Abstract
SQUAMOSA PROMOTER BINDING PROTEIN–box genes (SBP-box genes) encode plant-specific proteins that share a highly conserved DNA binding domain, the SBP domain. Although likely to represent transcription factors, little is known about their role in development. In Arabidopsis, SBP-box genes constitute a structurally heterogeneous family of 16 members known as SPL genes. For one of these genes, SPL8, we isolated three independent transposon-tagged mutants, all of which exhibited a strong reduction in fertility. Microscopic analysis revealed that this reduced fertility is attributable primarily to abnormally developed microsporangia, which exhibit premeiotic abortion of the sporocytes. In addition to its role in microsporogenesis, the SPL8 knockout also seems to affect megasporogenesis, trichome formation on sepals, and stamen filament elongation. The SPL8 mutants described help to uncover the roles of SBP-box genes in plant development.
INTRODUCTION
Transcription factors generally are believed to play key roles in the control of tissue-specific gene expression. Because this is a process central to the differentiation of multicellular organisms, determining the function of transcription factors can provide important insights into eukaryotic development. Analysis of the Arabidopsis genome reveals 29 classes of transcription factors, 16 of which appear to be unique to plants (Arabidopsis Genome Initiative, 2000). One of these is characterized by the presence of a DNA binding domain referred to as the SQUAMOSA PROMOTER BINDING PROTEIN (SBP) domain and encoded by the SBP-box, a feature characteristic of the Arabidop-sis SQUAMOSA PROMOTER BINDING PROTEIN–LIKE (SPL) gene family (Cardon et al., 1997, 1999). The 16 SPL genes found in the Arabidopsis genome form quite a heterogeneous family, but subfamilies can be recognized based on their genomic structures and the sequences of the deduced proteins (Cardon et al., 1999; Arabidopsis Genome Initiative, 2000; http://www.uni-frankfurt.de/fb15/botanik/mcb/AFGN/Huijser.htm).
The first SBP-domain proteins, isolated from snapdragon (Klein et al., 1996), showed in vitro binding to a sequence motif in the promoter region of the floral meristem identity gene SQUAMOSA (Huijser et al., 1992). A similar motif identified in the promoter region of APETALA1 (AP1), the presumed Arabidopsis ortholog of SQUAMOSA, was recognized in vitro by the SPL3 protein (Cardon et al., 1997). However, the precise role of SPL3 in normal development remains unsolved, although constitutive overexpression of SPL3 in transgenic Arabidopsis plants caused early flowering (Cardon et al., 1997), as did AP1 (Mandel and Yanofsky, 1995).
Despite the efforts of many groups to isolate and characterize an ever-increasing number of Arabidopsis mutants, none of the corresponding genes isolated and published to date has been found to represent an SBP-box gene. The only described SBP-box gene with a known mutant phenotype remains the LIGULELESS1 (LG1) gene of maize, isolated by Moreno and co-workers (1997). The lg1 mutation results in leaves that lack auricles and ligules (Becraft et al., 1990).
To obtain a better insight into the role of SBP domain transcription factors in plant development, we exploited available transposon-mutagenized populations of Arabidopsis to search for insertions in SBP-box genes. We identified and isolated three such transposon insertion alleles representing the SPL8 gene. The corresponding mutant plants all exhibited a strong reduction in fertility, primarily as a consequence of abnormal cell differentiation within the developing anthers. Only a few putative transcription factors that control this precisely ordered differentiation process (Scott et al., 1991; Goldberg et al., 1993; Sanders et al., 1999) have been isolated. Among these are the Arabidopsis proteins NOZZLE/SPOROCYTELESS (NZZ/SPL), whose single genetic locus is only distantly related to known transcription factor families (Schiefthaler et al., 1999; Yang et al., 1999), MALE STERILITY1 (MS1), a PHD-finger class member (Wilson et al., 2001), and ABORTED MICROSPORES (AMS), a MYC class basic helix-loop-helix domain protein (Sorensen et al., 2003). Whereas both MS1 and AMS are required primarily for postmeiotic pollen development, the early-acting NZZ/SPL gene is specific to the archesporial cells of the anther, which themselves give rise through successive divisions to both the sporogenic tissue and its surrounding cell layers. Interestingly, NZZ/SPL also affects early sporogenesis within developing ovules.
Here, we report the characterization of three loss-of-function mutant alleles representing the SBP-box gene SPL8, which, like NZZ/SPL, affects the early stages of microsporogenesis and megasporogenesis.
RESULTS
Identification and Isolation of Transposon-Mutagenized SPL8 Alleles
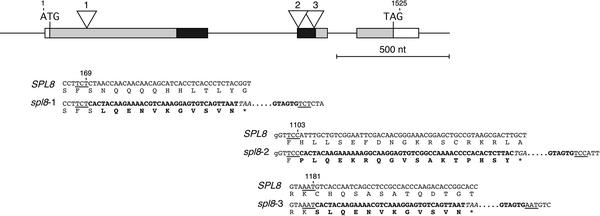
Through systematic screening using a PCR-based reverse genetics approach (Baumann et al., 1998), we searched for mutant alleles of SPL genes in an Arabidopsis Columbia-0 (Col-0) population mutagenized with the heterologous and autonomous transposable element En (Spm) from maize (ZIGIA population) (Wisman et al., 1998; Steiner-Lange et al., 2001). However, in the case of SPL8, we fortuitously identified a first insertion allele within this population through a forward genetics screen for photosynthetic mutants (Varotto et al., 2000; Pesaresi et al., 2001). The plant line 5ATA20 exhibited a marked reduction in the effective quantum yield of photosystem II (P. Pesaresi and D. Leister, unpublished data). Furthermore, this line was found to harbor multiple copies of the transposon, and sequencing of their flanking genomic DNA revealed one of these to be inserted at the SPL8 locus, 1101 bp downstream of the presumed translation start codon and residing in the SBP domain coding sequence of the second exon (Figure 1). However, subsequent segregation analysis revealed that this insertion allele of SPL8 (referred to below as spl8-2; note that the alleles described in this article are numbered according to the order of their positions within the locus) could not be responsible for the observed reduced photosynthetic performance. Additional PCR-based screening of the ZIGIA population allowed the isolation of a second SPL8 mutant allele (referred to as spl8-3). In the corresponding line 7AT39, the insertion also was found in the SBP-box, but this time in an opposite orientation and 78 bp downstream of the site of insertion found in line 5ATA20 (Figure 1).
Figure 1.
Genomic Organization of the SPL8 Locus and Its Mutant Alleles.
Partial nucleotide sequences and the encoded amino acid residues around the En transposon insertion sites of the three mutant alleles compared with the wild-type allele are shown below a scheme of the intron-exon structure of the SPL8 locus. The exons are depicted as boxes. The SBP-box is shown in black, and other coding parts are shaded. The transposon insertions found separately in the three different mutant alleles are marked by open triangles, with the corresponding allele indicated above. Numbers above the sequences indicate relative positions within the genomic sequence starting from the first nucleotide of the translational start codon. Nucleotides corresponding to the target site duplications are underlined. Boldface characters in the DNA sequence highlight the termini of the transposon, and below them, the corresponding encoded amino acid residues that differ from the wild-type allele also are shown in boldface. Asterisks mark in-frame translational stop codons (italic) within the En sequence. nt, nucleotides.
Finally, a third SPL8 insertion allele (spl8-1) was identified after screening the SINS database of transposon flanking sequences derived from the SLAT collection generated at the Sainsbury Laboratory (Norwich, UK) by Jones and co-workers (http://www.jic.bbsrc.ac.uk/sainsbury-lab/jonathan-jones/SINS-database/sins.htm). The SLAT collection represents pools of seeds obtained from a population of Arabidopsis plants mutagenized with a nonautonomous derivative, dSpm, of the maize transposable Spm/En element (Tissier et al., 1999). From the corresponding pool of seeds, we were able to identify two plants that both contained a transposon 167 bp downstream of the presumed start codon in the first exon but upstream of the SBP-box of SPL8 (Figure 1). DNA gel blot analysis, using right and left end border sequences of En, confirmed that one of these plants contained a unique transposon insertion in the SPL8 gene, whereas the other was found to harbor a second insertion in its genome. Selfing these plants resulted in progeny from which an spl8-1 homozygous mutant plant was selected to be the progenitor of the spl8-1 mutant lines 30213 and 30214.
The homozygous spl8 mutant lines obtained from the ZIGIA population and the SLAT collection all showed the same deviations, in particular reduced fertility, as described below, from the wild-type phenotype. Segregation analysis of heterozygous plants showed a 3:1 wild type:mutant ratio, reflecting the recessive nature of the spl8 mutant alleles. Furthermore, F1 plants obtained from allelic tests among the different spl8 mutant lines again displayed these deviations, proving that the spl8 mutations are responsible for the observed phenotype. In all three mutant alleles, the En/dSpm insertions were expected to cause a premature translational stop of the predicted SPL8 gene product, as depicted in Figure 1. In fact, reverse transcriptase–mediated PCR performed on RNA extracted from inflorescences did not allow the detection of SPL8-derived transcripts in homozygous spl8-1 mutants. However, very low levels of transcripts of wild-type length were detected in both spl8-2 and spl8-3 plants (data not shown). This finding could be explained by assuming a low frequency of excision of the autonomous En element residing at the SPL8 locus in these two lines.
In the absence of an activator to encode a transposase, the dSpm element in line 30213 (i.e., representing the spl8-1 allele) is expected to remain stably integrated (Tissier et al., 1999), whereas the autonomous En elements present in ZIGIA lines may transpose during development and thereby cause additional mutations. Furthermore, line 30213 carries a transposon insertion only at the SPL8 locus, in contrast to the ZIGIA lines, which carry multiple En elements in their genomes.
To avoid any influence of secondary site mutations on the analysis of the spl8 mutant phenotype, the spl8-1 mutant line was selected for further detailed histological analysis, as described below. However, we first characterized all three lines to confirm that they display the same major phenotypic aberrations.
Characterization of the spl8 Mutant Morphology
During the vegetative phase of growth, spl8 mutants were indistinguishable from the corresponding wild-type plants, and the time to flowering remained unaffected (Figure 2A). However, after anthesis, when petals and sepals start withering and wild-type siliques elongate strongly and develop embryo-containing seeds, the carpels of early spl8 mutant flowers did not elongate and remained without seeds. Only at later stages of (primary) inflorescence development did spl8 mutants form flowers with elongating siliques.
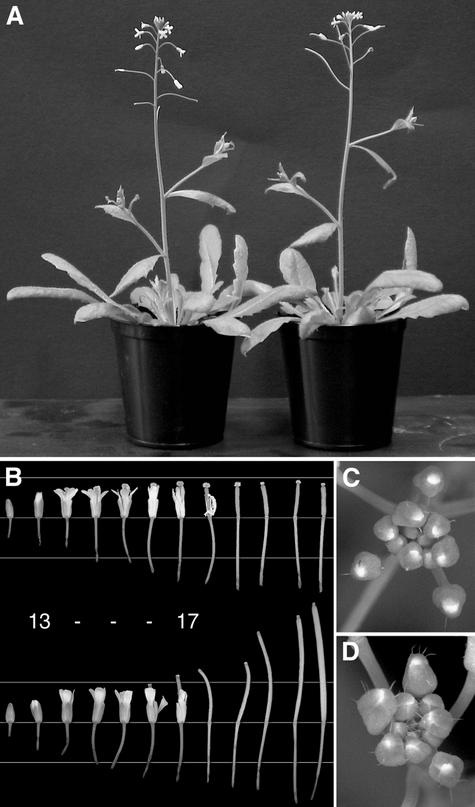
Figure 2.
Phenotype of the spl8 Mutant.
(A) A homozygous spl8-1 mutant plant (left) compared with the wild type. Both plants are the same age and were grown under the same greenhouse conditions.
(B) Flowers from a single spl8-1 mutant (top row) and a wild-type inflorescence (bottom row) at similar stages of development. The flowers are arranged in basipetal order with the first formed and oldest flower at right. Note the somewhat slender appearance of the spl8 mutant flowers before anthesis and of the petals after anthesis. In particular, note the long turgescent remaining stigma of the spl8-1 mutant and the failure of its silique to fully stretch after anthesis. Elongation of the pedicels remains unaffected. The horizontal lines behind the flowers are spaced 0.5 cm apart. Numbers between the rows of flowers indicate approximate developmental stages according to Smyth et al. (1990) and Bowman et al. (1991): stage 13, anthesis; stage 17, floral organ abscission.
(C) Close-up of an spl8-1 mutant inflorescence tip showing young floral buds with a reduced number of trichomes on their sepals compared with the wild type in (D).
(D) Close-up of a wild-type inflorescence tip.
The 12 earliest flowers and siliques from the main inflorescences of an spl8 mutant plant and a wild-type plant, both at the same developmental stage and comparable to the plants shown in Figure 2A, are lined up in Figure 2B. Up to stage 13 of flower development (Smyth et al., 1990), spl8 flowers were almost indistinguishable from wild-type flowers, except that they appeared somewhat more slender. However, whereas wild-type siliques after stage 16 clearly elongated, the gynoecia of early spl8 mutant flowers did not change in length.
A closer look with the scanning electron microscope revealed that in developing flowers, the spl8 anthers and their filaments remained smaller than those of the wild type (Figures 3A and 3B). As a result of their reduced size, the spl8 mutant stamens, four long and two short as in the wild type, did not overgrow the pistil, as is typical for wild-type flower development (i.e., at stage 14). At stage 13 to 14 of flower development, spl8 mutant anthers underwent dehiscence, but they always released fewer pollen grains than wild-type anthers (Figures 3C and 3D).
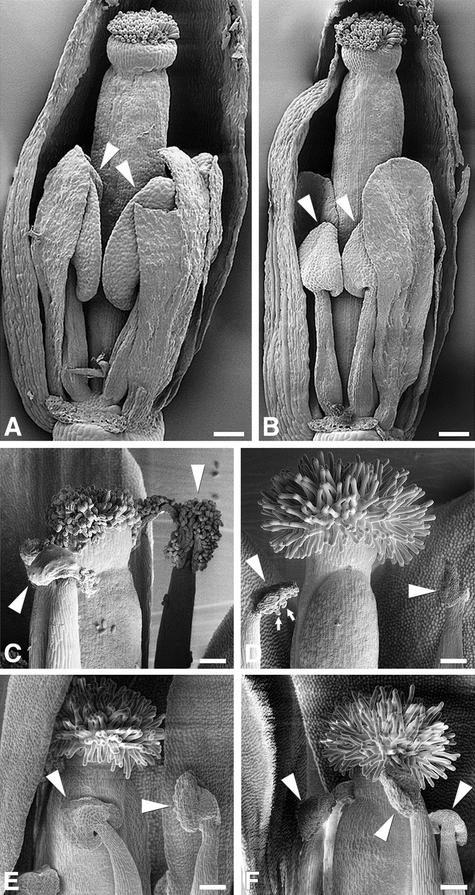
Figure 3.
Scanning Electron Microscopy Analysis of spl8 Mutant Flower Development.
(A) Wild-type flower at a developmental stage matching stage 11 to 12 (i.e., stigmatic papillae become visible, and petals and stamen have approximately the same height).
(B) spl8-1 mutant flower at a developmental stage comparable to that of the wild-type flower shown in (A). The flower is more slender, and anthers (arrowheads) remain smaller compared with the wild type.
(C) Wild-type anthers and stigma at anthesis. Ample pollen is released by the anthers (arrowheads) and adheres to the stigma surface.
(D) spl8-1 mutant anthers and stigma at anthesis. The anthers show dehiscence but release much less pollen (small arrows) compared with wild-type anthers. Note also that as a consequence of failing pollination, the papillae of the spl8-1 mutant stigma remain turgescent.
(E) and (F) A phenotype similar to that of spl8-1 (D) is displayed by spl8-2 (E) and spl8-3 (F) mutants at anthesis.
Sepals and petals have been removed in part to unveil the inner floral organ whorls. Bars = 100 μm.
To quantify the degree of semisterility in spl8 mutants, we determined seed set and pollen production compared with those of wild-type plants (Table 1). The first five to seven flowers in the primary inflorescence of spl8 mutants grown under long-day conditions generally did not set seed (in secondary inflorescences, the number of flowers remaining seedless was even higher). In the wild type, occasionally only the first formed flower remained seedless. Flowers formed at later stages of inflorescence development in spl8 did set seed but fewer of them compared with the wild type (Table 2). It should be noted that in the wild type during the final stage of inflorescence development (i.e., before the complete cessation of flower formation), seed production seemed to decrease again, probably as a result of a diminishing supply of nutrients.
Table 1.
First Seed Set, Pollen Production, and Trichome Formation of Wild-Type and spl8 Mutant Flowers
| Col-0
|
spl8-1
|
spl8-2
|
spl8-3
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | Mean | sd | n | Mean | sd | n | Mean | sd | n | Mean | sd | n |
| First flower to set seed | 1.1 | 0.2 | 36 | 5.9a | 1.4 | 43 | 5.3a | 1.1 | 12 | 7.2a | 1.3 | 6 |
| Pollen per anther | ||||||||||||
| 1st flower | 340.9 | 72.2 | 27 | 8.3a | 14.7 | 31 | 4.9a | 7.5 | 39 | 14.4a | 25.6 | 37 |
| 10th flower | 511.8b | 47.1 | 30 | 98.6a,b | 24.3 | 30 | 117.6a,b | 40.4 | 29 | 90.3a,b | 45.2 | 35 |
| Sepal trichomesc | ||||||||||||
| 1st flower | 22.3 | 4.5 | 10 | 3.3a | 1.3 | 10 | 4.5a | 1.4 | 6 | 4.6a | 2.6 | 6 |
| 10th flower | 4.7b | 1.3 | 10 | 0.4a,b | 0.5 | 10 | 0.7a,b | 0.8 | 6 | 0.6a,b | 0.8 | 6 |
Flowers formed in primary inflorescences of plants grown under long-day conditions. Flowers were numbered according to their appearance.
Significantly different from the wild type (t test; P < 0.001).
Significantly different from the first flower in the same inflorescence (t test; P < 0.001).
Total number of trichomes on the abaxial side of all first-whorl organs.
Table 2.
Seed Production of Wild-Type and spl8 Mutant Flowers
| Col-0
|
spl8-1
|
spl8-2
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Flower | Mean | sd | n | Mean | sd | n | Mean | sd | n |
| 1st flower | 28.2 | 21.0 | 12 | 0.0a | 0.0 | 12 | 0.0a | 0.0 | 12 |
| 10th flower | 52.6b | 3.8 | 12 | 13.1a,b | 10.4 | 12 | 17.0a,b | 10.4 | 12 |
| 10th flowerc | n.d. | 27.0a | 20.3 | 12 | 21.8a | 17.8 | 12 | ||
| 10th flowerd | n.d. | 19.9a | 16.8 | 12 | 30.7a | 14.9 | 12 | ||
Flowers formed in primary inflorescences of plants grown under long-day conditions. Flowers were numbered according to their appearance. n.d., not determined.
Significantly different from the nontreated wild-type flower at a similar position within the inflorescence (t test; P < 0.001).
Significantly different from the first flower in the same inflorescence (t test; P < 0.001).
After manual self-pollination without emasculation.
After manual cross-pollination with wild-type pollen without emasculation.
Seed set of spl8 flowers at defined positions in the primary inflorescence correlated with pollen production, with early flowers producing less pollen than later flowers (Table 1). This correlation also held true for the wild type, but significantly more pollen was produced compared with that in spl8 mutants.
As a consequence of the strong reduction in pollen production, possibly together with the reduced length of the filaments, the frequency of self-fertilization of spl8 mutant flowers was reduced dramatically or did not occur at all, as was generally the case for the first flowers formed (Table 1).
Manual pollination of homozygous spl8 mutant plants with their own pollen improved seed set. However, even when manually pollinated without emasculation, so as not to damage the flowers, seed set in the spl8 mutants remained low compared with that in the wild type (Table 2). That this is not simply the result of reduced numbers and/or viability of spl8 mutant pollen is suggested by the fact that cross-pollination with wild-type pollen also did not increase seed set to wild-type levels (Table 2). Furthermore, we determined the average number of seeds produced by emasculated wild-type flowers after cross-pollination with either mutant or wild-type pollen. This resulted in 46.5 (sd = 13.1, n = 6) seeds after pollination by the wild type and 31.3 (sd = 19.8, n = 6) and 43.5 (sd = 14.2, n = 6) seeds when pollinated by spl8-1 and spl8-2 mutants, respectively. These values do not differ significantly (t test; P > 0.1) from each other and strongly suggest that additional factors affect the overall fertility of spl8 mutant flowers (see Histological Analysis of spl8 Mutant Ovule Development below).
Histological Analysis of spl8 Mutant Anther Development
To elucidate the nature of the observed male sterility, transverse sections were prepared from anthers of early-formed and largely sterile spl8-1 mutant flowers (grown under long-day conditions). In the observations below, we refer to the staging of wild-type anther development as described by Sanders and co-workers (1999).
Soon after stamen initiation, at stage 2 of wild-type anther development, archesporial cells arise from four small groups of hypodermal (L2 layer) cells at the corners of the anther primordium. Further mitotic divisions of these cells result in an anther with two theca, each of which bears a pair of pollen sacs or microsporangia. By the time the wild-type anther has reached stage 5 of development, each microsporangium has differentiated into a group of sporocytes surrounded by a tapetum, a middle cell layer, and an endothecium. At stage 6, wild-type sporocytes enter meiosis and form tetrads of microspores surrounded by a callose wall (stage 7), and the anther increases notably in size.
Compared with that in the wild type, the initiation of microsporogenesis already was disturbed in the spl8-1 mutant. Archesporial cells failed to be formed at all four positions of the anther. As a consequence, spl8-1 mutant anthers sometimes developed with fewer than four differentiated pollen sacs (Figure 4A). Also, the subsequent histogenesis of the microsporangia that are formed in spl8 mutant anthers proceeded abnormally. The division of the outer secondary parietal cell layer remained incomplete, although sporadic, presumptive middle layer and endothecium cells were visible (Figure 4B).
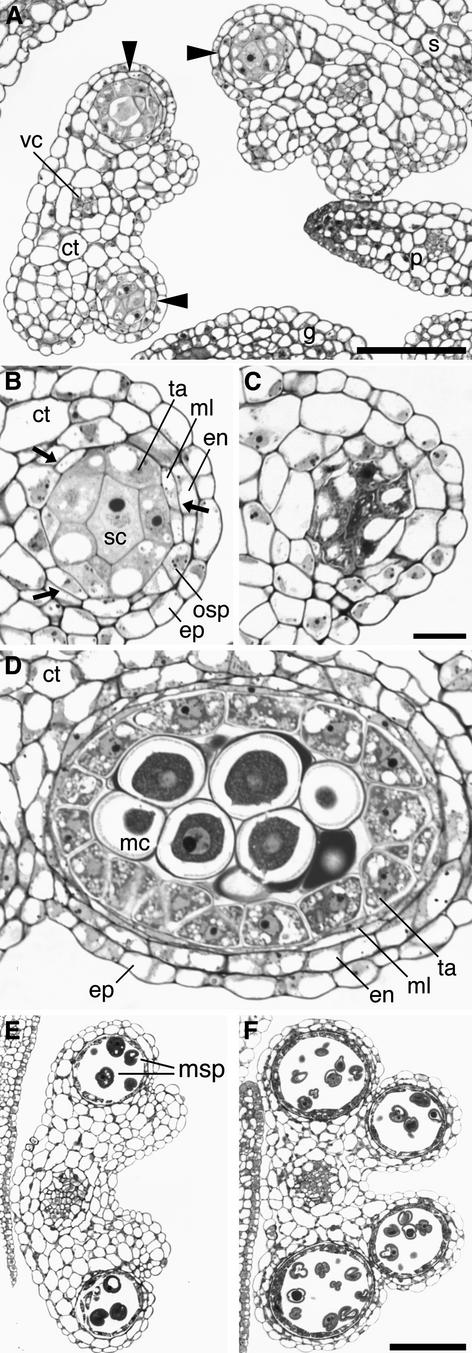
Figure 4.
Light Microscopic Analysis of Developing spl8 Mutant Anthers.
(A) Cross-section through spl8-1 mutant anthers comparable to stage 4 to 5 of wild-type anther development. spl8-1 mutant anthers often develop fewer than four pollen sacs (arrowheads).
(B) An spl8-1 mutant pollen sac at a higher magnification to illustrate the often-observed disturbed tissue differentiation. In particular, the outer secondary parietal cells may not undergo a further mitotic division and remain undifferentiated. Intermittently, in the developing outer secondary parietal layer, formation of presumptive endothecium and middle layer cells caused by periclinal divisions (arrows) can be seen. Furthermore, the cells of the inner secondary parietal layer or tapetum are less well differentiated, reduced in number, and have an appearance closer to that of the sporogenous cells. Sporogenous cell number also is reduced compared with that in the wild type.
(C) The failure of spl8-1 sporogenous cells to undergo meiosis, as observed after stage 5 of wild-type anther development, and the subsequent decay of the pollen sac is revealed by the dense staining of their cytoplasm and the vacuolated tapetal cells.
(D) Somewhat oblique cross-section through a wild-type pollen sac at stage 5 to 6 of anther development just before meiosis. All cell layers are well differentiated, and dark-staining callose surrounds the meiocytes.
(E) Cross-section through an spl8-1 mutant anther comparable to stage 9 to 10 of wild-type anther development (cf. [F]). This anther bears two pollen sacs with some free microspores.
(F) Cross-section through a wild-type anther at stage 9 to 10. All pollen sacs are well developed and contain free vacuolated microspores with exine walls.
ct, connective tissue; en, endothecium; ep, epidermis; g, gynoecium; mc, meiocyte; ml, middle layer; msp, microspore; osp, outer secondary parietal cell layer; p, petal; s, sepal; sc, sporogenous cells; ta, tapetum; vc, vascular cells. (B) to (D) are at the same scale, as are (E) and (F). Bars = 25 μm in (A), 10 μm in (C), and 50 μm in (F).
The failure of sporogenous cells to undergo meiosis, observed after stage 5, also may contribute to the strong reduction of spl8 male fertility. However, without understanding the mechanism of SPL8 action on anther differentiation and development, it is difficult to determine whether the disruption in the microsporangium wall layers causes sporogenous cell abortion or sporogenous cell abortion leads to disruption in the microsporangium wall layers. Whereas in the subsequent stages (i.e., 7 and upward) the wild-type microspores were released from the tetrads and continued differentiation, the spl8 sporogenic tissue degenerated, as deduced from the large vacuoles that appeared in the tapetal cells and the dense staining of the sporogenous cells (Figure 4C). As a result, the “mature” spl8 mutant anthers appeared shorter and narrower than the wild-type anthers. However, occasionally, callose deposition was seen in some developing pollen sacs of the spl8 mutant after stage 5 of anther development (cf. with the wild type in Figure 4D). This finding indicates that some sporocytes initiate meiosis and continue their development (Figure 4E), which agrees with the observation that spl8 mutant anthers may release some pollen.
Histological Analysis of spl8 Mutant Ovule Development
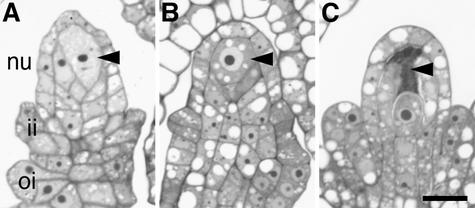
To determine if the spl8 mutation also can affect the entrance into meiosis during megasporogenesis, we examined some semithin sections from developing ovules. In Arabidopsis, megasporogenesis proceeds without the formation of parietal and sporogenous cells. Instead, one hypodermal archesporial cell per developing ovule gives rise directly to the megaspore mother cell (Schneitz et al., 1995; Bajon et al., 1999). Stage 2 of ovule development (according to Schneitz et al., 1995), which encompasses megasporogenesis, starts with the enlargement of the megaspore mother cell before the entrance into meiosis (Figure 5A), which is completed by the formation of a linear tetrad of four haploid megaspores, only one of which survives. The ovules also initiate the differentiation of the inner and outer integuments during this stage. Until the enlargement of the megaspore mother cell, ovule development in the spl8-1 mutant seemed to follow wild-type development (Figure 5B). However, occasionally, developing ovules were observed in which the megaspore mother cell did not enter meiosis; instead, a subsequent dense cytoplasmic staining suggested degeneration of the megaspore mother cell (Figure 5C).
Figure 5.
Light Microscopic Analysis of Developing spl8 Mutant Ovules.
(A) Longitudinal section through a developing wild-type ovule. An enlarged megaspore mother cell (arrowhead) and the initiation of the integuments can be recognized, indicating a stage just before the entrance into meiosis.
(B) Longitudinal section through a developing spl8-1 mutant ovule. Based on the degree of megaspore mother cell (arrowhead) enlargement and integument formation, this ovule is slightly earlier in development compared with the wild-type ovule shown in (A).
(C) Longitudinal section through a developing spl8-1 mutant ovule at a somewhat later stage of development compared with the wild-type ovule shown in (A). The integuments are more advanced in their development, but the megasporocyte seems to degenerate, as deduced from the dense staining of the cytoplasm and the loss of turgor.
ii, inner integument; nu, nucellus; oi, outer integument. (A) to (C) are at the same scale. Bar = 10 μm in (C).
As noted above, even after manual pollination, spl8 mutant flowers produced fewer seeds than wild-type flowers. A degeneracy of approximately half of the spl8 ovules may explain this discrepancy in seed set, assuming that degeneration of ovules in the wild type must be rare, because we did not observe this phenomenon in sections of wild-type flowers. However, the frequency with which abnormal ovule development was observed in spl8-1 mutant flowers seemed to vary, as deduced from the few spl8-1 mutant flower buds that were analyzed by sequential sectioning of the entire pistil. In the most extreme case, all of the ovules within the spl8-1 bud displayed a degenerate megaspore mother cell. In other samples, fewer than one-third of the ovules were affected. Like the production of pollen, this probably correlates with the position of the flower within the inflorescence. This aspect has not been investigated further.
Temporal and Spatial Expression Patterns of SPL8
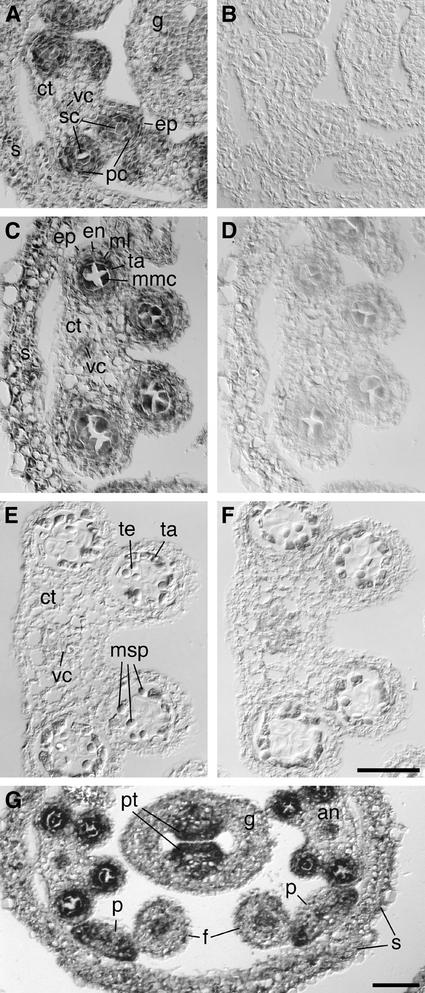
Previous RNA gel blot analysis performed on poly(A)+ RNA isolated from all aerial tissues of long-day-grown plants (Cardon et al., 1999) revealed low levels of SPL8 transcripts during vegetative growth. High levels were found in inflorescences formed after the floral transition. To determine the spatial expression pattern of SPL8 during this phase of reproductive growth, in situ hybridization analysis was performed on cross-sections of young flower buds (Figure 6). To exclude cross-hybridization to other SPL genes, only the transcribed sequence upstream of the SPB-box region of SPL8 was used to generate the antisense RNA probe. Comparison of the hybridization signals with those obtained with an SPL8 sense probe suggested that most if not all early floral tissues expressed SPL8. A renewed inspection of the floral buds of the three spl8 mutant lines revealed that the low expression of SPL8 in other floral tissues might be correlated with the presence of significantly fewer trichomes on the sepals compared with the wild type (Figures 2C and 2D). However, the developing pollen sacs, up to stage 6 of anther development, contained increased levels of the SPL8 transcripts (Figures 6A to 6D). At later stages of anther development, this signal disappeared (Figures 6E and 6F). This finding correlates with the deviations observed in the development of spl8 mutant anthers. Likewise, the stronger signal in the placental region of the carpels (Figure 6G) may correlate with the occasionally observed abnormal ovules visualized in spl8 mutant flowers.
Figure 6.
In Situ Localization of SPL8 Transcripts.
Cross-sections through wild-type anthers at different stages of development probed with digoxigenin-labeled SPL8 antisense or sense RNA.
(A) and (C) At premeiotic stages 4 to 5 (A) and 5 to 6 (C), SPL8 transcripts are detectable primarily in the parietal and sporogenous cell layers that form the four pollen sacs of the anther.
(E) At postmeiotic stage 7 to 8, SPL8 transcripts are no longer detectable in the anther.
(B), (D), and (F) Control hybridizations on consecutive sections with an SPL8 sense probe showing low or absent background signals.
(G) Overview of SPL8 expression in a cross-section of a young wild-type floral bud. In addition to the expression in the developing pollen sacs of the anthers, increased SPL8 transcript levels are detectable in the placental tissue of the developing gynoecium and at the margins of the petals.
an, anther; ct, connective tissue; en, endothecium; ep, epidermis; f, filament; g, gynoecium; ml, middle layer; mmc, microspore mother cell; msp, microspore; p, petal; pc, parietal cells; pt, placental tissue; s, sepal; sc, sporogenous cells; ta, tapetum; te, tetrad; vc, vascular cells. (A) to (F) are at the same scale. Bars = 50 μm.
DISCUSSION
We were able to identify and isolate three mutant spl8 alleles from two different transposon-tagged Arabidopsis populations. The phenotypes of the corresponding homozygous mutant plants all exhibited a strong reduction in male fertility. Together with the outcome of the heteroallelic tests and the segregation analyses after backcrossing to the wild type, these observations allowed the conclusion that the phenotype described is attributable to the mutation of the SPL8 gene and of SPL8 alone. This finding enabled us to determine a developmental role for a member of the Arabidopsis SBP-box gene family.
SPL8 Has a Major Effect on Sporogenesis
Our phenotypic analysis clearly showed that a major effect of SPL8 was on microsporogenesis and megasporogenesis within the anthers and ovules, respectively, where both spore types may fail to enter meiosis. In particular, SPL8 gene function is required (1) for the proper initiation of microsporangium formation at defined positions within the anther and (2) for the regular entrance of spore mother cells into meiosis. The role of SPL8 in the entrance into meiosis seems not to be specific to microsporogenesis. Although seemingly not as frequent, megasporocytes also encountered difficulties undergoing meiosis in the absence of SPL8. In the absence of a functional SPL8 gene, the sporocytes did not form free meiocytes and eventually degenerated. In addition to meiocyte differentiation, SPL8 also affected the formation of the parietal layers of the microsporangium. In the absence of SPL8 gene function, the outer secondary parietal cell layers did not properly differentiate to surround the meiocytes. The spl8 mutant shows evidence that the endothecium and the middle layer have a common origin and in this respect supports the suggested cell lineage in the anther wall differentiation of dicots (Owen and Makaroff, 1995; Yang et al., 1999).
To what extent the developmental fate of sporogenous cells and parietal layers are interdependent remains unclear, but SPL8 has been found to be transcriptionally active in both tissues. Furthermore, the observation that tapetal ablation (Mariani et al., 1990) leads to premeiotic sporocyte abortion, along with the observations in other sterility mutants such as nzz/spl (Schiefthaler et al., 1999; Yang et al., 1999) and gne2 (Sorensen et al., 2002), suggest that correct differentiation of both the sporogenous cells and the surrounding parietal cells is essential for the entry of the developing meiocytes into meiosis. Also, in the most severely affected anthers of spl8 in which early meiocyte abortion took place, no callose deposition was observed. This finding reiterates the importance of callose deposition for the entry into meiosis (Worrall et al., 1992). In addition, the tapetum cell layer has been proposed to provide nutrients to the developing microspores and is transcriptionally highly active, although callose deposition and the entrance into meiosis in gne2 is seen in the absence of an adjacent tapetal layer (Sorensen et al., 2002).
Another highly interesting phenotypic aspect of the spl8 mutant is the failure of initiation of all four microsporangia within the developing anther. Transverse sections through the early flower buds of spl8 mutants showed a complete absence of differentiated microsporangia in the greatly reduced anthers, whereas other transverse sections detected one or two partially developed microsporangia. This observation suggests that SPL8 somehow is involved in controlling the sensitivity of a yet unknown signal for pattern formation within the anther. In this context, it is interesting that SPL8 expression itself was found to be inducible by the phytohormone gibberellin (M. Schmid and D. Weigel, personal communication). Mutants known to be affected in either the biosynthesis or the perception of gibberellin often display severe fertility problems (Koornneef et al., 1985; Jacobsen and Olszewski, 1993; Fei and Sawhney, 1999; Goto and Pharis, 1999). Furthermore, trichome formation, which also was reduced on the sepals of spl8 mutant flowers, is known to be controlled by gibberellin (Chien and Sussex, 1996; Perazza et al., 1998).
spl8 mutants displayed a variable degree of expression, probably as a result of endogenous and environmental factors (e.g., the position of the flower in the inflorescence and photoperiod). However, and despite the fact that a complete loss of function will result in dramatically reduced progeny and thus a strong reduction in reproductive fitness within an Arabidopsis population, SPL8 function is not essential to Arabidopsis ontogenesis. All spl8 mutants remained capable of producing functional pollen and of setting viable seed. An obvious explanation would be a functional redundancy derived from another SBP-box gene.
As mentioned above, SPL8 is one of 16 SBP-box genes found in the Arabidopsis genome. However, sequence alignment of their deduced protein products (data not shown) did not reveal a homolog (i.e., a possible SPL8 paralog). In fact, outside of its SBP domain, SPL8 does not display any obvious sequence similarity to other Arabidopsis SBP domain proteins. Thus, the possibility of another Arabidopsis SBP-box gene compensating for an SPL8 loss of function seems unlikely, but as long as the mode of action of SBP domain proteins remains unknown, this possibility cannot be excluded.
SPL8 Displays Only Limited Sequence Conservation Outside of the SBP Domain
The SBP domain encoded by the SBP-box has been shown to bind defined DNA sequences in vitro and to harbor a bipartite nuclear localization signal (Klein et al., 1996; R. Birkenbihl and P. Huijser, unpublished data). Furthermore, a translational fusion of a SBP domain protein and green fluorescent protein was found to be directed toward the nucleus (Moreno et al., 1997; U. Unte, R. Birkenbihl, and P. Huijser, unpublished data). Therefore, it is assumed that the SBP-box gene SPL8 acts as a transcription factor, controlling as yet unknown target genes whose functions are, at least in part, necessary for normal sporogenesis and microsporangium wall formation. In addition, their functions may be required for floral organ growth and trichome formation.
Besides the DNA binding SBP domain, with its nuclear localization signal, a Basic Local Alignment Search Tool (BLAST) search of sequences available in the electronic databases did not indicate other known conserved domains that could be of help in understanding the mechanism of SPL8 function. However, almost immediately upstream of the SBP domain, SPL8 shares a short stretch of amino acid residues, RIGLNLGRTYF, with a few SBP domain proteins known from other species, such as AMSBPH3 from snapdragon and ZMSBP1, ZMSBP3, and ZMSBP4 from maize. Interestingly, part of this domain, GLNLGRTYF, also is conserved in the maize LG1 protein, the only other SBP domain protein with a known loss-of-function phenotype. LG1 controls the formation of the ligule, a structure typical of the leaves of grasses. In a search for the function of a possible LG1 ortholog in dicots, Mooney and Freeling (1997) have suggested that the stipules at the base of Arabidopsis leaves could represent structures homologous with the ligules of maize leaves. We have not determined if stipules in spl8 mutants are affected. Although a phylogenetic analysis based on the SBP domain sequence suggests that LG1 is related more closely to SPL8 than to any of the other Arabidopsis SBP domain proteins (data not shown), the sequence similarity outside of the SBP domain is very limited. The conserved stretch of amino acid residues mentioned above indicates the presence of at least a few other maize SBP domain proteins with greater overall similarity to SPL8, and these might be related more closely in function to SPL8 than is LG1.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana plants from ecotype Columbia-0 (Col-0) were grown in a phytochamber (CMP 3244; Conviron, Winnipeg, Canada) in plastic trays filled with ready-to-use commercial, prefertilized soil mixture (type ED73; Werkverband). For stratification, seeds were kept on wet filter paper for 4 days at 4°C in the dark before transferring to soil. Growing conditions were 22°C, 50% RH, and ∼150 μE·m−2·s−1 light (fluorescent Sylvania F72T12 cool-white light [75%] and incandescent Sylvania 100-W lamps [25%]). Cultivation was under a 16-h-light/8-h-dark (long-day) photoperiod regime.
Genomic DNA and RNA Isolation
Leaves from long-day-grown Arabidopsis plants were collected, and the DNA was isolated using a cetyl-trimethyl-ammonium bromide–based extraction method (Sommer et al., 1990). Pooled genomic DNA was used subsequently for transposon insertion analysis with the help of PCR, as described by Baumann et al. (1998).
Total RNA for the detection of SPL8 transcripts was isolated from young inflorescences using the Qiagen RNeasy kit (Valencia, CA) according to the manufacturer's protocol Plant+Fungi. Before use for reverse transcriptase–mediated (RT) PCR, the RNA was treated with DNaseI (Roche, Mannheim, Germany) and purified subsequently over RNeasy mini spin columns (Qiagen).
RT-PCR was performed using the Qiagen OneStep RT-PCR kit, according to the manufacturer's instructions, in combination with the following primer oligonucleotides: 5′-TCCATCGTTCTCTCCGGAGATGAAAG-3′ (GC420; SPL8 forward primer) and 5′-GAATTTGAAGACGAAGACGCTGACGTG-3′ (GC370; SPL8 reverse primer). Three-step cycling conditions were as follows: 26 PCR cycles at 94°C for 40 s, 65°C for 30 s, and 72°C for 1 min using 100 ng of RNA.
Mutant Isolation and Molecular Genetic Analysis
The different male-sterile spl8 mutants were identified in a reverse genetics screen of En-1– and dSpm-mutagenized Arabidopsis Col-0 populations. The generation of the En-1–mutagenized population of plants has been described by Wisman et al. (1998), whereas the generation of the dSpm-containing population has been described by Tissier et al. (1999).
The SLAT line 30213, which carries the spl8-1 allele, and the ZIGIA line 5AT20, which carries the spl8-2 allele, were back-crossed one time to the Col-0 wild type. The described mutant phenotype recurred only in F2 plants, segregating in a Mendelian fashion (i.e., 3:1 wild type:mutant). All of the mutants, but none of the wild-type-looking F2 plants, were found to be homozygous for the respective spl8 mutation.
For the allelic test cross, homozygous spl8-1 and spl8-3 were crossed together. The resulting F1 generation displayed the semisterile phenotype of its parents. To discriminate both mutant alleles and to confirm the heteroallelic nature of the F1 plants, PCR analysis with the following oligonucleotide primer combinations was used: 5′-AGAAGCACGACGGCTGTAGAATAGGA-3′ (En205) together with 5′-AACAACGACCGCCGTCACATCACC-3′ (GC326) to identify the spl8-1 allele and En205 together with 5′-AAGACAGAAGTGGATTTCACGTCCAAC-3′ (GC637) or 5′-GAGCGTCGGTCCCCACACTTCTATAC-3′ (En8130) together with 5′-GAATTTGAAGACGAAGACGCTGACG-3′ (GC370) to identify spl8-3.
Oligonucleotides used for the PCR-based reverse genetics screen were 5′-TTGTTCTCCTACGACCAGACAGGACC-3′ (GC633) and 5′-TTG-TCGAATTCCGACAGCAAATGGAAC-3′ (GC634).
Oligonucleotide Synthesis, DNA Sequencing, and Sequence Analysis
DNA sequences were determined by the Max Planck Institute for Plant Breeding Research DNA core facility on Applied Biosystems (Weiterstadt, Germany) Abi Prism 377 and 3700 sequencers using BigDye terminator chemistry. Premixed reagents were from Applied Biosystems. Oligonucleotides were purchased from Invitrogen (The Netherlands). DNA and protein sequence analysis was performed using the GCG analysis tools (Wisconsin Package version 10 for digital UNIX; Genetics Computer Group, Madison, WI) and the MacVector program (Oxford Molecular Group).
Microscopy and in Situ Hybridization
A Leica binocular microscope (Wetzlar, Germany) equipped with a digital camera was used to photograph floral buds and siliques at different developmental stages.
For scanning electron microscopy, freshly dissected flowers were mounted on aluminum specimen stubs using Tissue-Tek OCT compound (Sakura Finetek, Tokyo, Japan) and immediately shock frozen in liquid nitrogen. The samples were transferred subsequently to a Zeiss DSM 940 electron microscope (Jena, Germany) equipped with a cryo-chamber (Oxford Instruments). After sublimation of possible ice on their surfaces, the samples were sputter coated with gold and examined at an accelerating voltage of 5 kV.
For light microscopic examination of anthers, young flower buds ranging in length from 0.4 to 1.4 mm were fixed, dehydrated, and embedded as described by Sorensen et al. (2002). Semithin sections of 0.8 μm cut on a Reichert ultramicrotome were stained with toluidine blue.
Tissue preparation and in situ hybridization with digoxigenin-labeled RNA were performed according to the protocol published by Huijser et al. (1992) as modified by Samach et al. (1997). To avoid cross-hybridization with other SPL genes, only SPL8 sequences 5′ of the SBP-box were used to generate both sense and antisense probes for in situ hybridization. The fragment was amplified by PCR from the cloned SPL8 cDNA using the oligonucleotide primer combination 5′-GACTCTGACCCGACC-CGGTCATCCTTC-3′ and 5′-CGAGTTCGCCATCCCTGACTCACC-3′. The PCR product obtained was cloned subsequently in both orientations in the T7 RNA polymerase promoter containing TOPOTA2-1 vector (Invitrogen) according to the instructions of the manufacturer. After linearization with HindIII, the plasmids were used as templates for T7 RNA polymerase in vitro transcription and digoxigenin labeling according to the description with the Roche Nucleic Acid Labeling and Detection Kit.
Both the in situ hybridization slides and the semithin sections were examined and photographed with a Zeiss Axiophot microscope equipped with differential interference contrast optics and a digital camera (Intas).
Remaining Techniques and Methods
Standard molecular biology techniques were performed according to Sambrook et al. (1989). Digital photographic images were cropped and assembled using Adobe Photoshop 4.0 (Adobe Systems, Mountain View, CA). Color and contrast corrections were performed on entire images only.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
GenBank accession number for AMSBPH3 is AJ011621. GenBank accession number for ZMSBP1 is AJ011614. GenBank accession number for ZMSBP3 is AJ011616. GenBank accession number for ZMSBP4 is AJ011617.
Acknowledgments
We are grateful to Susanne Höhmann and Sandra Kröber for excellent technical support and to all members of the laboratory for helpful discussions. We also thank Markus Schmid and Detlef Weigel for providing information concerning the gibberellin inducibility of SPL8 transcription. Part of this work was supported by the Deutsche Forschungsgemeinschaft. M.G. was funded by an Alexander von Humboldt fellowship.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010678.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bajon, C., Horlow, C., Motamayor, J.C., Sauvanet, A., and Robert, D. (1999). Megasporogenesis in Arabidopsis thaliana L.: An ultrastructural study. Sex. Plant Reprod. 12, 99–109. [Google Scholar]
- Baumann, E., Lewald, J., Saedler, H., Schulz, B., and Wisman, E. (1998). Successful PCR-based reverse genetic screen using an En-1-mutagenised Arabidopsis thaliana population generated via single-seed descent. Theor. Appl. Genet. 97, 729–734. [Google Scholar]
- Becraft, P.W., Bongard-Pierce, D.K., Sylvester, A.W., Poethig, R.S., and Freeling, M. (1990). The Liguleless-1 gene acts tissue specifically in maize leaf development. Dev. Biol. 141, 220–232. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Cardon, G.H., Höhmann, S., Klein, J., Nettesheim, K., Saedler, H., and Huijser, P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237, 91–104. [DOI] [PubMed] [Google Scholar]
- Cardon, G.H., Höhmann, S., Nettesheim, K., Saedler, H., and Huijser, P. (1997). Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J. 12, 367–377. [DOI] [PubMed] [Google Scholar]
- Chien, J.C., and Sussex, I.M. (1996). Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 111, 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, H., and Sawhney, V.K. (1999). Role of plant growth substances in MS33-controlled stamen filament growth in Arabidopsis. Physiol. Plant. 105, 165–170. [Google Scholar]
- Goldberg, R.B., Beals, T.P., and Sanders, P.M. (1993). Anther development: Basic principles and practical applications. Plant Cell 5, 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, N., and Pharis, R.P. (1999). Role of gibberellins in the development of floral organs of the gibberellin-deficient mutant, ga1-1, of Arabidopsis thaliana. Can. J. Bot. 77, 944–954. [Google Scholar]
- Huijser, P., Klein, J., Lönnig, W.E., Meijer, H., Saedler, H., and Sommer, H. (1992). Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene SQUAMOSA in Antirrhinum majus. EMBO J. 11, 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Olszewski, N.E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5, 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, J., Saedler, H., and Huijser, P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 250, 7–16. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Elgersma, A., Hanhart, C.J., Loenen, M.E.P.V., Rijn, L.V., and Zeevaart, J.A.D. (1985). A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol. Plant. 65, 33–39. [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1995). A gene triggering flower formation in Arabidopsis. Nature 377, 522–524. [DOI] [PubMed] [Google Scholar]
- Mariani, C., De Beuckeleer, M., Truettner, J., Leemans, J., and Goldberg, R.B. (1990). Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature 347, 737–741. [Google Scholar]
- Mooney, M., and Freeling, M. (1997). Using regulatory genes to investigate the evolution of leaf form. Maydica 42, 173–184. [Google Scholar]
- Moreno, M.A., Harper, L.C., Krueger, R.W., Dellaporta, S.L., and Freeling, M. (1997). Liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize organogenesis. Genes Dev. 11, 616–628. [DOI] [PubMed] [Google Scholar]
- Owen, H.A., and Makaroff, C.A. (1995). Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185, 7–21. [Google Scholar]
- Perazza, D., Vachon, G., and Herzog, M. (1998). Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiol. 117, 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi, P., Varotto, C., Richly, E., Kurth, J., Salamini, F., and Leister, D. (2001). Functional genomics of Arabidopsis photosynthesis. Plant Physiol. Biochem. 39, 285–294. [Google Scholar]
- Samach, A., Kohalmi, S.E., Motte, P., Datta, R., and Haughn, G.W. (1997). Divergence of function and regulation of class B floral organ identity genes. Plant Cell 9, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sanders, P.M., Bui, A.Q., Weterings, K., McIntire, K.N., Hsu, Y.-C., Lee, P.Y., Truong, M.T., Beals, T.P., and Goldberg, R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11, 297–322. [Google Scholar]
- Schiefthaler, U., Balasubramanian, S., Sieber, P., Chevalier, D., Wisman, E., and Schneitz, K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz, K., Hülskamp, M., and Pruitt, R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: A light microscopic study of cleared whole-mount tissue. Plant J. 7, 731–749. [Google Scholar]
- Scott, R., Hodge, R., Paul, W., and Draper, J. (1991). The molecular biology of anther differentiation. Plant Sci. 80, 167–191. [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, H., Beltran, J.-P., Huijser, P., Pape, H., Lönnig, W.-E., Saedler, H., and Schwarz-Sommer, Z.S. (1990). Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 9, 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, A., Guerineau, F., Canales-Holzeis, C., Dickinson, H.G., and Scott, R.J. (2002). A novel extinction screen in Arabidopsis thaliana identifies mutant plants defective in early microsporangial development. Plant J. 29, 581–594. [DOI] [PubMed] [Google Scholar]
- Sorensen, A.-M., Kröber, S., Unte, U.S., Huijser, P., Dekker, K., and Saedler, H. (2003). The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33, 413–423. [DOI] [PubMed] [Google Scholar]
- Steiner-Lange, S., Gremse, M., Kuckenberg, M., Nissing, E., Schächtele, D., Spenrath, N., Wolff, M., Saedler, H., and Dekker, K. (2001). Efficient identification of Arabidopsis knock-out mutants using DNA-arrays of transposon flanking sequences. Plant Biol. 3, 391–397. [Google Scholar]
- Tissier, A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G., and Jones, J.D.G. (1999). Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto, C., Pesaresi, P., Maiwald, D., Kurth, J., Salamini, F., and Leister, D. (2000). Identification of photosynthetic mutants of Arabidopsis by automatic screening for altered effective quantum yield of photosystem 2. Photosynthetica 38, 497–504. [Google Scholar]
- Wilson, Z.A., Morroll, S.M., Dawson, J., Swarup, R., and Tighe, P.J. (2001). The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28, 27–39. [DOI] [PubMed] [Google Scholar]
- Wisman, E., Cardon, H.G., Fransz, P., and Saedler, H. (1998). The behaviour of the autonomous transposable element En/Spm in Arabidopsis thaliana allows efficient mutagenesis. Plant Mol. Biol. 37, 989–999. [DOI] [PubMed] [Google Scholar]
- Worrall, D., Hird, D.L., Hodge, R., Paul, W., Draper, J., and Scott, R. (1992). Premature dissolution of the microsporocyte callose wall causes male sterility in transgenic tobacco. Plant Cell 4, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W.-C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13, 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]