus Protein in Type-I and Type-II Seeds of Line 147-7 Compared with Wild-Type Seeds at 25 DAA.
(A) Cross-section of a type-I seed from the transgenic line treated with preimmune serum.
(B) A consecutive section of (A) treated with polyclonal antibody against cotton Sus. Note that Sus protein signals were much reduced in fibers compared with that of the wild type in (C).
(C) Cross-section of a wild-type seed showing strong Sus protein signals in the fibers.
(D) The same image in (A) viewed toward the inner side of the section.
(E) The same image in (B) viewed toward the inner side of the section showing that the Sus signals were reduced in the seed coat transfer cells but not in the embryos (i.e., cotyledon and hypocotyl) compared with those in the wild-type seed (F).
(F) The same image in (C) viewed toward the inner side of the section. Note the strong Sus signals in seed coat transfer cells and embryos. The cotyledons, which showed levels of Sus protein similar to that in (E), are not visible as a result of the different sectioning position of the seed. The residual endosperm also showed Sus protein signals.
(G) Longitudinal section of a type-II seed from the transgenic line treated with Sus antibody. Note that Sus protein was undetectable throughout the section and that the seed was deformed, with a gap formed between the outer and inner seed coats at the micropyle (arrowhead) and chalazal ends.
(H) Magnified view of the central area in (G). Note the absence of embryo and the presence of some residual abnormal endosperm cells that lacked any labeling of Sus protein. Also note that there was no transfer cell at the innermost layer of the inner seed coat (arrowheads) and that the epidermal cells of the outer seed coat were degenerated (arrows) compared with those of the wild type in (C) and (F).
The black signal represents Sus proteins. cha, chalaza; ct, cotyledon; en, endosperm; f, fiber; hp, hypocotyl; isc, inner seed coat; osc, outer seed coat; tc, transfer cell. Bars = 400 μm in (A), 800 μm in (G), and 200 μm in (H). The scale in (B) to (F) is the same as that in (A).
For the stunted type-II or type-III seeds at 25 DAA, Sus activity was undetectable throughout the seed. Consistently, Sus proteins were immunologically undetectable in either the seed coat or the endosperm (Figures 8G and 8H). The seeds were shrunken and deformed, with cellular degeneration at both the chalazal and micropylar ends and in the seed center (Figure 8G). Under high magnification, it is evident that the seed coat epidermis degenerated and the transfer cell layer was not formed at the innermost layer of the seed coat (Figure 8H). Notably, except for some distorted residual endosperm cells, there was no embryo at the center of the type-II seeds (Figure 8H). The cellular phenotype of type-III seeds was similar to that of type-II seeds, but with smaller seed coat cells and few fibers (data not shown).
Crosses were performed to obtain further genetic evidence that suppression of Sus only in the seed coat reduces fiber growth without affecting the remaining seed development, whereas suppression of Sus in the endosperm and embryo arrests seed development entirely. We pollinated emasculated hemizygous transgenic lines with pollen from wild-type plants. All of the seeds derived from these crosses would contain the Sus transgene in the maternal seed coat, whereas the embryos would be either wild type or hemizygous. The crosses with line 147-7, a four-copy line (Figure 1A), yielded a segregation ratio of 16:276 normal-sized to shrunken seeds. However, the crosses with a single-copy line, 101-1-22, using wild-type pollen yielded a segregation ratio of 150:136 normal-sized to shrunken seeds. In each case, the seeds were derived from eight fruit. The normal-sized seeds showed ∼70% reduction of fiber mass and normal embryos, resembling type-I seeds (Figures 7 and 8). Further reciprocal crosses were made between the wild type as the female parent and the single-copy line, 101-1-22, as the male parent. Such crosses produced a segregation ratio of 189:173 normal-sized seeds with normal growth of fibers to shrunken seeds from a total of 10 fruit. The shrunken seeds were stunted with few fibers, resembling the type-II seeds in Figure 7. Overall, these results are in agreement with the segregation ratio predicted from the transgene copy number. These data support the observation that repression of Sus in the endosperm and embryo blocked the development of the entire seed, whereas suppression of Sus in the seed coat affected only fiber development.
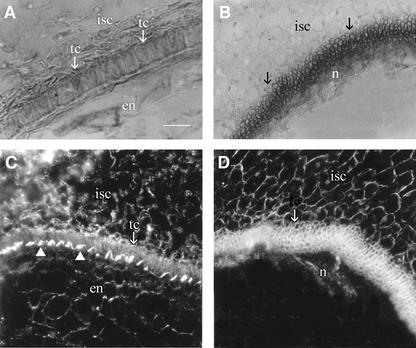
Finally, to assess the impact of embryo arrest on seed coat development independent of Sus suppression, we examined unfertilized wild-type ovules from emasculated flowers. Figure 9A shows the well-developed transfer cell layer at the innermost layer of the seed coat of a fertilized wild-type seed at 12 DAA. Remarkably, in unfertilized ovules, the transfer cell layer did not form; rather, this region became a disorganized mass of numerous tiny cells (Figure 9B). Dark-field imaging revealed the characteristic wall ingrowth of the transfer cells adjacent to the endosperm cells in the seed resulting from fertilization (Figure 9C). However, this feature was absent in the unfertilized ovule that did not form any endosperm cells (Figure 9D).
Figure 9.
Unfertilized Ovules Failed to Form Seed Coat Transfer Cells.
(A) Cross-section of a wild-type seed at 12 DAA showing the presence of transfer cells at the innermost layer of the inner seed coat.
(B) Unfertilized ovules had a disorganized mass of numerous small cells rather than transfer cells (arrows) by 12 DAA. Also note the residual nucellus and the absence of endosperm tissue.
(C) Cross-section of a wild-type seed at 12 DAA viewed by dark-field microscopy showing the wall ingrowth (arrowheads) of the transfer cells toward the cellularized endosperm.
(D) Cross-section of an unfertilized ovule viewed by dark-field microscopy showing numerous small cells at the innermost layer of the inner seed coat, which lacked the wall ingrowths characteristic of the transfer cells shown in (C). Also note the presence of the residual nucellus tissue and the absence of endosperm.
en, endosperm; isc, inner seed coat; n, nucellus; tc, transfer cell. Bar = 60 μm in (A). The scale in (B) to (D) is the same as that in (A).
DISCUSSION
Sus Plays a Crucial Role in Fiber Cell Initiation and Elongation
We present here several lines of direct evidence to demonstrate the crucial role of Sus in cotton fiber initiation and elongation. First, the transformation of cotton with Sus suppression gene constructs led to a fiberless phenotype in multiple independent lines, which was inherited and cosegregated with the transgene (Figures 1 and 2). By contrast, all T1 progeny that had lost the transgene through segregation showed a wild-type phenotype in fiber and seed growth. These data demonstrate that the presence of the Sus transgene is the causal basis of the observed fiber-repressed phenotype. Second, the expression of Sus protein was suppressed strongly in the ovule epidermis of the fiberless lines, specifically in cells destined to become fibers (Figure 3). Importantly, the degree of Sus suppression correlated well with the level of inhibition of fiber cell initiation (Figure 3). The fiberless phenotype can be attributed specifically to the reduction of Sus, because the activity of invertase, a functionally closely related enzyme, was not affected (Table 1). Finally, analysis of a range of transgenic lines established a close linear correlation between the reduction of Sus activity at 0 DAA and the repression of fiber elongation at 3 DAA (Figures 5 and 6). These results demonstrate that both cotton fiber cell initiation and elongation are highly sensitive to changes of Sus activity and that Sus has a high flux control coefficient for these processes.
The role of Sus has been studied previously in a range of sink tissues. In maize, which has two Sus genes, Sh1 and Sus1, mutation of Sh1 reduces endosperm Sus activity by >90% (Chourey and Nelson, 1976). This results in early endosperm cell degeneration, a 22% reduction of starch, and a shrunken seed (Chourey et al., 1991, 1998). It remains to be seen how sensitive the maize seed is to a smaller reduction of Sus. In potato, although antisense repression of Sus activity by 96% reduces tuber starch accumulation and dry weight, reduction of Sus by 50 to 75% has no or little effect on tuber development (Zrenner et al., 1995). Similarly, suppression of Sus activity by 99% in tomato fruit does not affect starch and sugar accumulation or fruit development (Chengappa et al., 1999). In light of this analysis, our results are of particular significance for two reasons. First, in contrast to the observation that a large proportion of Sus activity is dispensable in potato tubers and tomato fruit, our data show a rate-limiting role for Sus in cotton fiber cell initiation and elongation (Figures 3, 5, and 6). Indeed, the impact on fiber elongation can be seen with only a 15% reduction of Sus activity (Figure 6B). Second, previous studies showed that expression of Sus is involved mainly in the synthesis and storage of starch and protein, for example, in Vicia faba seeds (Weber et al., 1997) and potato tubers (Zrenner et al., 1995). However, the elongating cotton fibers are rapidly expanding single cells with no detectable storage activities for either starch or proteins (Basra and Malik, 1984). Thus, our study provides a remarkable example of the essential role of Sus in plant growth at the level of a well-defined metabolically active single cell. These data show that Sus can be used as a biochemical marker for the sink strength of fiber cell development.
Biochemical Basis for the Critical Role of Sus in Fiber Initiation and Elongation
The suppression of Sus expression reduced hexose levels in transgenic ovules (Figure 3, Table 1). This could have several negative effects on fiber initiation and elongation. Hexoses are the major osmotically active solutes in the fibers (Ruan et al., 1997, 2001). The reduction of hexose thus would decrease the osmotic potential, and hence the turgor, of the fiber initials (Ruan and Patrick, 1995). Consequently, the initial spherical protrusion and the subsequent elongation of the fiber cells would be difficult to achieve. Suppression of Sus also may impair the cell wall integrity of the fiber initials by reducing the supply of UDP-glucose, the substrate for the synthesis of both cellulose and many noncellulose cell wall compounds (Buchala, 1999). Both of these possibilities would account for the shrunken and collapsed fiber initials that form (Figure 4) and for the greatly reduced fiber elongation (Figure 5).
Alternatively, hexoses could serve as signals to control the activities of some promoters and transcription factors (Loreti et al., 2001). Therefore, the reduced hexose levels may disrupt the function of regulatory genes that trigger fiber initiation.
Sus Has Different Roles in the Maternal and Filial Tissues of Seeds
Another important observation in this study is the differential impact on seed development of the presence or absence of a viable embryo in the Sus-suppressed lines (Figures 7 and 8). In this study, the transgenic seed population was morphologically homogenous at 0 DAA but began to show heterogeneity at ∼6 DAA, which became more evident at 25 DAA (Figures 2 and 7). The lines of evidence described below demonstrate that suppression of Sus in the seed coat reduces only fiber growth (i.e., type I), whereas additional repression of Sus in the endosperm and embryo arrests seed development entirely (i.e., types II and III in Figure 7).
First, when the suppression of Sus was evident only in the seed coat, fiber growth was reduced but the remaining seed development was normal (type-I seeds in Figures 7 and 8). Second, both the normal-sized and the shrunken seeds in a given transgenic line originated from the phenotypically identical young seed population (Figure 2). They all carry the transgene in the maternal tissue and have the same level of Sus suppression in their maternal fiber cells at 0 DAA (Figure 3). This finding shows that Sus suppression in the seed coat is not responsible for the shrunken-seed phenotype. Third, in contrast to the type-I seed, which had normal levels of Sus expression in the endosperm and embryo, the shrunken seed showed undetectable levels of Sus protein in its abnormal endosperm and had no embryo (Figure 8). Fourth, the percentage of shrunken seeds correlated approximately with the copy number of the transgene in the transgenic lines. In other words, the higher the copy number, the higher the number of shrunken seeds as a result of the presence of the active transgene in the endosperm and embryo (Figure 1). Fifth, crosses between hemizygous transgenic lines and wild-type pollen confirmed that the suppression of Sus in the seed coat repressed only fiber development without any impact on the remaining seed development. Reciprocal crosses between the wild type as the female parent and a hemizygous transgenic line as the male parent produced the expected percentage of shrunken seeds with few fibers. This finding confirms the notion that the suppression of Sus in the endosperm and embryo led to inviable stunted seeds.
A closer examination of the shrunken seeds revealed that the failure of endosperm and embryo development inhibited the formation of the adjacent transfer cells of the seed coat (Figure 8). Consistently, this inhibition was mimicked by a lack of fertilization (Figure 9). These results show that Sus plays a critical role in embryogenesis (Figure 8), which in turn controls seed coat development by regulating the formation of transfer cells that encircle the filial tissue (Figures 8 and 9). To our knowledge, such control of maternal cell development mediated by Sus in the endosperm and embryo has not been reported previously. A similar observation comes from a study of the maize miniature mutant. In that mutant, the deficiency of invertase activity in the endosperm correlates with the withdrawal of the maternal tissue, the pedicel, from the endosperm (Miller and Chourey, 1992). The mechanism of the modulation of seed coat transfer cell development by embryonic tissues is unknown at present, but it probably involves sugar and hormone signaling and regulation by proteins secreted from the embryo (Farley et al., 2000; Thompson et al., 2001).
In conclusion, our results provide direct evidence that the expression of Sus in the ovule epidermis is crucial for the initiation and elongation of single-celled fibers. We also found that the suppression of Sus in the seed coat only reduces fiber growth without affecting embryo development and seed size. However, repression of Sus in the endosperm and embryo arrests seed development entirely. These results provide novel insights into the role of Sus in controlling plant cell and seed development and offer opportunities to modify fiber and seed development through genetic engineering of Sus expression.
METHODS
Plant Material
Cotton (Gossypium hirsutum var Coker 315) plants were grown in soil mixture under controlled conditions as described previously (Ruan et al., 2001). Cotton fruit age was determined by tagging each pedicel on the day of anthesis. For DNA and RNA extraction, enzyme assay, and carbohydrate measurement, samples were frozen in liquid N2 and stored at −70°C until analysis. Fresh samples were used for immunolocalization, fluorescence microscopy, and electron microscopy.
Sus Suppression Gene Constructs and Plant Transformation
The 390-bp Sus cDNA fragment was amplified from the 3′ end of the SS3 cDNA clone (Ruan et al., 1997) by means of PCR, with primers designed to generate a product ranging from 211 bp upstream to 179 bp downstream of the stop codon. The oligonucleotide sequences, with EcoRI restriction sites incorporated at the 5′ ends of the primers, were as follows: YLSusE-a, 5′-GCGGAATTCCTGGGATAAGATCTCCCAAG-3′; and YLSusE-b, 5′-CGCGAATTCAAGTCACCATATTTAACTGG-3′. The amplified fragment was digested with EcoRI, purified with the Wizard PCR Preps DNA purification system (Promega), and cloned into the pPLEX 521 vector downstream of the S7 promoter of the Subterranean clover stunt virus (Schünmann et al., 2003) in the sense or antisense orientation.
The Sus suppression constructs were transformed into cotton var Coker 315 by Agrobacterium tumefaciens infection of cotyledon pieces and selection on kanamycin sulfate–containing medium as described previously (Murray et al., 1999). After ∼10 months of in vitro culture and selection, the putative transgenic plants were transferred to a glasshouse for further screening and analysis.
Genomic DNA Isolation, PCR, and DNA Gel Blot Analysis
Genomic DNA was isolated from young leaves or developing embryos of putative transgenic lines and wild-type plants according to Paterson et al. (1993). The following primers were used to detect the presence of the neomycin phosphotransferase II (NPTII) gene: L (5′-GAGGCTATTCGGCTATGA-3′) and R (5′-ACTTCGCCCAATAGCAG-3′), which span the region from 202 to 440 bp of the gene. A second set of primers was used to determine the presence of the S7 promoter and the Sus transgene: S7-280F (5′-CTCAGGATCAGTGATGCTAG-3′) and YLSusE-a for the antisense and YLSusE-b for the sense constructs. This primer set generated a PCR product at 690 bp, which included 300 bp of the S7 promoter plus 390 bp of the Sus transgene.
For DNA gel blot analysis, 15 μg of DNA from each sample was digested with one of the selected restriction enzymes, electrophoresed on a 0.75% agarose gel, and blotted to nylon membranes using standard molecular protocols. The membranes were hybridized with a 32P-labeled 1-kb NPTII fragment or the 390-bp Sus insert used to make the constructs (see above) at 65°C overnight and washed in 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and then 0.1× SSC for 30 min each at the same temperature before exposure to x-ray film.
RNA Gel Blot Analysis
RNA extraction and RNA gel blot analysis were performed according to Ruan et al. (2001).
Immunolocalization
Immunolocalization was conducted as described previously (Ruan and Chourey, 1998) using the Histogold System for Immuno-Histological Staining (Zymed Laboratories, San Francisco, CA). Briefly, paraffin-embedded samples were cut (10 μm), affixed to slides, deparaffined, rehydrated, and washed with PBS. Thereafter, slides were incubated with serum-blocking solution for 10 min, followed by incubation for 30 min with cotton Sus polyclonal antibodies diluted 1:1500 (Ruan et al., 1997). After washing with PBS, slides were incubated for 30 min in a solution of secondary antibody (goat anti-rabbit IgG linked to colloidal gold). Slides then were washed thoroughly with PBS, incubated for 4 min with freshly prepared silver enhancement reagents, and washed with excess distilled water. Slides were dehydrated in an ethanol series and mounted permanently in Permount (Fisher Scientific) for microscopic examination.
Enzyme Assay
Frozen samples (∼150 mg each) were ground into fine powder in liquid nitrogen before extraction on ice in 2 mL of grinding buffer, which contained 50 mM Na2HPO4, pH 8.0, 1 mM EDTA, 25 mM ascorbic acid, 5 mM DTT, 20 mM β-mercaptoethanol, 10% glycerol, 1% polyethylene glycol 6000, 0.1 mM phenylmethylsulfonyl fluoride, 1 μg/mL E64, 1 μg/mL pepstatin, and 2 μg/mL leupeptin. The extract was centrifuged for 5 min at 12,000g. A 0.5-mL supernatant was desalted by passage through a 4-mL Sephadex G-25 column (Pharmacia) preequilibrated with the extraction buffer minus polyethylene glycol. A 100-μL aliquot was used for protein assay using BSA as the standard. The activities of soluble Sus, acid invertase, and alkaline invertase were measured immediately in a 250-μL reaction mixture for 0 and 30 min at pH 6.5, 5.0, and 7.5, respectively (for details, see King et al., 1997). The control for the Sus assay lacked UDP. The resulting fructose was determined by its reduction reaction with triphenyltetrazolium chloride (TTC). The TTC solution contained 0.25% (w/v) TTC, 1 M NaOH, and 0.08% Triton X-100. It was initially colorless but turned red on reduction with fructose or glucose. Briefly, 200 μL of each enzyme reaction product and fructose standard was mixed with an equal volume of TTC solution followed by incubation at 40°C for 8 min. The samples then were transferred to a 96-well microplate for automatic absorbance measurement at 495 nm using the Spectra Max 340 PC Plate Reader (Molecular Devices, Sunnyvale, CA).
We also checked the possibility of the plasma membrane association of Sus in fibers (Amor et al., 1995) using the EGTA elution method (Haigler et al., 2001) and electron microscopy immunolocalization. We found no membrane association of Sus until the onset of secondary cell wall synthesis at ∼16 to 18 days after anthesis (DAA). Therefore, only soluble Sus activities are presented here.
Carbohydrate Measurement
Approximately 50 mg of seed tissue was ground in liquid nitrogen. The samples were extracted with 1 mL of preheated 80% ethanol for 5 min at 80°C. After cooling, they were centrifuged at 12,000g for 10 min. The supernatants were collected, whereas the pellets were resuspended in 0.5 mL of 50% ethanol and respun as described above. The resulting pellet was reextracted with 0.5 mL of water and recentrifuged. The total 2-mL supernatant collected was mixed with an equal volume of chloroform and shaken vigorously. The aqueous phase was collected, dried in a vacuum, and redissolved in 0.5 mL of water. The sucrose, glucose, and fructose contents were measured enzymatically on a spectrophotometer at 340 nm as described previously (King et al., 1997). The final pellet from the extraction was used for the measurement of starch as glucose equivalents after degradation with amyloglucosidase and α-amylase.
Scanning Electron Microscopy
Scanning electron microscopy of cotton ovules at 0 DAA was performed according to Craig and Beaton (1996).
Fiber Length Measurements
Free-hand sections were cut from freshly harvested cotton seeds at 3 DAA and viewed immediately by means of bright-field microscopy. Digital images were taken for subsequent length measurements. The fiber length from seeds at 25 DAA was measured according to Schubert et al. (1973). In both cases, the measurements were performed from the chalazal ends of the seeds.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Number
The GenBank accession number for the SS3 cDNA clone is U73588.
Acknowledgments
We gratefully acknowledge excellent technical assistance from Ping Hua He for cotton transformation, Catherine Jones for the enzyme assay, and Celia Miller for electron microscopy. We thank Colin Jenkins and Jeff Ellis for critical reading of the manuscript. The polyclonal antibody against cotton Sus was a gift from Deborah P. Delmer. This work was supported by the Australian Cotton Research and Development Corporation and by Aventis CropScience.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010108.
References
- Amor, Y., Haigler, C.H., Johnson, S., Wainscott, M., and Delmer, D.P. (1995). A membrane-associated form of Sus and its potential role in synthesis of cellulose and callose in plants. Proc. Natl. Acad. Sci. USA 92, 9353–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basra, A., and Malik, C.P. (1984). Development of the cotton fiber. Int. Rev. Cytol. 89, 65–113. [Google Scholar]
- Buchala, A.J. (1999). Noncellulosic carbohydrates in cotton fibers. In Cotton Fibers: Developmental Biology, Quality Improvement and Textile Processing, A.S. Basra, ed (New York: Food Products Press), pp. 113–136.
- Chengappa, S., Guilleroux, M., Phillips, W., and Shields, R. (1999). Transgenic tomato plants with decreased sucrose synthase are unaltered in starch and sugar accumulation in the fruit. Plant Mol. Biol. 40, 213–221. [DOI] [PubMed] [Google Scholar]
- Chourey, P.S., Chen, Y.C., and Miller, M.E. (1991). Early cell degeneration in developing endosperm is unique to the Shrunken mutation in maize. Maydica 36, 141–146. [Google Scholar]
- Chourey, P.S., and Nelson, O. (1976). The enzymatic deficiency conditioned by the shrunken 1 mutations in maize. Biochem. Genet. 14, 1041–1055. [DOI] [PubMed] [Google Scholar]
- Chourey, P.S., and Nelson, O. (1979). Interallelic complementation at the sh locus in maize at the enzyme level. Genetics 91, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey, P.S., Taliercio, E.W., Carlson, S.J., and Ruan, Y.-L. (1998). Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol. Gen. Genet. 259, 88–96. [DOI] [PubMed] [Google Scholar]
- Craig, S., and Beaton, C.D. (1996). A simple cryo-SEM method for delicate plant tissues. J. Microsc. 182, 102–105. [Google Scholar]
- Farley, S.J., Patrick, J.W., and Offler, C.E. (2000). Functional transfer cells differentiate in cultured cotyledons of Vicia faba L. seeds. Protoplasma 214, 102–117. [Google Scholar]
- Geigenberger, P., and Stitt, M. (1993). Sucrose synthase catalyses a readily reversible reaction in vivo in developing potato tubers and other plant tissues. Planta 189, 329–339. [DOI] [PubMed] [Google Scholar]
- Haigler, C.H., Ivanova-Datcheva, M., Hogan, P.S., Salnikov, V.V., Hwang, S., Martin, K., and Delmer, D.P. (2001). Carbon partitioning to cellulose synthesis. Plant Mol. Biol. 47, 29–51. [PubMed] [Google Scholar]
- John, M.E. (1999). Genetic engineering strategies for cotton fiber modification. In Cotton Fibers: Developmental Biology, Quality Improvement and Textile Processing, A.S. Basra, ed (New York: Food Products Press), pp. 271–292.
- King, S.P., Lunn, J.E., and Furbank, R.T. (1997). Carbohydrate content and enzyme metabolism in developing canola (Brassica napus L.) siliques. Plant Physiol. 114, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti, E., Bellis, L.D., Alpi, A., and Perata, P. (2001). Why and how do plant cells sense sugars? Ann. Bot. 88, 803–812. [Google Scholar]
- Miller, M.E., and Chourey, P.S. (1992). The maize invertase-deficient miniature-1 seed mutation is associated with aberrant pedicel and endosperm development. Plant Cell 4, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, F., Llewellyn, D., McFadden, H., Last, D., Dennis, E.S., and Peacock, W.J. (1999). Expression of the Talaromyces flavus glucose oxidase gene in cotton and tobacco reduces fungal infection, but is also phytotoxic. Mol. Breeding 5, 219–232. [Google Scholar]
- Nolte, K.D., Hendrix, D.L., Radin, J.W., and Koch, K.E. (1995). Sucrose synthase localization during initiation of seed development and trichome differentiation in cotton ovules. Plant Physiol. 109, 1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A.H., Brubaker, C.L., and Wendel, J.F. (1993). A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. Rep. 11, 122–127. [Google Scholar]
- Pear, J.R., Kawagoe, Y., Schreckengost, W.E., Delmer, D.P., and Stalker, D.M. (1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA 93, 12637–12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, Y.-L., and Chourey, P.S. (1998). A fiberless seed mutation in cotton is associated with lack of fiber cell initiation in ovule epidermis and alterations in sucrose synthase expression and carbon partitioning in developing seeds. Plant Physiol. 118, 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, Y.-L., Chourey, P.S., Delmer, P.D., and Perez-Grau, L. (1997). The differential expression of sucrose synthase in relation to diverse patterns of carbon partitioning in developing cotton seed. Plant Physiol. 115, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, Y.L., Llewellyn, D.J., and Furbank, R.T. (2000). Pathway and control of sucrose import into initiating cotton fibers. Aust. J. Plant Physiol. 27, 795–800. [Google Scholar]
- Ruan, Y.L., Llewellyn, D.J., and Furbank, R.T. (2001). The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 13, 47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, Y.-L., and Patrick, J.W. (1995). The cellular pathway of postphloem sugar transport in developing tomato fruit. Planta 196, 434–444. [Google Scholar]
- Schubert, A.M., Benedict, C.R., Berlin, J.D., and Kohel, R.J. (1973). Cotton fiber development: Kinetics of cell elongation and secondary wall thickening. Crop Sci. 13, 704–709. [Google Scholar]
- Schünmann, P.H.D., Llewellyn, D., Surin, B., Boevink, P., Defeyter, R.C., and Waterhouse, P.M. (2003). A suite of novel promoters and terminators for plant biotechnology. Funct. Plant Biol., in press. [DOI] [PubMed]
- Thompson, R.D., Hueros, G., Becker, H.-A., and Maitz, M. (2001). Development and functions of seed transfer cells. Plant Sci. 160, 775–783. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Wang, M.B., and Finnegan, E.J. (2001). Role of short RNAs in gene silencing. Trends Plant Sci. 6, 297–301. [DOI] [PubMed] [Google Scholar]
- Weber, H., Buchner, P., Borisjuk, L., and Wobus, U. (1997). Sugar import and metabolism during seed development. Plant J. 9, 841–850. [DOI] [PubMed] [Google Scholar]
- Zrenner, R., Salanoubat, M., Willmitzer, L., and Sonnewald, U. (1995). Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). Plant J. 7, 97–107. [DOI] [PubMed] [Google Scholar]