Abstract
Mycobacterium tuberculosis produces a large number of structurally diverse lipids generated from the carboxylation products of acetyl-CoA and propionyl-CoA. A biotin-dependent acyl-CoA carboxylase was purified from M. tuberculosis H37Rv by avidin affinity chromatography, and the three major protein components were determined by N-terminal sequencing to be the 63-kDa α3-subunit (AccA3, Rv3285), the 59-kDa β5-subunit (AccD5, Rv3280), and the 56-kDa β4-subunit (AccD4, Rv3799). A minor protein of about 24 kDa that co-purified with the above subunits was identified by matrix-assisted laser desorption/ionization-time of flight mass spectrometry to be the product of Rv3281 that is located immediately downstream of the open reading frame encoding the β5-subunit. This protein displays identity over a short stretch of amino acids with the recently discovered ε-subunits of Streptomyces coelicolor, suggesting that it might be an ε-subunit of the mycobacterial acyl-CoA carboxylase. To test this hypothesis, the carboxylase subunits were expressed in Escherichia coli and purified. Acyl-CoA carboxylase activity was successfully reconstituted for the first time from purified subunits of the acyl-CoA carboxylase of M. tuberculosis. The reconstituted α3-β5 showed higher activity with propionyl-CoA than with acetyl-CoA, and the addition of the ε-subunit stimulated the carboxylation by 3.2- and 6.3-fold, respectively. The α3-β4 showed very low activity with the above substrates but carboxylated long chain acyl-CoA. This ε-subunit contains five sets of tandem repeats at the N terminus that are required for maximal enhancement of carboxylase activity. The Rv3281 open reading frame is co-transcribed with Rv3280 in the mycobacterial cell, and the level of ε-protein was highest during the log phase and decreased during the stationary phase.
Biotin-dependent carboxylases are involved in the synthesis of malonyl-CoA and methylmalonyl-CoA. Most biotin-dependent enzymes contain three functional components: the biotin carboxylase (BC),2 the biotin carboxyl carrier protein (BCCP), and the carboxyltransferase (CT) (1). The acyl-CoA carboxylation reaction occurs in two steps in two separate subsites of the enzyme. The first partial reaction involves the fixation of CO2 on biotin and requires the cooperation of BC and BCCP components; the biotin group is moved to interact with the BC component, resulting in the formation of carboxyl biotin. This carboxyl biotin then swings out to the carboxyltransferase component, resulting in the formation of the carboxylated product (2).
Acyl-CoA carboxylases from Mycobacterium tuberculosis and Mycobacterium bovis BCG have been purified previously and shown to have both propionyl-CoA carboxylase (PCC) and acetyl-CoA carboxylase (ACC) activities (3). Mycobacterium smegmatis PCC was reported to be composed of two subunits with the biotin being associated with the heavier subunit (4, 5), as found also in M. tuberculosis (3). M. tuberculosis has three accA genes annotated as an α-subunit that contains the BC and BCCP domains and six accD genes annotated as β-subunits that comprise the CT domain (6). It is not known which of these genes are expressed in M. tuberculosis. The acyl-CoA selectivity of the CT domain in the different β-subunits also remains unclear. Isolation of the various subunits and reconstitution of active carboxylase would be required to examine the catalytic activities of the various gene products. However, there are no reports of reconstitution of active acyl-CoA carboxylase from purified subunits of the M. tuberculosis acyl-CoA carboxylase.
Recently, a new group of acyl-CoA carboxylases was identified in S. coelicolor that contained a third type of subunit, designated ε. The ε-subunit considerably enhanced the basal activity obtained with the α-and β-subunits from this organism (7, 8). It is not known whether the mycobacterial carboxylase belongs to this new group. In the genome of M. tuberculosis, an open reading frame (ORF) corresponding to an ε-subunit has not been identified, and the purified carboxylase from the pathogen has not been thoroughly examined for subunit composition. We report the identification of an ε-like ORF adjacent to the β5 ORF (Rv3280) and present evidence for the expression of this ORF in M. tuberculosis. MALDI-TOF MS analysis of a 24-kDa protein that was purified with the native carboxylase by avidin-affinity chromatography showed it to be the ε-subunit of the carboxylase. We also showed that ACCA3, ACCD4, and ACCD5 are the major subunits of the native carboxylase. Reconstitution of active carboxylase from expressed and purified subunits showed that the ε-subunit stimulates carboxylation by α3-β5, demonstrating that M. tuberculosis has a novel carboxylase similar to that in S. coelicolor.
EXPERIMENTAL PROCEDURES
Materials
Sodium [14C]bicarbonate (50 Ci mol−1) was purchased from American Radiolabeled Chemicals Inc., and all other chemicals and reagents were of the highest grade and were purchased from Sigma and Fisher. Nucleotide primers were synthesized by Integrated DNA Technologies, Inc.
Bacterial Strains, Culture, and Transformation Conditions
Escherichia coli strain DH5α was used for routine subcloning and was transformed according to Sambrook et al. (9). E. coli BL21 Star (DE3) and E. coli Rosetta (DE3) were used for expression of recombinant proteins (10). All media were purchased from Difco. Host strains for cloning and expression experiments were grown on Luria Bertani (LB) broth or agar. Ampicillin, chloramphenicol, and kanamycin were added when required at final concentrations of 100, 34, and 50 μg ml−1, respectively.
Growth Conditions, Protein Production, and Preparation of Cell-free Extracts
For expression of heterologous proteins, E. coli strains harboring the appropriate plasmids were grown at 37 °C in LB medium in the presence of the corresponding antibiotics for plasmid maintenance. Overnight cultures were diluted 1:100 in fresh medium, grown to A600 0.8, and induced with 1 mm isopropyl-β-d-thiogalactopyranoside for 4 h at the same temperature with shaking. E. coli BL21 transformed with pCY216 (11), which expresses the E. coli biotin ligase (BirA), was grown under the same conditions as above and induced by the addition of arabinose to a final concentration of 0.5%. The cells were harvested, washed, and resuspended in 1× equilibration/wash buffer, pH 7.0, containing 50 mm sodium phosphate and 300 mm NaCl and were disrupted by sonication using a Branson Sonifier 450 (Branson Ultrasonics Corp., Danbury, CT). Cell debris was removed by centrifugation, and the supernatant was used as the cell-free extract.
Gel Electrophoresis and Western Blot Analysis
Cell-free extracts and purified proteins were analyzed by SDS-PAGE (12) using a Bio-Rad minigel apparatus. Protein concentration was determined by the method of Bradford (13). To detect His-tagged protein and biotin-containing proteins, Western blot analyses were carried out with His probe (H-3) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and peroxidase-conjugated avidin (Pierce), respectively. For Western blotting, cell extracts or purified proteins were subjected to SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane by using Trans-blot Cell from Bio-Rad.
Antibody Production and ε-Protein Expression in M. tuberculosis
The purified 177-residue ε-subunit was resolved on SDS-PAGE and used for raising antibodies in rabbit by Harlan Bioproducts for Science, Inc (Madison, WI) according to their standard 73-day protocol. M. tuberculosis H37Rv cultures were grown aerobically in roller cultures in 7H9 medium at 37 °C. Equal quantities of cell lysates from 3-, 5-, 7-, and 9-day-old cultures were resolved on 12% SDS-PAGE and were transferred to Immobilon polyvinylidene difluoride membrane. The blot was probed with antisera raised as described above, and the bands were quantitated on an AlphaImager Gel Doc system.
Avidin Affinity Chromatography and Electrophoresis
Acyl-CoA carboxylase was purified by avidin affinity chromatography as described previously (3). M. tuberculosis H37Rv inactivated by γ irradiation (obtained from Colorado State University, TB Research Materials and Vaccine Testing Contract NIAID, National Institutes of Health, NO1 AI-753230) were disrupted by a French pressure cell operating at 15,000 p.s.i. (passed four times) in 100 mm potassium phosphate (pH 7.0) containing 1.0 mm dithio-erythritol, 1.0 mm EDTA, and 10% glycerol. Just before each passage, 50 μl of 0.1 m phenylmethylsulfonyl fluoride in isopropyl alcohol was added to inhibit proteases. The homogenate was centrifuged at 30,000 × g for 30 min, and the supernatant was then centrifuged at 105,000 × g for 90 min. The final supernatant was mixed with a monomeric avidin-Sepharose affinity resin; the mixture was stirred slowly for 1 h, and the gel was poured into a column. The gel was washed with 5 bed volumes of the same buffer at 0.5 ml/min, and a linear gradient of 0–0.2 mm d-biotin in the phosphate buffer in a total volume of 300 ml was applied. The fractions (5.0 ml) containing the highest carboxylase activity were pooled and concentrated by ultrafiltration using Amicon PM-3 membrane. To determine the N-terminal protein sequence, the separated proteins from 7.5% SDS-PAGE were transferred on Immobilon polyvinylidene difluoride membrane (Millipore) in 10 mm CAPS buffer (pH 11.0) with 10% methanol. After staining with Coomassie Blue, N-terminal sequence of each protein band was determined by automated Edman degradation on a protein sequencer (Cleveland Clinic, Cleveland, OH). The ε-like proteins were resolved on 15% SDS-PAGE and analyzed by MALDI-TOF MS following tryptic digestion.
Gene Cloning and Plasmid Constructions
DNA corresponding to each carboxylase subunit ORF was amplified using Pfu Turbo Hotstart DNA polymerase (Stratagene), and expression was performed using the pET expression vector (Novagen). The primers used for amplification of the genes Rv3285 (α3), Rv3799c (β4), Rv3280 (β5), and Rv3281 (ε) are listed in Table 1. All of the PCR products were cloned using the PCR-BluntII TOPO vector into E. coli TOP10 competent cells. These inserts were released by digestion as described by Table 1 and subcloned in pET16b or pET28a (+) vectors. Rv3285, Rv3799c, and Rv3280 were expressed in pET16b, and Rv3281 was expressed in pET28a (+). In these vectors, the ORFs were directionally cloned and expressed as His-tagged fusion proteins in E. coli BL21 Star (DE3) or E. coli Rosetta (DE3).
TABLE 1.
Primers used for this study
| Primer name | Sequencea(5′→3′ direction) | Comments |
|---|---|---|
| AccA3F | GGCATATGGCTAGTCACGCCGGCT | Upstream of accA3, NdeI site added |
| AccA3R | GGAGATCTTTACTTGATCTCGGCG | Downstream of accA3, BglII site added |
| AccD4F | GGCATATGACCGTCACCGAGCCG | Upstream of accD4, NdeI site added |
| AccD4R | GGCTCGAGTCAGGCCGTGCTTGC | Downstream of accD4, XhoI site added |
| AccD5F | GGCATATGACAAGCGTTACCGAC | Upstream of accD5, NdeI site added |
| AccD5R | GGCTCGAGTCACAGGGGCACGTT | Downstream of accD5, XhoI site added |
| 1ENF | CATATGGGAACGTGCCCCTGTG | Upstream 1M of Rv3281, NdeI site added |
| 1ECR | GGATCCTCGGCGCATGTGCGTC | Downstream of Rv3281, BamHI site added |
| 102EF | GGCATATGACCGAGAAGCCGCTGC | Positions 102V of Rv3281, NdeI site added |
| 102ER | GCTCGAGTCATCGGCGCATGTGCGTC | Downstream of Rv3281, XhoI site added |
| D5–227F | CTTCGTGTACCGCCAGCAGCTGG | Internal primer of accD5, RT-PCR |
| E-139R | CTGCCGGATTGTTCGTCTCGTTC | Internal primer of Rv3281, RT-PCR |
Endonuclease restriction sites added to sequences are underlined.
Purification of Expressed Proteins
The E. coli strain BL21 star (DE3) containing the plasmids pTJA3 (α3), pTJD4 (β4), and pTJD5 (β5) were induced with isopropyl-β-d-thiogalactopyranoside for protein overexpression as described above. Following lysis, the 16,000 ×g supernatants were mixed with TALON resin (BD Biosciences) at room temperature for 30 min to allow the polyhistidine-tagged protein to bind the resin. The His-tagged fusion proteins were recovered by elution with 100 mm imidazole for the α3- and β4-subunits or 500 mm imidazole for the β5-subunits. For biotinylation of the cloned α3-subunit, E. coli cell lysate containing the expressed α3-subunit was incubated with an extract of E. coli cells expressing the cloned E. coli biotin ligase (BirA) from the pCY216 plasmid at 30 °C for 3 h (11). Biotinylation of the α3-subunit was confirmed by Western blotting with avidin-horseradish peroxidase conjugate (Santa Cruz Biotechnology). The E. coli strain Rosetta (DE3) was used for expression of pTJE1 and pTJE102. The His-tagged ε-proteins were insoluble and were solubilized with 6m guanidine-HCl from the 16,000 ×g pellet obtained from cell lysate. The solubilized proteins were bound to TALON resin, which was pre-equilibrated in 6 m guanidine-HCl, and were purified under denaturing conditions according to the manufacturer’s protocol. Following elution with 150 mm imidazole, the guanidine-HCl was removed, and the purified ε-proteins were refolded by stepwise dialysis. The first step of dialysis was performed against a solution containing 2 m guanidine-HCl, 100 mm potassium phosphate (pH 8.0), and 10% glycerol for 12 h at 4 °C followed by two rounds of dialysis against 100 mm potassium phosphate (pH 8.0) and 10% glycerol for 12 h at 4 °C.
Reconstitution of Carboxylase Activity
The purified ε-subunit in dilute form (100 μg/ml) was initially combined with a dilute (100 μg/ml) preparation of either the β4- or the β5-subunits, after which the β-ε subunits were combined with a dilute preparation of the biotinylated α3-subunit. Following this, the reconstituted enzyme was concentrated 5–7-fold by ultrafiltration in Centriprep YM-10 filters (Millipore) at 700 ×g for 6 h at 4 °C.
Acyl-CoA Carboxylase Assay
Enzyme activities of the in vitro reconstituted subunits obtained by mixing purified proteins were measured by following the incorporation of label from H14VCO3− into acid-stable reaction material (14). The reaction mixture contained 100 mm potassium phosphate, pH 8.0, 300 μg of bovine serum albumin, 2 mm ATP, 10 mm MgCl2, 5 mM NaH14CO3 (50 Ci mol−1), 2 mm acetyl-CoA, 2 mm propionyl-CoA, and 200–600 μg of reconstituted enzyme in a total reaction volume of 100 μl. The reaction was initiated by the addition of NaH14CO3, allowed to proceed at 30 °C for 3 h, and stopped with 150 μl of 6 m HCl. Following evaporation, the residue was resuspended in 100 μl of water, and the 14C radioactivity was determined by liquid scintillation counting. To determine the effect of inhibitors on acyl-CoA carboxylase activity, the reconstituted enzyme was incubated with the inhibitor for 30 min at room temperature prior to the assay described above.
RESULTS
Avidin Purification and Identification of the Native Carboxylase Subunits
The carboxylases purified from M. tuberculosis H37Rv by avidin affinity chromatography showed activity with both acetyl-CoA (12.3 nmol/mg/min) and propionyl-CoA (20.6 nmol/mg/min) as substrates. When the purified native carboxylase preparation from M. tuberculosis was resolved on 7.5% SDS-PAGE, three proteins with apparent molecular masses of 63, 59, and 56 kDa were observed. The N-terminal amino acid sequence determination revealed them as being the products of Rv3285 (AccA3, α3; ASHAGSRIARISK, lacking the terminal methionine), Rv3280 (AccD5, β5; TSVTDRSAHSAER, lacking the terminal methionine), and Rv3799c (AccD4, β4; VTEPVL, lacking 2 N-terminal amino acids) (Fig. 1A).
FIGURE 1. Determination of subunit composition and domain organization of M. tuberculosis acyl-CoA carboxylase.
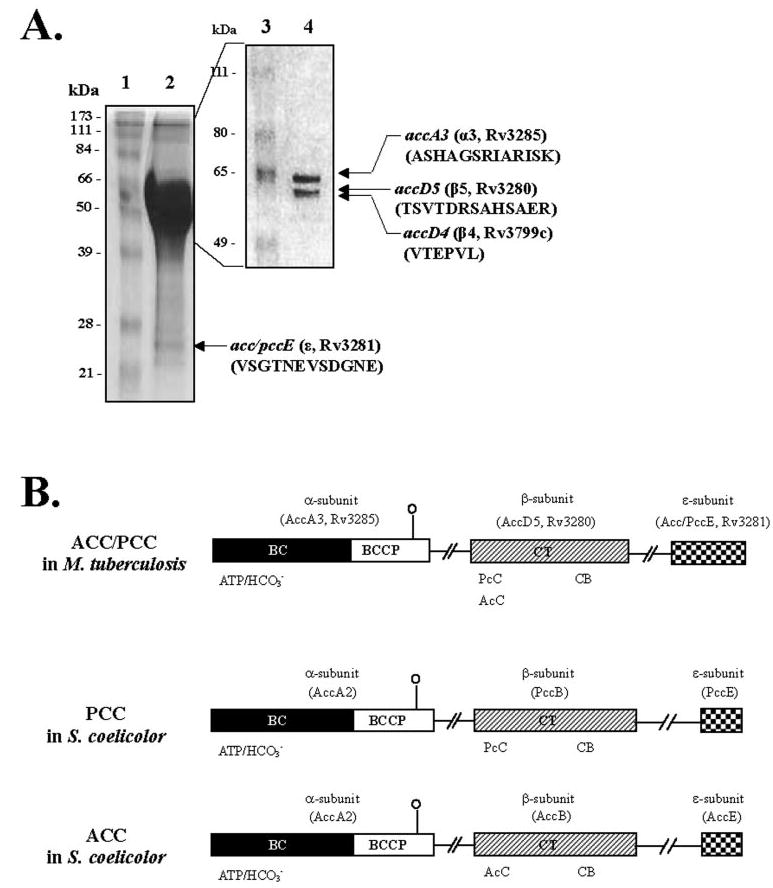
A, identification of the subunits of the biotin-dependent acyl-CoA carboxylase. Acyl-CoA carboxylase was purified by single-step avidin affinity chromatography, and the purified complex was resolved on 7.5% SDS-PAGE. The N-terminal amino acid sequences of the 63- (α3), 59- (β5), and 56- (β4) kDa bands were determined by Edman degradation. The 24-kDa band (ε) was resolved on 15% SDS-PAGE, and the amino acid sequence was determined by MALDI-TOF MS following tryptic digestion. Lane 1, molecular weight protein marker; lane 2, avidin-purified acyl-CoA carboxylase complex on 15% SDS-PAGE; lane 3, molecular weight protein marker; and lane4, 7.5% SDS-PAGE of avidin-purified acyl-CoA carboxylase. B, domain organization and amino acid sequence similarity of M. tuberculosis and S. coelicolor acyl-CoA carboxylase. Sequence similarities are shown by similarly shaded boxes. AccA3, Rv3285, acyl-CoA carboxylase α-subunit from M. tuberculosis; AccD5, Rv3280, propionyl-CoA carboxylase β-subunit from M. tuberculosis; Acc/PccE, Rv3281, acyl-CoA carboxylase ε-subunit from M. tuberculosis; AccA2, acyl-CoA carboxylase α-subunit from S. coelicolor; AccB, acetyl-CoA carboxylase β-subunit from S. coelicolor; PccB, propionyl-CoA carboxylase β-subunit from S. coelicolor; AccE, acetyl-CoA carboxylase ε-subunit from S. coelicolor; PccE, propionyl-CoA carboxylase ε-subunit from S. coelicolor; ATP/CO3−, biotin carboxylation domain that binds ATP and CO2 fixation, respectively; CB, putative carboxybiotin-binding domain; AcC and PcC, carboxyltransferase domain that binds acetyl-CoA and propionyl-CoA, respectively.
We expressed the α3-, β4-, and β5-subunits as His-tagged fusion proteins in E. coli. The biotinylated α3-subunit was purified and mixed with the purified β4- or β5-subunit and assayed for carboxylation of acetyl-CoA and propionyl-CoA. The reconstituted enzyme showed only very low activity but appeared to support the conclusion that the β5-subunit preferred propionyl-CoA. The low activity observed with the purified subunits made us wonder whether we were missing another component that might be required for effective carboxylation. At this time, a novel class of carboxylase that requires an additional ε-subunit for activity was described in S. coelicolor (7, 8). We used the most conserved segments of the four ε-subunits found in Streptomyces to search the mycobacterial genome. We discovered that a segment of a hypothetical protein downstream of accD5 showed homology to the conserved region of the ε-subunit of Streptomyces. To determine whether the native carboxylase contained an ε-subunit protein, the purified native carboxylase preparation was resolved by 15% SDS-PAGE. In addition to α3, β4, and β5, a 24-kDa band was observed (Fig. 1A). MALDI-TOF MS analysis following trypsin digestion revealed a sequence, VSGT-NEVSDGNE, which we identified as the product of the Rv3281 ORF lacking 17 N-terminal amino acids. The presence of a trypsin cleavage site immediately upstream of this peptide may explain the absence of the N-terminal end. Interestingly, Rv3281 is located immediately downstream of the ORF encoding the AccD5 (β5, Rv3280), as shown in Fig. 1B. Since this suggested a potential role for Rv3281 gene product in carboxylation involving the β5-subunit, we examined the role of Rv3281 in acyl-CoA carboxylation.
ε Transcript and Protein Expression in M. tuberculosis
To test whether the putative ε-subunit of M. tuberculosis is expressed in the pathogen, semiquantitative RT-PCR analysis of the transcripts was done. RT-PCR data clearly showed that the ORF encoding the ε-subunit is transcribed in the mycobacterial cell. RT-PCR analysis with primers spanning the β5-ε junction yielded a product expected if both ORFs were transcribed as one unit (Fig. 2A). We assayed the ε-protein expression level in cultures of M. tuberculosis using rabbit antisera raised against the ε-protein expressed in E. coli. The protein level was maximal at 5 days of growth when the culture was in its exponential growth phase and declined when the culture entered late log and stationary phase (Fig. 2B).
FIGURE 2. ε transcript and protein expression level in M. tuberculosis cultures grown in vitro.
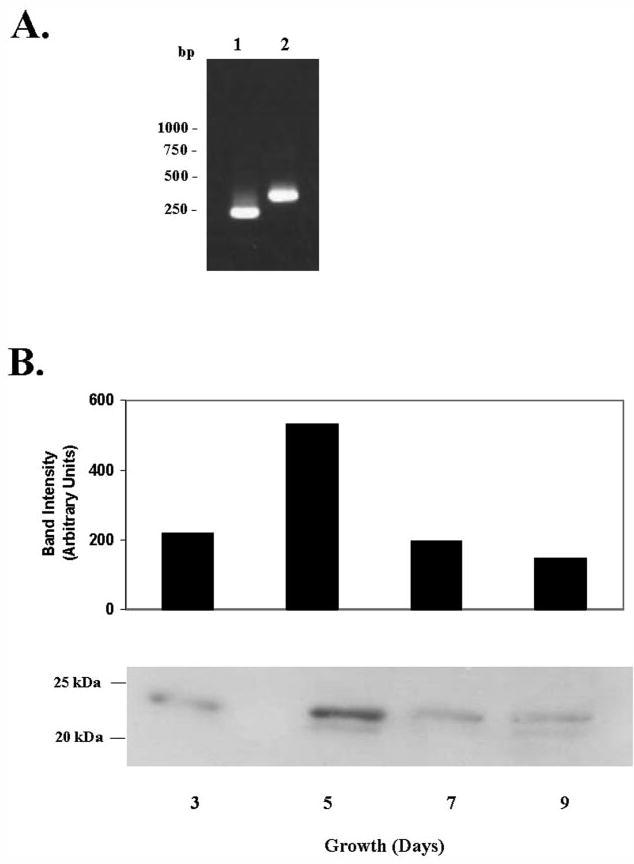
A, semiquantitative RT-PCR showing expression of the ε ORF in M. tuberculosis. Lane 1, PCR product obtained with internal primers 102EF and 1ECR (Table 1); lane 2, PCR product obtained with primers spanning the β5-ε ORF junction (D5-227F and E-139R, Table 1). B, Western blot of total cell lysates from M. tuberculosis H37Rv cultures grown at 37 °C. Equal quantities of lysates from each time point were resolved on 12% SDS-PAGE, transferred to Immobilon polyvinylidene difluoride membrane, and probed with rabbit antisera raised against purified ε-subunit.
In Vitro Reconstitution of Carboxylase Activity with the Purified Subunits
To examine the possible role of Rv3281 in the carboxylation reaction involving the α3-, β4-, and β5-subunits, we expressed α3-, β4-, β5-, and ε-subunits as His-tagged fusion proteins in E. coli and purified them (Fig. 3A). Biotinylation of the expressed α3-subunit was performed with the expressed E. coli biotin ligase, and successful biotinylation of the α3-subunit was confirmed by Western blotting using the avidin-horseradish peroxidase conjugate (data not shown). The biotinylated α3-subunit was purified and mixed with the purified β4- or β5-subunit and assayed for carboxylation of acetyl-CoA or propionyl-CoA. In our initial reconstitution experiments, the expressed and purified subunits were mixed together after concentration of individual subunits. These preparations showed very low acyl-CoA carboxylase activities. When we analyzed the concentrated subunits by Superose-12 gel filtration chromatography, we found that the individual concentrated subunits were in aggregated forms; aggregation would prevent interaction with other subunits and thus hinder successful reconstitution. Attempts to mix the aggregated forms under denaturing conditions in 6m urea followed by renaturation of the subunits by stepwise dialysis yielded extremely low carboxylase activity. Therefore, individual subunits were purified by cobalt affinity chromatography, and the dilute subunit preparations were mixed prior to concentration. The purified ε-subunit in dilute form was initially combined with the dilute solution of either the β4- or the β5-subunits before the addition of a dilute preparation of the biotinylated α3-subunit followed by concentration by ultrafiltration. Such a reconstitution protocol yielded high levels of carboxylase activity with the α3-β5-ε when acetyl-CoA or propionyl-CoA was used as the substrate. As shown in Fig. 3B, the reconstituted α3-β5 showed nearly 3-fold higher activity with propionyl-CoA when compared with acetyl-CoA. However, when reconstituted with the ε-subunit, the α3-β5-ε gave 6.3-fold increased ACC activity. The PCC activity increased only 3.2-fold with the inclusion of the ε-subunit. The reconstituted α3-β4 and α3-β4-ε showed very low carboxylase activity with either substrate. α3-β4 and α3-β5 catalyzed carboxylation of long chain fatty acyl-CoA, and avidin completely inhibited this carboxylation. This carboxylation was not affected by the addition of the ε-subunit (Fig. 3, B and C). These results indicate that the ε-subunit interacts with only the β5-subunit and modulates its carboxylase activity for acetyl-CoA and propionyl-CoA.
FIGURE 3. Purification of M. tuberculosis acyl-CoA carboxylase subunits expressed in E. coli and reconstitution of acyl-CoA carboxylase activity with purified subunits.
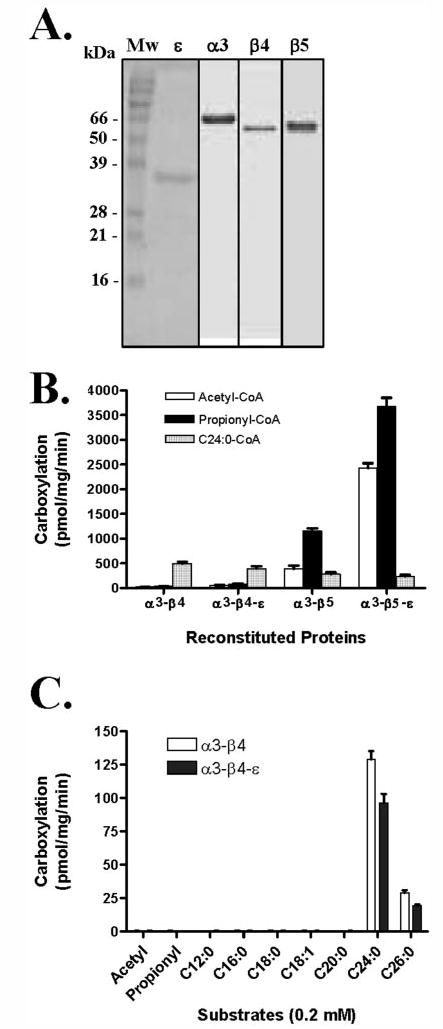
A, each subunit was expressed in E. coli and purified by cobalt affinity chromatography. The α3-subunit was biotinylated using biotin ligase (pCY216) prior to purification. Lane 1, Mw, molecular weight protein marker; lane 2, purified α-subunit (full-length) from E. coli; lane 3, purified α3-subunit from E. coli; lane 4, purified β4-subunit from E. coli; and lane 5, purified β5-subunit from E. coli. B, influence of ε on the carboxylase activities of α3-β4 and α3-β5 with acetyl-CoA (white bars), propionyl-CoA (black bars), and long chain acyl-CoA (shaded bars) substrates was measured under optimal conditions. C, carboxylase activities of α3-β4 (white bars) and α3-β4-ε (black bars) with acetyl-CoA, propionyl-CoA, and long chain acyl-CoA substrates.
Characterization of the ε-Subunit (Rv3281)
An analysis of the amino acid sequence of the ε-subunit revealed a low level of identity at the C terminus with the reported ε-subunit of S. coelicolor (7, 8). As shown in Fig. 4A, the M. tuberculosis ε-subunit has five sets of the repeated amino acids ((N/A)EVSDGNETNNP) at the N terminus that are not found in the S. coelicolor ε-subunits (CAC21625 and CAA19984). To determine whether the N-terminal repeated amino acids were important for activity, we cloned a truncated version of the ε-subunit that contained only the C-terminal portion that showed homology with S. coelicolor ε-subunit. When the full-length (ε) or truncated (εmut) versions of the ε-subunit were used for reconstitution with the α3- and β5-subunits and assayed for acyl-CoA carboxylase activities with acetyl-CoA or propionyl-CoA, the truncated ε-subunit showed very much less stimulation of the carboxylase activity when compared with the full-length ε-subunit, indicating that N-terminal repeats in the M. tuberculosis ε-subunit are important for carboxylase activity (Fig. 4B).
FIGURE 4. Primary structure of M. tuberculosis ε-subunit (Rv3281) and activity of full-length and truncated ε-subunits.
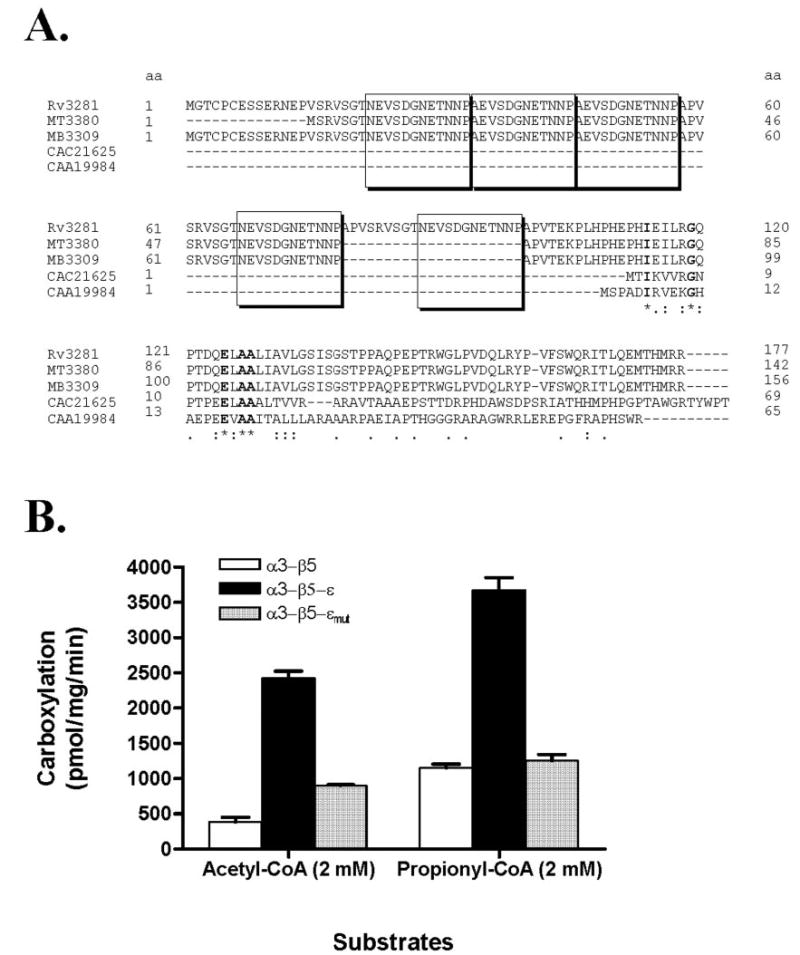
A, amino acid sequence of Rv3281 showing N-terminal repeats and sequence similarity with S. coelicolor ε-subunits and other putative mycobacterial ε-like proteins. Amino acid sequences of Rv3281 (M. tuberculosis H37Rv), MT3380 (M. tuberculosis CDC1551), MB3309 (M. bovis subsp. Bovis AF2122/97), CAC21625 (S. coelicolor), and CAA19984 (S. coelicolor A3 (2)) are shown. Boxes indicate the repeated amino acids ((N/A)EVSDGNETNNP). Identical amino acids are indicated by an asterisk below, and conserved residues are indicated by a period or colon. B, the effects of full-length (ε) and N-terminal truncated versions (εmut, 102 amino acids deleted at N-terminal) on acyl-CoA carboxylase activity were measured by mixing the purified α3- and β5-subunits with the respective ε-subunits using acetyl-CoA and propionyl-CoA as substrates. Activity obtained under optimal substrate concentration is represented as mean ± S.D. from three independent measurements. Acyl-CoA carboxylase activity was measured in α3-β5 (white bars), α3-β5-ε (black bars), and α3-β5-εmut (shaded bars).
Determination of Optimal Stoichiometry of Expressed Subunits
The optimal stoichiometric combination of the α3-, β5-, and ε-subunits was determined by following the reconstitution procedure described above with various molar concentrations of each subunit followed by the standard carboxylation assay with acetyl- or propionyl-CoA. The highest carboxylation activity was obtained when the molar ratios of α3, β5, and ε were 1:1:2 (Fig. 5).
FIGURE 5. Determination of optimal stoichiometry for reconstitution of acyl-CoA carboxylase activity.
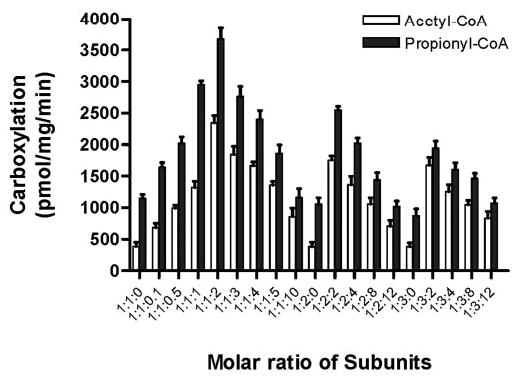
Purified subunits were mixed in the indicated ratios prior to concentration. The molar ratios of subunits are indicated as A:B:C where A represents the ratio of the α3-subunit, B represents the ratio of the β5-subunit, and C represents the ratio of the ε-subunit. Acyl-CoA carboxylase activity with acetyl-CoA (white bars) and propionyl-CoA (black bars) substrates were measured after concentration. Results presented are the average of at least three independent experiments with a standard error.
Kinetic Characterization of α3-β5 and α3-β5-ε
The time course of acyl-CoA carboxylation with the reconstituted α3-β5 and β3-β5-ε was determined at 30 °C. As shown in Fig. 6, acetyl-CoA and propionyl-CoA carboxylation increased with incubation times up to 3 h. The dependence of the acyl-CoA carboxylase activities of the reconstituted enzyme on the concentration of acetyl-CoA, propionyl-CoA, bicarbonate, ATP, and MgCl2 was investigated as shown in Fig. 7. The reconstituted carboxylase exhibited typical Michaelis-Menten kinetics as shown Table 2. The α3-β5 and α3-β5-ε showed specificity constants (SC, Vmax/Km) of 1.3 and 7.2 for acetyl-CoA substrate and 10.4 and 14.8 for propionyl-CoA substrate, respectively (Table 2). These results confirmed that ε-subunit increased the SC of the α3-β5 for acetyl-CoA by 5.5-fold, whereas the SC for propionyl-CoA was increased only 1.4-fold.
FIGURE 6. Time course of acyl-CoA carboxylase activity with the reconstituted subunits.
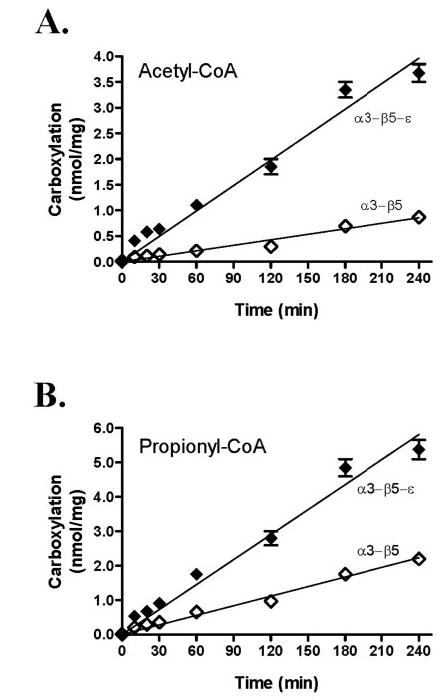
A, time dependence of ACC measured with α3-β5 (⋄) and α3-β5-ε (♦). B, time dependence of PCC measured with α3-β5 ⋄) and α3-β5-ε (♦). Results presented are the average of at least three independent experiments with standard error.
FIGURE 7. Substrate concentration dependence of acetyl-CoA and propionyl-CoA carboxylase activities of α3-β5 and α3-β5-ε.
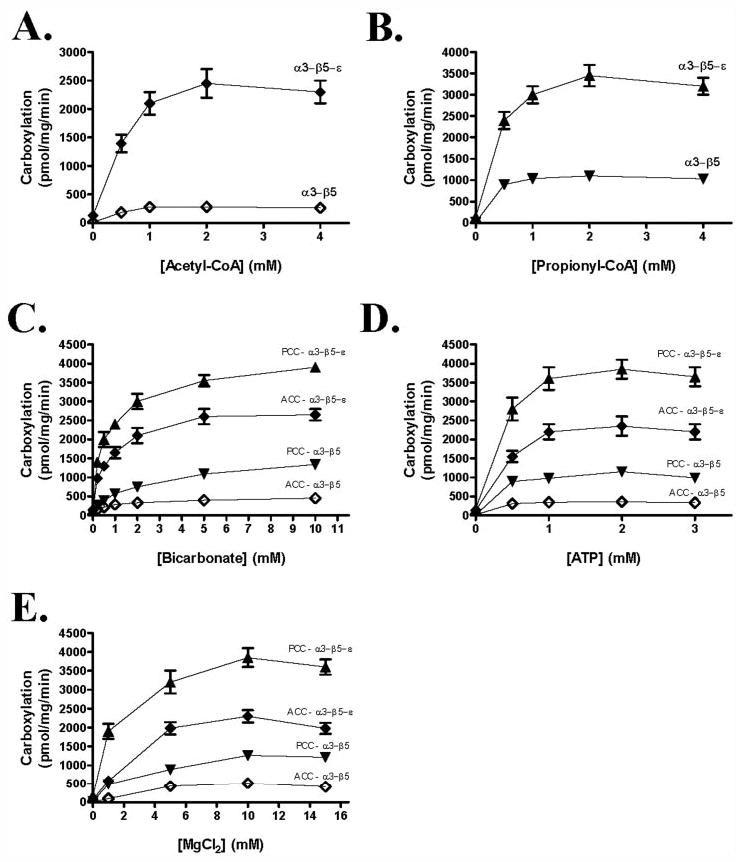
Acetyl-CoA (A), propionyl-CoA (B), bicarbonate (C), ATP (D), and MgCl2 (E) dependence of the carboxylation was measured with reconstituted α3-β5 and α3-β5-ε. ⋄ and ♦, ACC activity of α3-β5 (⋄) and α3-β5-ε (♦). ▾ and ▴, PCC activity of α3-β5 (▾) and α3-β5-ε (▴). Results presented are the average of at least three independent experiments with a standard error.
TABLE 2. Kinetic parameters for purified and reconstituted acyl-CoA carboxylase subunits of M. tuberculosis expressed in E. coli.
Km (μm), Vmax (pmol/mg/min), and SC (Vmax/Km) are shown.
|
Complexes |
|||||||
|---|---|---|---|---|---|---|---|
| α3β5 |
α3β5ε
|
||||||
| Substrates | Km | Vmax | SC | Km | Vmax | SC | |
| ACC | Acetyl-CoA | 232.0 | 302.9 | 1.3 | 377.7 | 2709.0 | 7.2 |
| Bicarbonate | 584.4 | 438.5 | 0.8 | 631.0 | 2669.0 | 4.2 | |
| ATP | 75.2 | 339.3 | 4.5 | 289.8 | 2431.0 | 8.4 | |
| PCC | Propionyl-CoA | 105.4 | 1095.0 | 10.4 | 233.0 | 3442.0 | 14.8 |
| Bicarbonate | 1752.0 | 1493.0 | 0.9 | 545.6 | 3802.0 | 7.0 | |
| ATP | 120.7 | 1099.0 | 9.1 | 215.3 | 3923.0 | 18.2 | |
Effect of Inhibitors on the Reconstituted Carboxylase
Incubation of the purified native carboxylase with avidin completely inhibited the acetyl-CoA and propionyl-CoA carboxylation reactions, showing that it is a biotin-dependent acyl-CoA carboxylase (data not shown). When the purified reconstituted α3-β5 and α3-β5-ε were incubated with avidin, the ACC and PCC activities were also completely inhibited as expected (Fig. 8). Acetyl-CoA carboxylation by α3-β5 and α3-β5-ε was inhibited by 90% with 100 μm diclofop when compared with the solvent (Me2SO) control. In contrast, propionyl-CoA carboxylase activity showed only 30% inhibition (Fig. 8).
FIGURE 8. Effect of inhibitors on reconstituted carboxylases.
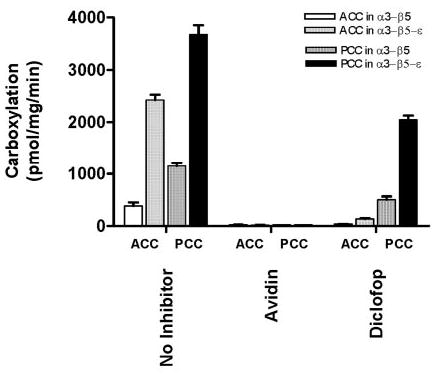
Inhibition of acyl-CoA carboxylase activity was measured by preincubating the purified α3-β5 and α3-β5-ε with avidin (1 unit) and diclofop (100 μm). Activity obtained under optimal substrate concentrations is represented as mean ± S.D. from three independent measurements. ACC activity was measured in α3-β5 (white bars) and α3-β5-ε (light shaded bars), and PCC activity was measured in α3-β5 (dark shaded bars) and α3-β5-ε (black bars).
DISCUSSION
Malonyl-CoA and methylmalonyl-CoA are products of the carboxylation of acetyl-CoA and propionyl-CoA, respectively, by an acyl-CoA carboxylase. Such acyl-CoA carboxylases, in most organisms, are composed of the α-subunit, which contains the BC and BCCP domains and the β-subunit, which contains the CT domain. In the M. tuberculosis genome, where the genes involved in lipid metabolism are abundant, there are three genes encoding the putative α-subunits and six genes encoding the putative β-subunits of the acyl-CoA carboxylases (6). It is not known which ORFs encode the subunits of the native acyl-CoA carboxylase. We identified the three major proteins in our avidin-purified M. tuberculosis acyl-CoA carboxylase preparation as the α3-, β4-, and β5-subunits by N-terminal sequencing. Recently, a new group of acyl-CoA carboxylases was discovered in S. coelicolor, which contained a third type of subunit designated as ε in addition to the α- and β-subunits (7, 8). Adjacent to the ORF encoding the β5-subunit, an ORF that has not been annotated in the mycobacterial genome (Rv3281) was found, the product of which showed homology with the recently identified ε-subunit of Streptomyces. We also identified a novel 24-kDa protein in the native carboxylase preparation and determined that it was the product of Rv3281.
To understand the nature of the biotin-dependent enzymes, we analyzed the amino acid sequence similarity between each similarly functioning domain of the M. tuberculosis carboxylase with other reported ACC and PCC protein sequences. As the biotin-dependent enzymes share similar reaction mechanisms, these enzymes also share a high degree of amino acid sequence similarity (data not shown). As shown in Fig. 1B, the domain organization of M. tuberculosis ACC/PCC is similar to that of the PCC and ACC of S. coelicolor (7, 8). The first half of M. tuberculosis AccA3 is composed of the BC that catalyzes the ATP-driven carboxylation of biotin to form carboxybiotin, and the amino acid sequence displays a high degree of homology with the N-terminal domain of the α-subunit (AccA2) of ACC and PCC from S. coelicolor. The other half of M. tuberculosis AccA3 is the BCCP domain that is found in all biotin-dependent enzymes, and this domain functions in transferring CO2 from one subsite to another, allowing carboxylation, and the amino acid sequence shows high homology with the C-terminal domain of the α-subunit (AccA2) of ACC and PCC from S. coelicolor. The amino acid sequence of M. tuberculosis AccA3 showed 68% identity with AccA2 of S. coelicolor. The M. tuberculosis AccD5 is most similar to the S. coelicolor PccB (67% identity), and the M. tuberculosis AccD4 also has similarity to PccB (49% identity) and AccB (44% identity) of S. coelicolor.
There are significant differences between the M. tuberculosis ε-subunit and the S. coelicolor ε-subunits. The M. tuberculosis genome contains only one ORF encoding an ε-subunit, whereas in the case of S. coelicolor, there are two ε-subunits, AccE and PccE, which interact specifically with the AccB and PccB, respectively. The M. tuberculosis ε-subunit is larger (177 amino acids) than its counterparts in S. coeli-color (65–69 amino acids) and contains a set of five tandem repeats of 12 amino acids each at the N terminus, which is not seen in the S. coelicolor ε-subunits. We determined that the N-terminal tandem repeats were important for activity and probably stabilize the protein-protein interactions between the α3-, β5-, and ε-subunits. Although the α3-β5-ε of M. tuberculosis belongs to the new group of acyl-CoA carboxylases found in S. coelicolor, there were substantial differences in the kinetic parameters between the two. The SC of the M. tuberculosis α3-β5-ε for acetyl-CoA was nearly 2-fold lower than the value for propionyl-CoA (Table 2). In contrast, the SC of the S. coelicolor ACC for acetyl-CoA was nearly 3-fold lower than that of the PCC for propionyl-CoA, and no activity was detected with the PCC when acetyl-CoA was used as substrate (8). Furthermore, the reconstituted M. tuberculosis acyl-CoA carboxylase yielded the highest activity when the ratio of the α3-, β5-, and ε-subunits was 1:1:2, whereas in the case of S. coelicolor, near maximal velocity was obtained when the ratio of the subunits was 1:1:3 for AccE and 1:1:9 for PccE (8).
Mutagenesis studies on related bacteria suggest that accD4 is involved in carboxylation of long chain acids required for mycolic acid synthesis, consistent with the finding that it was an essential gene in M. smegmatis and M. tuberculosis (15–17). In agreement with these reports, the reconstituted α3-β4 of M. tuberculosis carboxylated long chain fatty acyl-CoA in an ε-independent manner. The mycobacterial cell wall is rich in unusual lipids that are known to play critical roles in pathogenesis. Methylmalonyl-CoA is the unique precursor of the multiple methyl-branched lipids that have been shown to function as virulence factors, and its synthesis is a potential drug target. It is expected that the mycobacterium may require higher levels of n-fatty acids during its exponential growth phase and, as it enters the stationary phase, higher levels of branched fatty acids may be required for modifications of the cell wall. Our data indicated that expression of the ε-protein in the mycobacterial cell reaches its highest level during the exponential growth phase and decreases during the stationary phase. During the exponential phase, when n-acyl lipids are synthesized from malonyl-CoA, the ACC activity of the carboxylase needs to be increased, and that may be done by increased levels of the ε-subunit, which appears to favor the ACC activity. When branched lipids are required during the stationary phase, the PCC activity of the carboxylase may be favored by decreasing the expression of the ε-subunit.
Commercial herbicides such as diclofop, haloxyfop, and sethoxydim are potent inhibitors of ACC in plants and have been shown to act on the CT domain (18). Our studies with the reconstituted α3-β5-ε showed that diclofop inhibited the ACC activity almost completely. Since there is no report on the inhibition of PCC activity with such inhibitors, we examined whether the PCC activity of our reconstituted carboxylase was sensitive to inhibition by diclofop. We found that the PCC activity of the α3-β5-ε was inhibited by a much lesser degree by diclofop, suggesting that the inhibitor may distinguish the binding sites for acetyl-CoA and propionyl-CoA on the CT domain of the β5-subunit. Further studies with inhibitors of the α3-β5-ε may provide leads for drug development.
M. tuberculosis and M. bovis have three α-subunit genes and six β-subunit genes, but Mycobacterium leprae, which is known to have lost a substantial part of the original mycobacterial genome, has one α-subunit gene, three β-subunit genes, and one ε-subunit gene (19). The organization of the ORFs for the α3-, β5-, and ε-subunits and that of the biotin ligase (birA) in the genomes of M. tuberculosis H37Rv, M. tuberculosis CDC1551, M. bovis, M. leprae, and M. smegmatis is nearly identical in all these organisms and, in each case, the ε-subunit is located immediately downstream of the β5-subunit (Fig. 9). This conservation indicates that the ε-subunit probably plays an important role in the carboxylation reaction in all these species. Therefore, further study of its role may lead to deeper understanding of the acyl-CoA carboxylases in mycobacteria.
FIGURE 9. Organization of the carboxylase gene cluster in M. tuberculosis H37Rv and other mycobacterial species.
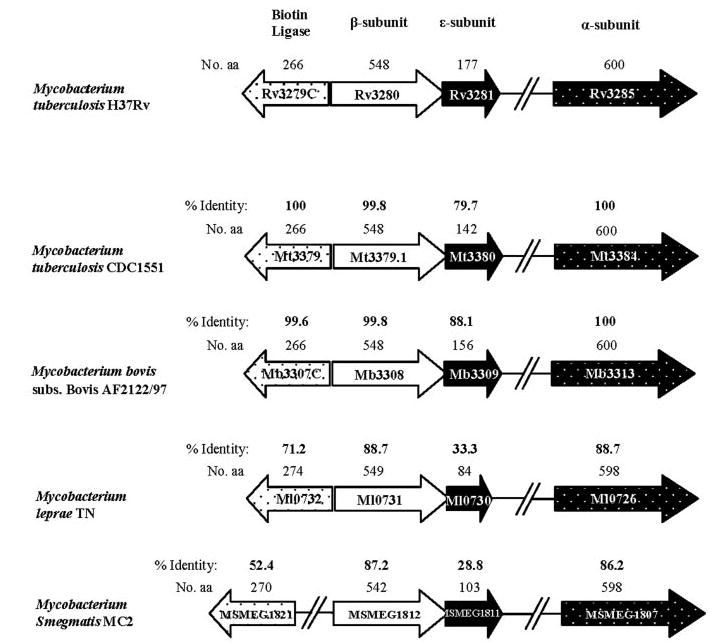
The percentages of amino acid (aa) identity of M. tuberculosis H37Rv subunits with other respective mycobacterial subunits are indicated in bold. The sizes in amino acids are given in regular numerals above the respective subunit.
Acknowledgments
We thank Prof. John E. Cronan, Jr. for kindly providing us the pCY216 plasmid.
Footnotes
This work was supported by Grants AI46582 and AI35272 from the National Institutes of Health.
The abbreviations used are: BC, biotin carboxylase; BCCP, biotin carboxyl carrier protein; CT, carboxyltransferase; PCC, propionyl-CoA carboxylase; ACC, acetyl-CoA carboxylase; ORF, open reading frame; MALDI-TOF MS, matrix-assisted laser desorption/ ionization time-of-flight mass spectrometry; RT, reverse transcriptase; SC, specificity constant; CAPS, 3-(cyclohexylamino)propanesulfonic acid.
References
- 1.Cronan JE, Jr, Waldrop GL. Prog Lipid Res. 2002;41:407–435. doi: 10.1016/s0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- 2.Attwood PV, Wallace JC. Acc Chem Res. 2002;35:113–120. doi: 10.1021/ar000049+. [DOI] [PubMed] [Google Scholar]
- 3.Rainwater DL, Kolattukudy PE. J Bacteriol. 1982;151:905–911. doi: 10.1128/jb.151.2.905-911.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henrikson KP, Allen SHG. J Biol Chem. 1979;254:5888–5891. [PubMed] [Google Scholar]
- 5.Hasse FC, Henrikson KP, Treble DH, Allen SHG. J Biol Chem. 1982;257:11994–11999. [PubMed] [Google Scholar]
- 6.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 7.Rodríguez E, Banchio C, Diacovich L, Bibb MJ, Gramajo H. Appl Environ Microbiol. 2001;67:4166–4176. doi: 10.1128/AEM.67.9.4166-4176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diacovich L, Peirú S, Kurth D, Rodríguez E, Podestá F, Khosla C, Gramajo H. J Biol Chem. 2002;277:31228–31236. doi: 10.1074/jbc.M203263200. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 10.Studier FW, Moffatt BA. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 11.Chapman-Smith A, Turner DL, Cronan JE, Jr, Morris TW, Wallace J. Biochem J. 1994;302:881–887. doi: 10.1042/bj3020881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 14.Hunaiti AR, Kolattukudy PE. Arch Biochem Biophys. 1982;216:362–371. doi: 10.1016/0003-9861(82)90222-3. [DOI] [PubMed] [Google Scholar]
- 15.Portevin D, de Sousa-D’Auria C, Montrozier H, Houssin C, Stella A, Laneelle MA, Bardou F, Guilhot C, Daffe M. J Biol Chem. 2005;280:8862–8874. doi: 10.1074/jbc.M408578200. [DOI] [PubMed] [Google Scholar]
- 16.Gande R, Gibson KJ, Brown AK, Krumbach K, Dover LG, Sahm H, Shioyama S, Oikawa T, Besra GS, Eggeling L. J Biol Chem. 2004;279:44847–44857. doi: 10.1074/jbc.M408648200. [DOI] [PubMed] [Google Scholar]
- 17.Sassetti CM, Boyd DH, Rubin EJ. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Tweel B, Tong L. Proc Natl Acad Sci. 2004;101:5910–5915. doi: 10.1073/pnas.0400891101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole ST, Eiglmeir K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honore N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward JR, Barrell BG. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]


