Abstract
Plant microtubules are organized into specific cell cycle-dependent arrays that have been implicated in diverse cellular processes, including cell division and organized cell expansion. Mutations in four Arabidopsis genes collectively called the PILZ group result in lethal embryos that consist of one or a few grossly enlarged cells. The mutant embryos lack microtubules but not actin filaments. Whereas the cytokinesis-specific syntaxin KNOLLE is not localized properly, trafficking of the putative auxin efflux carrier PIN1 to the plasma membrane is normal. The four PILZ group genes were isolated by map-based cloning and are shown to encode orthologs of mammalian tubulin-folding cofactors (TFCs) C, D, and E, and associated small G-protein Arl2 that mediate the formation of α/β-tubulin heterodimers in vitro. The TFC C ortholog, PORCINO, was detected in cytosolic protein complexes and did not colocalize with microtubules. Another gene with a related, although weaker, embryo-lethal phenotype, KIESEL, was shown to encode a TFC A ortholog. Our genetic ablation of microtubules shows their requirement in cell division and vesicle trafficking during cytokinesis, whereas cell growth is mediated by microtubule-independent vesicle trafficking to the plasma membrane during interphase.
Keywords: Arabidopsis, microtubules, cell division, actin filaments, tubulin-folding
The microtubule cytoskeleton plays important roles in both nondividing and dividing cells of eukaryotes, assisting in vesicle trafficking and mediating proper segregation of daughter chromosomes to opposite poles during mitosis (for review, see Cole and Lippincott-Schwartz 1995; Straight and Field 2000). In fission yeast, microtubules have also been implicated in maintaining cell shape and polarity (Sawin and Nurse 1998). Higher-plant cells form their own specific microtubule arrays, such as cortical hoops of interphase microtubules that organize cell elongation (Whittington et al. 2001), and two arrays related to the plant-specific mode of cell division (for review, see Lloyd and Hussey 2001). The preprophase band forms transiently at the onset of mitosis and marks the cortical division site (for review, see Smith 2001). The phragmoplast forms at the center of the division plane in late anaphase and guides the delivery of Golgi-derived vesicles. Their fusion results in the formation and lateral expansion of the cell plate that matures into a cell wall and flanking plasma membranes (for review, see Staehelin and Hepler 1996). Both preprophase band and phragmoplast also contain actin filaments; however, their specific role in plant cell division is not understood at present (Smith 2001).
Microtubules are polymerized from α/β-tubulin heterodimers (for review, see Nogales 2000). Newly synthesized α- and β-tubulin polypeptides undergo a sequence of folding steps catalyzed by chaperones. Initially, the tubulins are associated with the hexameric prefoldin complex that passes them on to the cytosolic chaperonin complex (Geissler et al. 1998; Vainberg et al. 1998; Hansen et al. 1999; for review, see Leroux and Hartl 2000; Llorca et al. 2000). Whereas fully functional actin and γ-tubulin polypeptides are released from the cytosolic chaperonin complex, α-tubulin and β-tubulin polypeptides are further processed by tubulin-folding cofactors (TFCs; for review, see Lewis et al. 1997; Nogales 2000). TFCs were originally identified by their tubulin-folding activity in extracts from mammalian cells (Gao et al. 1994; Llosa et al. 1996; Melki et al. 1996; Tian et al. 1996, 1997). In vitro folding assays suggest that α-tubulin binds to TFCs B and E, whereas β-tubulin binds to TFCs A and D (Lewis et al. 1997). α-tubulin/TFC E and β-tubulin/TFC D are bound by TFC C, forming a supercomplex from which α/β-tubulin heterodimers are released by GTP hydrolysis of β-tubulin. The small G-protein Arl2 appears to play a regulatory role, binding to and sequestering cofactor D (Bhamidipati et al. 2000). Homologs of most TFCs have been identified by genetic screens in yeast. Budding yeast mutants lacking TFC homologs have only conditional effects and are normally not lethal (Fleming et al. 2000, and references therein). In contrast, null mutations in fission yeast TFC genes cause abnormal cell shapes and mostly result in lethality (Hirata et al. 1998; Radcliffe and Toda 2000; Radcliffe et al. 1999, 2000a,b). Genetic analysis of TFC function in fission yeast has led to a different model of tubulin folding than proposed for mammals; an essential pathway of α-tubulin folding involves, successively, TFCs B, E, and D, with the Arl2 homolog acting upstream of TFC D, whereas a nonessential pathway of β-tubulin folding involves TFC A that passes β-tubulin on to TFC D to associate with α-tubulin (Radcliffe et al. 1999, 2000a,b). It is not clear whether the two models for tubulin folding reflect differences between multicellular and unicellular species or whether the in vitro assay does not fully represent the in vivo process. Thus, it would be desirable to analyze the in vivo functions of TFCs in a multicellular organism.
In the course of our analysis of cytokinesis in Arabidopsis, we have identified cell division-defective mutations in four genes collectively called the PILZ group because of their generic mutant embryo phenotype (Mayer et al. 1999). The mushroom-shaped mutant embryos consist of only one or a few grossly enlarged cells with one or more big nuclei. They are surrounded by abnormal endosperm that fails to cellularize and contains only a few enlarged nuclei. Although embryonic development is arrested, the mutant cells stay alive and continue to express cell cycle regulators. However, the cytokinesis-specific syntaxin KNOLLE is not properly localized in mitotic cells, which correlates with the absence of microtubule arrays (Mayer et al. 1999).
Here, we show that the Arabidopsis PILZ group genes encode orthologs of mammalian TFC C, D, and E, and the associated small G-protein Arl2. Another gene with a more variable but related mutant phenotype, KIESEL, was found to encode a TFC A ortholog. pilz mutants lack microtubule arrays, whereas actin filaments are present, suggesting that the PILZ gene products are specifically involved in a pathway leading to the synthesis of α/β-tubulin polymers. Because no close homolog of mammalian TFC C has been described for either yeast species, we examined the TFC C ortholog PORCINO in more detail. Our results suggest that in Arabidopsis, TFCs C–E, and Arl2 are stringently required for microtubule formation, whereas TFCs A and B may play accessory roles, thus resembling the mammalian in vitro tubulin-folding pathway. We also provide evidence that microtubules mediate vesicle trafficking to the division plane during cytokinesis but not to the cell surface during interphase.
Results
KIESEL gene mutations cause related, but weaker defects than PILZ group gene mutations
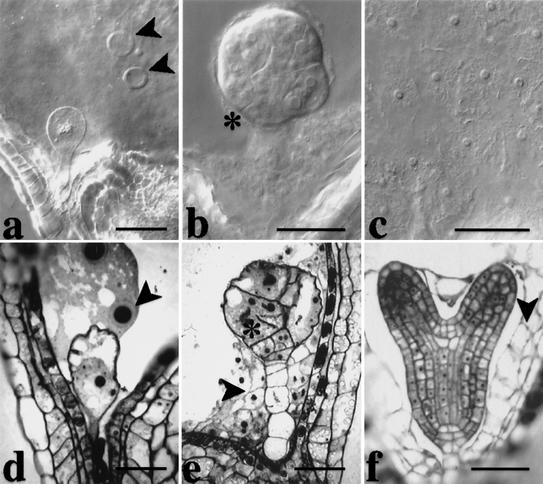
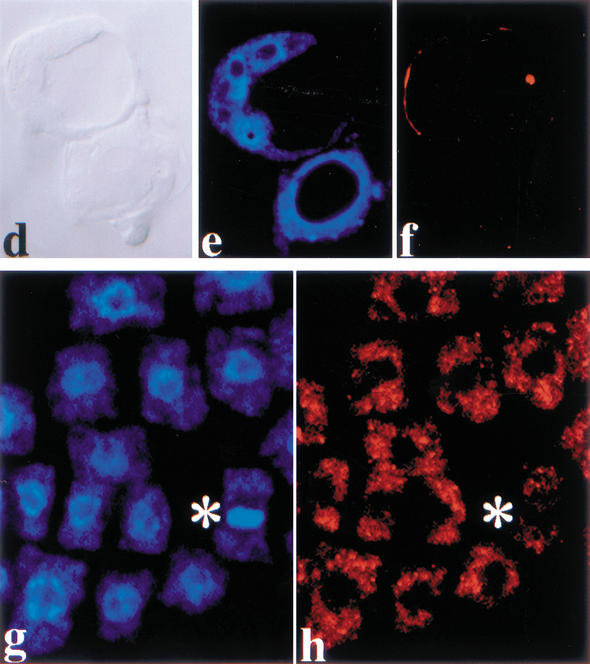
Mutations in four PILZ group genes, PORCINO (POR), HALLIMASCH (HAL), CHAMPIGNON (CHO), and PFIFFERLING (PFI) give nearly identical mutant phenotypes (Mayer et al. 1999). The mutant embryos consist of one or a few grossly enlarged cells, surrounded by an endosperm that fails to cellularize and contains but a few very big nuclei (Fig. 1a,d). A large-scale screen for mutations affecting cell division in embryogenesis led to the isolation of 12 new mutants with the characteristic pilz phenotype. They were all shown by complementation analysis to be additional alleles of the four PILZ group genes. In total, each PILZ group gene is represented by eight mutant alleles on average (see Table 1), suggesting saturation, that is, all Arabidopsis genes that mutate to the characteristic pilz mutant phenotype have been identified. In addition, we isolated three mutant lines with a related, although weaker embryo phenotype (Fig. 1b,e). These mutants are alleles of a new gene that we have named KIESEL (KIS). kis mutant embryos consist of variably enlarged cells with cell-wall stubs, which is reminiscent of the cytokinesis-defective mutants knolle and keule (Fig. 1b,e; Waizenegger et al. 2000). The phenotype of kis embryos could also be interpreted as a weaker variant of the pilz phenotype that is variably penetrant within a given embryo (Fig. 1, cf. b and e with a, d, and f). In contrast to pilz mutant endosperm (Fig. 1a), however, kis mutant endosperm is indistinguishable from wild-type endosperm in regard to nuclear multiplication and subsequent cellularization (Fig. 1c,e,f). These observations suggest that the KIS gene is predominantly required in the developing embryo.
Figure 1.
Comparison of embryo and endosperm development between por and kis mutants. (a,d) por. Note that the embryo is single celled and that the endosperm is not cellularized and contains only a few grossly enlarged nuclei with oversized nucleoli (arrowheads in a and d). (b,c,e) kis. The mutant embryo contains enlarged cells with incomplete cell walls (asterisks, b and e), whereas the endosperm contains many small nuclei (c) and cellularizes (arrowhead, e). (f) Wild-type heart stage with cellularized endosperm (arrowhead). (a–c) Nomarski-optics images of whole-mount preparations; (d–f) light micrographs of sections. Bar, 50 μm.
Table 1.
Arabidopsis tubulin-folding cofactors (TFCs) and their homologs in other organisms
|
|
TFC A
|
TFC
B
|
TFC C
|
TFC D
|
TFC
E
|
Arl2
|
|
|---|---|---|---|---|---|---|---|
| A.t. | Gene | Kis | AtTFC B | POR | CHO | PFI | HAL |
| AGI no. | At2g30410 | At3g10220 | At4g39920 | At3g60740 | At1g71440 | At2g18390 | |
| Number of alleles | 3 | (none) | 3 | 13 | 10 | 9 | |
| Protein size | 113 aa | 243 aa | 345 aa | 1254 aa | 531 aa | 185 aa | |
| H.s. | Protein size | 108 aa | 244 aa | 346 aa | 1192 aa | 527 aa | 184 aa |
| Percent ident./sim. | 44/64% (87) | 36/51% (240) | 42/60% (157) | 31/49% (1191) | 25/44% (491) | 60/75% (185) | |
| S.p. | Gene | ALP31 | ALP11 | SPAC328.08c | ALP1 | ALP21 | ALP41 |
| Phenotype | nonessential | cell lethal | (not analyzed) | cell lethal | cell lethal | cell lethal | |
| Protein size | 119 aa | 234 aa | (259 aa) | 1121 aa | 511 aa | 186 aa | |
| Percent ident./sim. | 30/50% (112) | 31/49% (229) | 26/48% (150) | 22/36% (610) | 21/38% (317) | 45/64% (177) | |
| S.c. | Gene | RBL2 | ALF1 | (CIN2) | CIN1 | PAC2 | CIN4 |
| Phenotype | conditional | conditional | conditional | conditional | conditional | conditional | |
| Protein size | 106 aa | 254 aa | (268 aa) | 1014 aa | 518 aa | 191 aa | |
| Percent ident./sim. | 34/59% (83) | 25/40% (225) | 31/47% (79) | 24/43% (456) | 34/55% (189) | ||
(A.t.) Arabidopsis thaliana; (H.s.) Homo sapiens; (S.p.) Schizosaccharomyces pombe; (S.c.) Saccharomyces cerevisiae; aa, amino acid residues. (AGI no.) Gene identification number of Arabidopsis Genome Initiative (www.arabidopsis.org). (% ident./sim.) Percent identical or similar amino acids (of total number in brackets) as compared with Arabidopsis protein (BlastP, blossom 62 with low sequence complexity filter). (SPAC328.08c) Conceptual translation of cosmid ORF (Genbank accession no. CAB96003) was identified as a potential fission yeast TFC C homolog by searching the NCBI database with the POR sequence. (CIN2) Functionally related to CIN1 and CIN4 (Fleming et al. 2000); 14% id./32% sim. with HsTFC C over 60% of their length (Bhamidipati et al. 2000) but not found by BLASTP search of NCBI database with the POR sequence.
The microtubule, but not the actin cytoskeleton fails to form in pilz group mutants
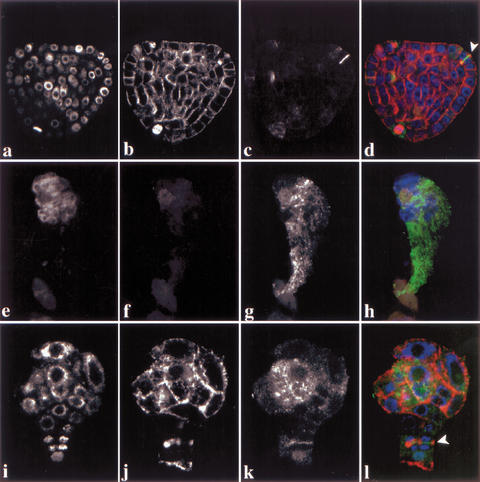
Plant cells display specific microtubule arrays in a cell cycle-dependent manner and accumulate the cytokinesis-specific syntaxin KNOLLE at the plane of division during mitosis (Fig. 2a–d; Lauber et al. 1997). pilz group mutant embryos did not display microtubule staining and failed to localize KNOLLE in mitotic cells (Fig. 2e–h). In contrast, kis mutants showed microtubule arrays (Fig. 2i–l). However, enlarged cells of kis mutant embryos were slightly abnormal in their microtubule organization. kis mutant cells undergoing cytokinesis divided abnormally, although they displayed phragmoplast microtubules and accumulated the KNOLLE syntaxin in the forming cell plate (Fig. 2k,l). Taken together, there was a close correlation between microtubule defects and abnormal embryo development.
Figure 2.
Immunolocalization of α-tubulin and KNOLLE proteins in por and kis mutant embryos. (a–d) Wild-type; (e–h) por; (i–l) kis heart-stage embryos. (a,e,i) DAPI staining; (b,f,j) α-tubulin; (c,g,k) KNOLLE; (d,h,l) overlays of DAPI (blue), α-tubulin (red), and KNOLLE (green). Note absence of microtubules in f; arrowheads mark phragmoplasts and cell plates of abnormally dividing cells in d and l. CLSM images.
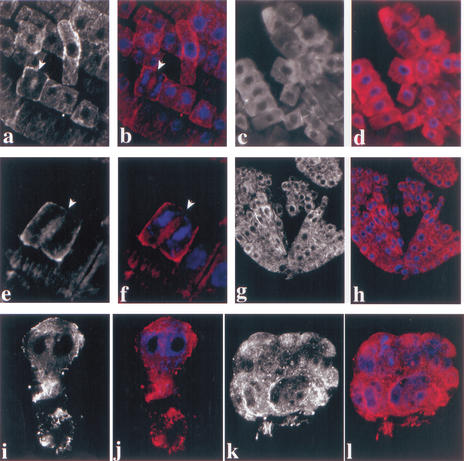
In plant cells, microtubules and actin filaments form closely associated cytoskeletal arrays during cell division (Staehelin and Hepler 1996). We therefore wondered whether actin filaments were also affected in pilz group mutant embryos. To test this possibility, we examined the actin cytoskeleton in por mutants by immunofluorescence staining with a monoclonal anti-actin antibody (Fig. 3). As a test for staining specificity, we treated wild-type root cells with the actin-depolymerizing drug latrunculin B, which abolished the immunofluorescence signal (Fig. 3a–d; see also Baluska et al. 2001). In addition, we observed phragmoplast-specific actin staining in wild-type cells undergoing cytokinesis, confirming the specificity of our staining procedure (Fig. 3e,f). Wild-type embryos displayed a fine mesh of actin filaments (Fig. 3g,h). Actin filaments were not noticeably affected in por mutant embryos, although their interphase cells were grossly enlarged as compared with wild-type (Fig. 3i,j). Actin filaments were also observed in other pilz mutant embryos (data not shown). In addition, kis mutant embryos displayed actin filaments as well (Fig. 3k,l). Thus, the cytoskeletal defect of por and kis mutant cells was confined to microtubules, whereas actin filaments were present.
Figure 3.
Immunolocalization of actin in por and kis embryos. (a–f) Wild-type root: (a,b) untreated control; note early phragmoplast (arrowheads); (c,d) latrunculin B treatment for 90 min, note absence of detectable actin filaments; (e,f) dividing cell, phragmoplast (arrowhead) flanked by forming daughter nuclei. (g–l) Heart-stage embryos: (g,h) wild-type; (i,j) por; (k,l) kis. (a,c,e,g,i,k) Actin staining; (b,d,f,h,j,l) actin (red) and DAPI (blue) double labeling. Note that embryonic cells are cytoplasm rich and lack a large vacuole, and thus display less pronounced actin filaments than the large and highly vacuolated root cells. CLSM images.
Ultrastructural analysis of por mutant embryos
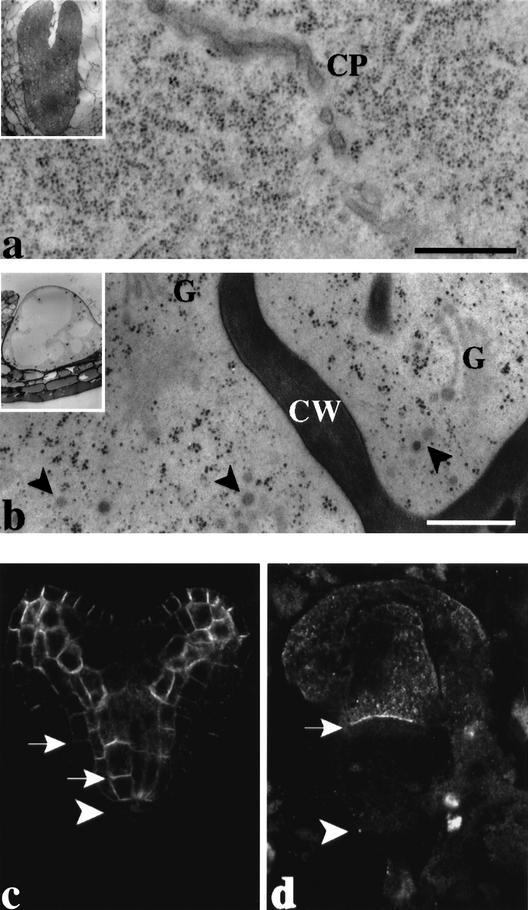
The development of pilz group mutant embryos is arrested because they are unable to undergo cell division. However, the mutant cells stay alive, increase in size, and express cell-cycle regulators until late in embryogenesis (Mayer et al. 1999). To determine whether their subcellular organization is affected, we analyzed por mutants by high-pressure-freezing/freeze-substitution electron microscopy (Fig. 4a,b). The Golgi stacks were normal, suggesting that the endomembrane system was not markedly affected. Electron-dense membrane vesicles accumulated in the cytoplasm and were often in close association with cell-wall fragments that form when cytokinesis is blocked (Fig. 4b). In contrast, dividing cells of wild-type embryos did not accumulate such vesicles in the vicinity of the forming cell plate (Fig. 4a). This result suggests that the absence of microtubules interferes with the delivery of membrane vesicles to the division plane.
Figure 4.
Vesicle trafficking in pilz mutant embryos. (a,b) Electron micrographs of ultrathin sections of high-pressure-frozen/freeze-substituted (a) wild-type and (b) por mutant torpedo-stage embryos. Note that electron-dense vesicles (arrowheads) accumulate near cell-wall stub in por (b) but not at the forming cell plate in wild type (a). (Insets) Overviews of embryos. (CP) Cell plate; (CW) cell wall; (G) Golgi stacks. Bar, 500 nm. (c,d) Immunolocalization of plasma-membrane protein PIN1 in (c) wild-type and (d) hal mutant heart-stage embryos. Note accumulation of PIN1 (arrows) at the basal ends of vascular cells in c and apical cell in d; arrowheads indicate boundary between derivatives of apical and basal daughter cells of the zygote.
Subcellular localization of the plasma-membrane protein PIN1 in pilz group mutant embryos
The pilz mutant embryos consist of one or a few grossly enlarged cells, indicating that cell growth continues in the absence of cell division. To determine whether or not vesicle trafficking to the cell surface was affected in mutant embryos, we examined the subcellular localization of the putative auxin efflux carrier, PIN1, an integral plasma-membrane protein (Gälweiler et al. 1998). PIN1 is expressed from very early stages of Arabidopsis embryogenesis and starts to accumulate at the basal ends of vascular precursor cells from the early-globular stage on (Steinmann et al. 1999). This polar localization of PIN1 persists in the cells of the vascular strands throughout wild-type embryogenesis (Fig. 4c). In hal mutant embryos, PIN1 protein was expressed in the large apical cell (Fig. 4d). This result is consistent with the expression of PIN1 in derivatives of the apical, but not the basal daughter cell of the zygote in wild-type embryos (Fig. 4c; Steinmann et al. 1999). Moreover, PIN1 accumulated at the basal end of the large apical cell in the hal mutant embryo, indicating comparable targeting as in wild-type embryos (Fig. 4c,d). Thus, expression and subcellular targeting of PIN1 protein were normal in the absence of microtubules.
The PILZ group and KIS genes encode orthologs of mammalian proteins involved in tubulin folding
To determine their role in microtubule formation, the five genes, POR, HAL, PFI, CHO, and KIS were isolated by map-based cloning. This procedure involved mapping against molecular markers in individual mapping populations to narrow down the relevant genomic intervals, followed by sequencing of PCR-amplified and subcloned genomic fragments from wild-type and mutant alleles (see Materials and Methods). The task was simplified by a candidate gene approach after cloning of the POR gene. The deduced POR protein shows 42% amino acid sequence identity to human TFC C (Fig. 5a). In addition, the C-terminal 150 amino acids of POR are 30% identical to the N-terminal half of the X-linked Retinitis pigmentosa (XRP2) gene product of human (Schwahn et al. 2001). Furthermore, a database search with the POR sequence identified a fission yeast ORF that may encode a TFC C homolog (Table 1). The three por alleles have different mutations in the highly conserved C-terminal one-third of the protein, although their phenotypes are nearly indistinguishable (Fig. 5a; data not shown; see below). The POR protein is predicted to form six α-helices, as indicated in Figure 5a. The S238L mutation of allele por-UU3025 is predicted to result in the formation of a supernumerary helix 4a, whereas the truncated protein of allele por-UU1087 lacks helix 6, and the allele por-DEI284 makes a variant protein with an internal deletion that shifts helix 6 toward helix 5.
Figure 5.
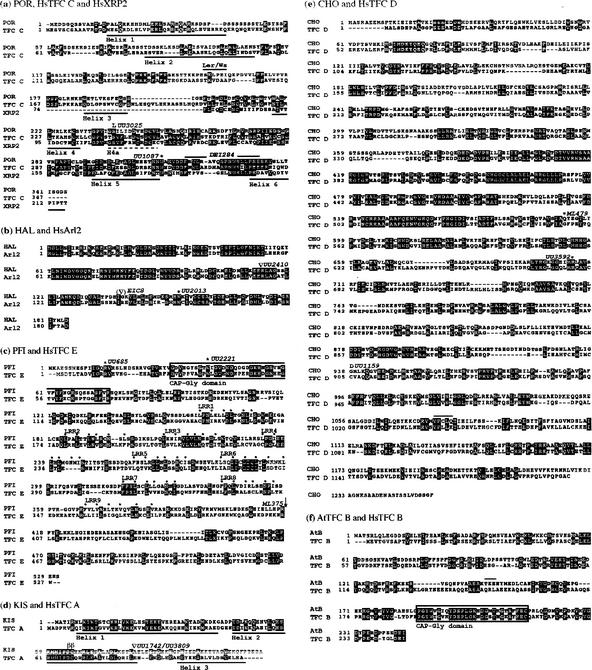
Sequence comparisons between Arabidopsis and human orthologs of tubulin-folding cofactors. The deduced amino acid sequences of the Arabidopsis proteins are based on comparisons between genomic and cDNA sequences. The published annotation (www.arabidopsis.org) was corrected as indicated below. Identical amino acids are highlighted by black, similar ones by gray shading. Genomic sequence changes in mutant alleles (designations indicated above the sequences) are shown as amino acid replacements, asterisks (stop codons) or open triangles (splice sites affected). (a) POR/HsTFC C/HsXRP2. Ler/Ws, ecotype-specific deletion of five amino acid residues (SKLPE; overlined); six helices predicted by MacVector program are underlined; 18 amino acids deleted in allele DEI284 are overlined; the predicted supernumerary helix 4a (H4a) of allele UU3025 is indicated by a broken line. Only the homologous region of HsXRP2 is shown. (b) HAL/HsArl2. Allele EIC8 has a deletion of 15 bp including a splice donor site (∇). (c) PFI/HsTFC E. Exons 1, 5, and 14–16 (corresponding to amino acids 1–103, 226–250, and 448–531, respectively) have been added and intron 2 inserted (between amino acids161 and 162); CAP–Gly domain is boxed (amino acids 32–76); nine leucine-rich repeat (LRR 1–9) motifs are indicated by dots above critical residues (amino acids 159–384); Q(207) → E and V(347) → L, Ler polymorphism; arrow indicates break point of 10-Mb inversion in allele ML375. (d) KIS/HsTFC A. The second exon has been shortened by 66 bp, now corresponding to amino acids 52–80, and a third exon added, corresponding to amino acids 81–113; three helices (determined by structure analysis of homologous Rbl2p from yeast; Steinbacher 1999) are underlined; (ββ) conserved residues DC that influence β-tubulin binding in TFC A (Llosa et al. 1996); splice site mutations insert 12 amino acid residues (INYIFHCYCLLS) between helix 2 and 3; allele MM240 (not indicated) has a deletion of the entire gene and 80 kb of flanking DNA. (e) CHO/HsTFC D. A donor splice site shifted by 15 bp adds amino acid residues 1077–1081, VPCLQ (overlined); R(237) → G, Ler polymorphism. (f) AtTFC B/HsTFC B. CAP–Gly domain is boxed (amino acids 187–214); an acceptor splice site shifted by 9 bp adds amino acid residues 147–149, TKE (overlined); S(75) → F, Ler polymorphism.
The deduced amino acid sequence of HAL protein is 60% identical to that of the small G-protein Arl2 that interacts with TFC D in mammalian cells (Fig. 5b; Bhamidipati et al. 2000). The three hal alleles sequenced have an internal deletion, a splice-site mutation and a stop-codon mutation, respectively (Fig. 5b). The HAL gene is identical with the TITAN5 gene isolated by McElver et al. (2000). These authors, however, interpreted the gene product as an ARF-type G-protein involved in vesicle trafficking.
The deduced amino acid sequence of the PFI protein is only 25% identical to that of human TFC E but shares the N-terminal CAP-Gly (formerly called CLIP170) domain involved in tubulin binding and a central region of nine loosely conserved leucine-rich repeat (LRR) motifs (Fig. 5c; Kobe and Deisenhofer 1995; Scheel et al. 1999). The X-ray-induced allele pfi-ML375 is associated with a 10-Mb inversion that disrupts the ORF, and two other alleles have early stop-codon mutations truncating the protein at amino acid residues 14 and 40, respectively (Fig. 5c).
The CHO gene product displays 30% amino acid sequence identity to human TFC D (Fig. 5e). Because the CHO gene extends over 6 kb of genomic sequence, only three cho alleles were subcloned and sequenced to identify mutations. Two cho alleles have stop-codon mutations that truncate the protein at 591 and 705 of 1254 amino acid residues, respectively (Fig. 5e). Thus, with the possible exception of POR, at least one mutant allele of each PILZ group gene truncates the protein and, thus, most likely eliminates gene function, suggesting that the mutant phenotypes are caused by the absence of functional gene product.
The KIS gene product resembles human TFC A, sharing 44% of the amino acid sequence (Fig. 5d). The strongest conservation is seen in the amino acid segments corresponding to the three α-helical domains originally described for the budding yeast TFC A homolog, Rbl2p (Steinbacher 1999). The kis mutant phenotype is variable, regardless of the allele (see Fig. 1). Two EMS alleles affect the same splice site, whereas the X-ray-induced allele kis-MM240 deletes the entire gene and 81.8 kb of flanking DNA sequence (see Materials and Methods), indicating that the variable mutant phenotype reflects the complete loss of gene function.
By searching the Arabidopsis database (www.arabidopsis.org), we detected an ORF for a predicted protein that shares 36% of the amino acid sequence and a C-terminal CAP–Gly domain with human TFC B (Fig. 5f). However, no mutation in the Arabidopsis TFC B gene (AtTFCB) with an embryo phenotype comparable with pilz or kis mutants was identified in our screens (see Discussion). In summary, the products of the PILZ group and KIS genes represent five of the six Arabidopsis orthologs of mammalian proteins involved in tubulin folding.
The POR gene encodes a cytosolic 38-kD protein that is found in higher molecular weight complexes
We analyzed the POR gene product in more detail because it was not clear whether any of the por mutant alleles abolished gene function completely. In addition, no close TFC C homolog had been described in yeast, although we identified an ORF for a POR-related predicted protein in fission yeast (Table 1).
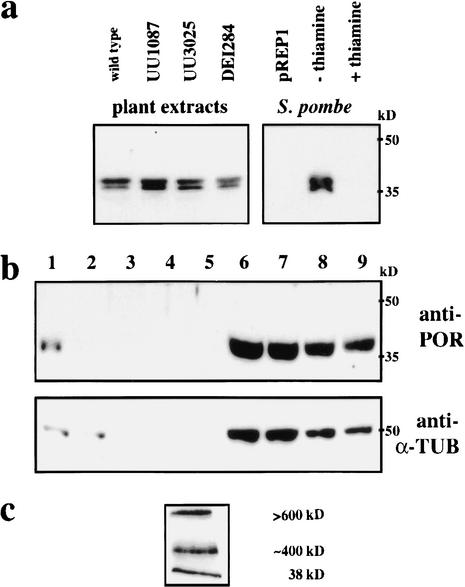
To analyze POR gene expression, we raised a rabbit polyclonal antiserum against his-tagged POR recombinant protein (see Materials and Methods). The anti-POR serum detected a 38-kD double band in protein extracts from several plant organs, suggesting ubiquitous expression (data not shown). The same doublet was also observed in extracts from Schizosaccharomyces pombe that expressed POR full-length cDNA from a thiamine-repressible promoter, but not in extracts from cells grown on thiamine or transformed with the empty vector (Fig. 6a). The double band was also detected with purified anti-POR serum and may thus be due to post-transcriptional processing. Arabidopsis plants heterozygous for any one of the three por alleles displayed the double band (Fig. 6a). Only after very long exposure of the protein blots did we observe additional very faint bands of 34 and 35 kD in the extracts from heterozygous plants for the two alleles UU1087 and DEI284, respectively, as predicted from the DNA sequence analysis (data not shown; see Fig. 4a). Under-representation of these bands suggests that the truncated proteins may be unstable. Thus, the por mutant phenotype results from severe loss or complete lack of gene function. Cell fractionation experiments indicated that POR protein was cytosolic, accumulating in the supernatant of 100,000g ultracentrifugation, as did α-tubulin (Fig. 6b).
Figure 6.
Immunodetection of POR protein. (a–c) Extracts from wild-type and por heterozygous plants and transformed yeast cells. (a, left) Comparison of leaf extracts from wild-type and heterozygous mutant seedlings. Note double bands at 38 kD. After long exposure, additional faint bands of 34 kD (UU1087) and 35 kD (DEI284) became visible. (Right) Expression of POR cDNA from the thiamine-repressible nmt1 promoter in S. pombe grown on thiamine-free medium also gives a double band. Protein extracts were separated by 15% SDS-PAGE and blots probed with anti-POR serum (1:5000). pREP1, empty vector control. (kD) Molecular weight markers. (b) Cell fractionation of extracts from wild-type callus. (Lane 1) Extract before centrifugation; (lanes 2–5) pellets of 500, 10,000, 40,000, and 100,000g centrifugations; (lanes 6–9) supernatants of 500, 10,000, 40,000, and 100,000g centrifugations. Samples were separated by 15% SDS-PAGE and blots probed with anti-POR serum (1:5000) or anti-α tubulin mAb (1:1000), respectively. (kD) Molecular weight markers. (c) Protein blot of the supernatant of 100,000g centrifugation after separation by nondenaturing 2%–19% acrylamide gradient micro-gel electrophoresis. The approximate sizes of the higher molecular weight bands were determined by comparison with size markers separated on a parallel gel stained with Coomassie blue. (d–h) Subcellular localization of POR protein. (d–f) por mutant and (g,h) wild-type torpedo-stage embryos stained with affinity-purified anti-POR serum (1:50). Note metaphase cell (asterisks) lacking association of POR with mitotic spindle. (d) Nomarski-optics image; (e,g) DAPI staining; (f,h) anti-POR staining. CLSM images.
Mammalian TFC proteins have been reported to form a large supercomplex (Tian et al. 1999). We investigated by nondenaturing micro-gel electrophoresis whether POR protein exists in cytosolic higher molecular weight complexes. Supernatants from 100,000g ultracentrifugation were subjected to 2%–19% PAGE gradient electrophoresis, and protein blots probed with anti-POR serum. POR protein was not only detected as a monomer of 38 kD, but also in two high molecular weight bands, of which one corresponded to ∼400 kD, whereas the other was >600 kD (Fig. 6c). Thus, POR protein appears to exist in large cytosolic protein complexes.
To determine the subcellular localization of POR, whole-mount preparations of embryos were stained by immunofluorescence with affinity-purified anti-POR serum (see Materials and Methods). The purified antiserum did not stain por mutant embryos (Fig. 6d–f). All three mutant alleles gave the same result (data not shown), indicating that the mutant proteins do not stably accumulate, which is consistent with the Western blot analysis described above. In wild-type embryos, a disperse signal was detected in the cytoplasm, which was especially clear in mitotic cells (Fig. 6g,h). Thus, the immunolocalization suggests that POR protein does not associate with microtubular arrays.
Discussion
Arabidopsis TFC orthologs encoded by the PILZ group genes are essential for microtubule formation
Tubulin-folding cofactors (TFCs) from mammalian cells promote, in vitro, the correct folding of tubulin monomers and the release of α/β-tubulin heterodimers (Tian et al. 1996). These extensive studies have led to a model of tubulin folding in which TFCs C, D, and E act as a tubulin dimer-making machine, whereas TFCs A and B serve as reservoirs for partially folded β- and α-tubulin intermediates, respectively (Tian et al. 1999). The in vivo requirements of mammalian TFCs and the associated small G-protein Arl2 are not known because there are no loss-of-function mutants (Tian et al. 1997).
Using Arabidopsis as a multicellular genetic model, we have identified loss-of-function mutations in the four PILZ group genes encoding orthologs of TFCs C, D, and E, and Arl2. All pilz mutations are embryo lethal and give nearly identical mutant phenotypes. Although the mutant embryos consist of only one or a few grossly enlarged cells, these cells stay alive and continue to express cell cycle regulators (Mayer et al. 1999). Thus, these Arabidopsis TFC functions are not essential for cell viability. All pilz mutant embryos lack microtubules, whereas actin filaments are present, thus revealing a specific requirement of TFCs for microtubule formation.
Our results suggest that the Arabidopsis orthologs of TFC C, D, and E as well as the small G-protein Arl2 ortholog define a common pathway for, or act at the same step in microtubule formation. Using a specific antiserum against the TFC C ortholog POR, we detected two high molecular weight forms, of which one was ∼400 kD in size, whereas the other was >600 kD large. Whereas the composition of the larger complex is entirely unclear, the estimated 400-kD complex would be consistent with a supercomplex containing TFCs C, D, and E, and α- and β-tubulin, as described for mammalian TFCs in vitro (Tian et al. 1999). Immunolocalization of POR protein gave a disperse signal in the cytosol, with no indication of an association with microtubule arrays. In addition, overexpression of POR in Arabidopsis (B. Priester and U. Meyer, unpubl.) or fission yeast had no adverse effects, suggesting that POR does not bind directly to tubulin monomers. This result is very similar to the lack of noticeable effects of TFC C overexpression in transfected HeLa cells (Bhamidipati et al. 2000). In summary, the in vivo requirements of Arabidopsis TFC orthologs for microtubule formation suggest that tubulin folding in plants may follow the mammalian pathway inferred from in vitro assays.
In vivo requirements of TFCs differ between Arabidopsis and fission yeast
Yeast TFC orthologs, with the exception of TFC C, were originally identified by their loss-of-function mutant phenotypes. Specifically, fission yeast orthologs of TFCs B, D, and E, and Arl2, are essential for cell viability, whereas TFC A is not (Radcliffe et al. 2000a). Overexpression studies in fission yeast suggested a linear pathway of TFC action in α-tubulin folding, with TFC B acting upstream of TFC E, which in turn, together with Arl2, acts upstream of TFC D (Radcliffe et al. 2000b). An additional nonessential pathway of β-tubulin folding was proposed to involve TFC A and to converge on TFC D. Thus, tubulin folding may be executed differently between unicellular and multicellular organisms. However, it is also conceivable that the in vitro folding assays may not reflect the in vivo requirements of mammalian TFCs accurately.
In contrast to the nonessential function of TFC A in fission yeast, elimination of the Arabidopsis TFC A ortholog encoded by the KIESEL (KIS) gene results in embryo lethality. However, the kis phenotype is weaker and more variable than the pilz group mutant embryo phenotype, and the surrounding endosperm appears morphologically normal, in contrast to the arrested pilz group mutant endosperm. KIS is a single-copy gene and there is no other sequence-related gene in the Arabidopsis genome, suggesting a less stringent requirement for TFC A in plant development than for TFCs C, D, and E. One possible explanation would be that KIS protein, which has a conserved β-tubulin binding site, may function as a reservoir of partially folded β-tubulin, as has been suggested for HsTFC A (Tian et al. 1996). In this view, the normal early development of the kis mutant endosperm might reflect that all newly synthesized tubulin is readily utilized to form spindles during the rapid synchronous mitotic divisions and pseudophragmoplasts during cellularization (Otegui and Staehelin 2000).
The fission yeast TFC B gene function is essential for cell viability, like the TFC D and TFC E gene functions. In comparison, the Arabidopsis gene encoding the TFC B ortholog was not affected in any of the pilz group and cytokinesis-defective embryo mutants, although our screens generated, on average, more than seven alleles per gene, including three alleles of the very small KIS gene. This allele frequency suggests that saturation for pilz-like and cytokinesis-defective embryo mutants has been achieved. Consistent with this interpretation, the 12 newly isolated pilz mutants were alleles of previously defined complementation groups (Mayer et al. 1999), whereas all three mutants with weaker phenotypes represented one new complementation group, the KIS gene. In addition, McElver et al. (2000) reported the isolation of the TTN5 gene, which is identical to HAL. Another mutant with a pilz-like phenotype, ttn1, was mapped to the genomic region of CHO (Liu and Meinke 1998) and may affect the same gene, although no complementation test has been performed. It is theoretically possible that AtTFC B gene mutations were not recovered because they might have earlier developmental defects, such as gametophyte lethality or zygotic arrest. However, elimination of any other TFC ortholog does not affect gametophyte development or function (see below). In addition, the strongest pilz mutant phenotype is represented by por embryos often consisting of a single grossly enlarged cell (Fig. 1; Mayer et al. 1999). Thus, the most plausible interpretation is that AtTFC B gene mutations may give an indistinct or no mutant embryo phenotype, implying that TFC B be less stringently required than TFC A. Consistent with this, human TFC B is not required for in vitro tubulin folding and has been proposed to sequester α-tubulin folding intermediates (Tian et al. 1997).
In summary, the in vivo requirements for TFCs A and B differ between Arabidopsis and fission yeast. Taking also into account the parallels between in vivo requirements of Arabidopsis TFCs for microtubule formation and in vitro requirements of mammalian TFCs for α/β-tubulin heterodimer formation, our observations suggest that multicellular and unicellular modes of tubulin folding may have diverged in evolution.
Genetic ablation of microtubules reveals their functional requirements in plant development
The current view of the roles of the plant microtubule cytoskeleton is largely based on drug interference studies that analyzed short-term effects in cell cultures or seedling roots. However, drugs have potential side effects and are not suitable for the analysis of functional requirements in a developmental context, such as embryogenesis, due to their limited access to the target tissue. These problems are circumvented by specific genetic elimination of microtubules, as indicated by the nearly identical effects caused by mutations in several TFC genes. This analysis was possible because each TFC is encoded by a single-copy gene, in contrast to the gene families encoding α- and β-tubulins in Arabidopsis (Kopczak et al. 1992; Snustad et al. 1992). The pilz group mutants thus enable us to distinguish between microtubule-dependent and microtubule-independent processes. In the absence of microtubules, cell divisions in the developing embryo as well as nuclear divisions and cellularization of the syncytial endosperm are blocked, except for one or two divisions. It is very likely that prefertilization stores of α/β-tubulin heterodimers and/or TFCs enable the zygote and the primary endosperm nucleus to undergo the very first division(s) before tubulin heterodimers become limited.
It may seem surprising that the absence of microtubules does not prevent the formation of pilz group mutant embryos altogether because plants have a haploid phase of the life cycle between meiosis and fertilization. However, the meiospores produce few-celled gametophytes, embryo sac, and pollen, by cleavage divisions that lack preprophase bands and the cytokinesis-specific syntaxin KNOLLE (Otegui and Staehelin 2000; Waizenegger et al. 2000). These cell divisions may be sustained by a premeiotic store of α/β-tubulin heterodimers and/or TFCs. For fertilization to occur, the immobile sperm cells must be carried by the tip-growing pollen tube to the ovule. This process is also not affected in the pilz group mutants, as there was no segregation distortion that would suggest reduced transmission through the haploid gametophytes. Thus, tip growth of the pollen tube does not require microtubules, whereas actin filaments appear to play an essential role (Vidali et al. 2001).
Both cell division and cell growth require delivery of membrane material by vesicle trafficking. In pilz group mutants, KNOLLE-positive membrane vesicles fail to traffic to the plane of cell division (Mayer et al. 1999), and electron-dense vesicles accumulate in close association with cell-wall fragments in por mutants, indicating that microtubules organize vesicle trafficking during cytokinesis. Although blocked in cell division, the cells of pilz mutant embryos stay alive, express cell-cycle regulators, and maintain their apical-basal cell fates, as indicated by the differential expression of PIN1 (Mayer et al. 1999; B. Priester and U. Mayer, unpubl.). The mutant cells are grossly enlarged and abnormally shaped. The abnormal cell shape is most likely due to the absence of cortical microtubule arrays that would constrain the direction of cell growth (Whittington et al. 2001). However, cell enlargement implies that vesicle trafficking to the plasma membrane occurs in the absence of microtubules, as evidenced by the proper targeting of PIN1. A role for actin filaments in this trafficking has been inferred from the inhibitory effects of actin-depolymerizing drugs (Geldner et al. 2001). Thus, plant cells appear to use microtubules in cytokinetic vesicle trafficking but actin filaments in interphase vesicle trafficking, in contrast to a predominant role of microtubules in mammalian cells (Cole and Lippincott-Schwartz 1995; Hamm-Alvarez and Sheetz 1998; Kreitzer et al. 2000).
In summary, we have described for the first time in any multicellular organism that genetic inactivation of tubulin-folding cofactors results in the specific elimination of microtubules. For plants, in particular, our results reveal an essential role of microtubules in dividing cells as well as a nonessential role in interphase cells. Controlled inactivation of TFCs during plant development thus provides a powerful tool for assessing the temporal and spatial requirements of microtubules in developmental processes.
Materials and methods
Plant strains, growth conditions, and genetic analysis
Twelve new pilz and three kiesel mutants were isolated from EMS-mutagenized or X-rayed Landsberg erecta (Ler) seed populations or identified from among embryo-lethal lines of the Versailles collection of T-DNA insertion lines (ecotype Wassilewskija; Bechtold et al. 1993), and tested for complementation with pilz group mutant alleles and among each other, as described previously (Mayer et al. 1999).
Map-based cloning, molecular gene identification, and sequence analysis
Mapping populations were established by crossing Ler plants heterozygous for mutant alleles with Columbia (Col) wild-type plants. Ler (mutant)/Col (wild-type) heterozygous F1 plants were identified by inspection of ovules and used to generate the F2-mapping populations. por-UU1087 was mapped to the bottom end of chromosome 4 between CAPS markers 60 (TaqI) on BAC T19P19 and DSH1 (MboII) on BAC T5J17 among 2036 gametes, hal-UU2013 to chromosome 2 between CAPS markers 52 (Tsp509I) on BAC T30D6 and 76 (HinfI) on BAC F24H14 among 1892 gametes, kis-UU3809 to chromosome 2 between CAPS markers 52 (VspI) on BAC T9D9 and 85 (RsaI) on BAC T6B20 among 772 gametes, cho-UU1159 to the bottom end of chromosome 3 at SSLP marker FUS6 on BAC T20K12 among 80 gametes, and pfi-UU685 to the bottom of chromosome 1 <1-cM north of CAPS marker PAB5 (MboII) on BAC F14O23 (www.arabidopsis.org) among 638 gametes.
For molecular identification of gene mutations, DNA from Ler (mutant)/Col (wild-type) heterozygous plants was subjected to two or three independent PCR amplifications with gene-specific primers and subcloned into Bluescript pKS+ (Stratagene) or pGEM (Promega). Mutant clones were identified by ecotype-specific polymorphisms and sequenced with an ABI310 sequencer. Sequence alterations of mutant alleles predicted to affect restriction sites were checked for allele-specific polymorphisms in PCR-amplified DNA from wild-type and heterozygous plants. DNA from the X-ray allele, pfi-MM375, was amplified by vectorette PCR (Devon et al. 1995), using nested genomic primers E7IIa (GTGAGAAGAAGGATTCT GAGATCC) and Es1184 (AGTCTGGGGAGATTGAGCT GCTC), and vectorette primers Vec1 (CGAATCGTAACG GTTCGTACGAGAA) and Vec2 (TCGTACGAGAATCGCT GTCCTCTCC) ligated to VspI restriction sites. The extent of the kis-MM240 deletion was determined by reiterated PCR amplification and sequencing. Following the analysis of Ler/Col polymorphic restriction sites flanking the KIS gene, DNA from the X-ray allele, kis-MM240, was amplified by vectorette PCR, using nested genomic primers KIS-12001F (CTGCAAG CAAGCCGATTAAGAAA) and KIS-12500F (TCTCTCTTT GAATCCCACTTTTTA), and the vectorette primers Vec1 and Vec2 ligated to HpaII restriction sites. Sequence information from the PCR product was used to design primer KIS-96301R (CTCCAAGTAGCAAATGCAAAAGTT). PCR amplification with primers KIS-12500F and KIS-96301R yielded a 1.6-kb product that was sequenced across the breakpoint at position 13,466/95,729 bp on BAC T6B20 (http://mips.gsf.de). This 82.2-kb deletion encompasses 1.2 kb upstream and 80.6 kb downstream of the KIS-coding sequence. cDNA clones were isolated from a flower-specific library (Weigel et al. 1992) or an embryo-enriched library (Grebe et al. 2000).
Sequence analysis, including secondary structure prediction, was performed using the MacVector and Vector NTI programs (Kodak). The BCM Search Launcher (http://dot.imgen.bcm.tmc.edu:9331) was used for sequence alignments of Arabidopsis and human orthologs.
Generation and purification of rabbit polyclonal anti-POR serum
Expression of full-length POR cDNA from His-tag expression vector pQE 30, purification of recombinant protein by Ni-NTA chromatography and acrylamid gel electrophoresis, rabbit immunization and antiserum purification were done as described (Lauber et al. 1997). In Western blot analysis, purified antiserum gave the same signal as unpurified antiserum.
Preparation of protein extracts and Western blot analysis
Preparation of plant material, SDS-PAGE, and Western blot analysis were as described (Lauber et al. 1997), except that 15% acrylamide gels were used, and the anti-POR serum was diluted 1:5000. POR cDNA was expressed from the thiamine-repressible nmt1 promoter of pREP1 vector in S. pombe wild-type strain 513 h−-leu 1/ura 4 after lithium acetate transformation (Moreno et al. 1991; Maundrell 1993; Hansen et al. 1998). Absence of thiamine increases the level of nmt1 promoter activity 80 × relative to 0.5 μM thiamine (Basi et al. 1993). For Western blot analysis, pelleted cells were disrupted with liquid nitrogen, suspended in Laemmli buffer, and boiled at 95°C for 5 min. Equal amounts from transformed cells carrying the empty vector or nmt1∷POR were loaded onto the gel.
Cell fractionation and nondenaturing gradient micro gels
Arabidopsis liquid callus culture (Fuerst et al. 1996) was filtered through Miracloth (Calbiochem), frozen in liquid nitrogen, and ground with mortar and pestle. After resuspending in homogenization buffer (50 mM potassium phosphate, 50 mM potassium chloride, 10% (v/v) glycerol, 10 mM mercaptoethanol, 1 mM phenylmethyl sulfonyl fluoride, 0.5 mM aluminium fluoride, protease inhibitor cocktail [P 9599, Sigma]), the homogenate was filtered through Miracloth and subsequently centrifuged at 500g (20 min), 10,000g (20 min), 40,000g (30 min), and 100,000g (100 min). An aliquot of the 100,000g supernatant was loaded onto a nondenaturing 2%–19% acrylamide gradient micro gel (Poehling and Neuhoff 1980).
Microscopy, immunolocalization, and image processing
Whole-mount preparations of ovules and plastic sections for light microscopy were prepared and analyzed as described (Mayer et al. 1999). Immunofluorescence localization of α-tubulin, KNOLLE, PIN1, and POR (with affinity-purified antiserum diluted 1:50) was done as described (Lauber et al. 1997). The protocol for actin immunolocalization with the mouse anti-actin monoclonal antibody C4 (ICN; diluted 1:500 in 5% BSA–PBS; Cy3-conjugated goat anti-mouse secondary antibody from Dianova, Hamburg, FRG, diluted 1:600) was adapted from Van Gestel et al. (2001) and Collings et al. (2001) to analyze whole-mount preparations of embryos as described (Lauber et al. 1997). Actin staining of seedling roots was done the same way, their treatment with 1 μM latrunculin B (Calbiochem) was as described (Geldner et al. 2001). Nuclei were stained with 1 mg/mL DAPI at 37°C for 20 min., followed by washing with H2O for 30 min. After mounting in Citifluor (Agar, Amersham) containing DABCO (25 mg/mL; Sigma), specimens were analyzed with a Leica confocal laser-scanning microscope (CLSM) with Leica TCS-NT software. The CLSM standard objective was 63× (water immersion), scanning was done with electronic magnification. For EM analysis, ovules were cryofixed by high-pressure freezing (Bal-Tec HPM 010, Balzers), freeze substituted in acetone containing 2% osmium tetroxide, 1% glutaraldehyde, and 0.5% uranyl acetate, and embedded in Spurr's resin. Ultrathin sections were stained with ethanolic uranyl acetate and lead citrate. Digital images were processed with Photoshop 5.5 and Illustrator 9.0 (Adobe).
Accession numbers
cDNA sequences and corrected annotations of genomic sequences have been submitted to Genbank under accession numbers AF486848, AF486849, AF486850, AF486851, AF486852, and AF486853.
Acknowledgments
We thank K. Hingst, J. Riemer, C. Gruber, and S. Richter for help with the molecular mapping of the genes and immunostaining; H. Steigele for EM assistance; B. Lattemann for rabbit immunization; M. Caboche for his constant support to L.L.; G. Pelletier for the access to the Versailles collection of mutants; M. Devic for the hal allele EIC8; T. Toda for fission yeast strains and advice; K. Palme for providing anti-PIN1 serum; and E. Benková, K. Cornelis, J. Friml, N. Geldner, K. Schrick, and H. Knaut for critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft through SFB446 and the ECCO project of the EU Biotechnology Program Framework 5.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Notes added in proof
While this paper was under review, Tzafrir et al. (2002, Plant Physiol. 128: 38–51) described the cloning of the TITAN1 gene, which is identical to the CHO gene analyzed here.
Re-annotations of genomic DNA sequences from chromosomes I and II (GenBank accession nos. NC_003070 and NC_003071), released on January 10, 2002, give the correct splicing patterns of PFI (At1g71440) and KIS (At2g30410).
Footnotes
E-MAIL ulrike.mayer@zmbp.uni-tuebingen.de; FAX 49-7071-295797.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.221702.
References
- Baluska F, Jasik J, Edelmann HG, Salajova T, Volkmann D. Latrunculin B-induced plant dwarfism: Plant cell elongation is F-actin-dependent. Dev Biol. 2001;231:113–124. doi: 10.1006/dbio.2000.0115. [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thalianaplants. C R Acad Sci Série III. 1993;316:1194–1199. [Google Scholar]
- Bhamidipati A, Lewis SA, Cowan NJ. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J Cell Biol. 2000;149:1087–1096. doi: 10.1083/jcb.149.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Lippincott-Schwartz J. Organization of organelles and membrane traffic by microtubules. Curr Opin Cell Biol. 1995;7:55–64. doi: 10.1016/0955-0674(95)80045-x. [DOI] [PubMed] [Google Scholar]
- Collings DA, Zsuppan G, Allen NS, Blancaflor EB. Demonstration of prominent actin filaments in the root columella. Planta. 2001;212:392–403. doi: 10.1007/s004250000406. [DOI] [PubMed] [Google Scholar]
- Devon RS, Porteous DJ, Brookes AJ. Splinkerettes—Improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res. 1995;23:1644–1645. doi: 10.1093/nar/23.9.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JA, Vega LR, Solomon F. Function of tubulin binding proteins in vivo. Genetics. 2000;156:69–80. doi: 10.1093/genetics/156.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst RA, Soni R, Murray JA, Lindsey K. Modulation of cyclin transcript levels in cultured cells of Arabidopsis thaliana. Plant Physiol. 1996;112:1023–1033. doi: 10.1104/pp.112.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K. Regulation of polar auxin transport by AtPIN1 in Arabidopsisvascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Gao Y, Meli R, Walden PD, Lewis SA, Ampe C, Rommelaere H, Vandekerckhove J, Cowen NJ. A novel cochaperonin that modulates the ATPase activity of cytoplasmic chaperonin. J Cell Biol. 1994;125:989–996. doi: 10.1083/jcb.125.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Siegers K, Schiebel E. A novel protein complex promoting formation of functional α- and γ-tubulin. EMBO J. 1998;17:952–966. doi: 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof Y-D, Jürgens G, Palme K. Auxin-transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld J-U, Salchert K, Koncz C, Jürgens G. A conserved domain of the ArabidopsisGNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell. 2000;12:343–356. doi: 10.1105/tpc.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm-Alvarez SF, Sheetz MP. Microtubule-dependent vesicle transport: Modulation of channel and transporter activity in liver and kidney. Physiol Rev. 1998;78:1109–1129. doi: 10.1152/physrev.1998.78.4.1109. [DOI] [PubMed] [Google Scholar]
- Hansen K, Birse CE, Proudfoot NJ. Nascent transcription from the nmt1 and nmt2 genes of Schizosaccharomyces pombeoverlaps neighbouring genes. EMBO J. 1998;17:3066–3077. doi: 10.1093/emboj/17.11.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen WJ, Cowan NJ, Welch WJ. Prefoldin-nascent chain complexes in the folding of cytoskeletal proteins. J Cell Biol. 1999;145:265–277. doi: 10.1083/jcb.145.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata D, Masuda H, Eddison M, Toda T. Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. EMBO J. 1998;17:658–666. doi: 10.1093/emboj/17.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- Kopczak SD, Haas NA, Hussey PJ, Silflow CD, Snustad DP. The small genome of Arabidopsiscontains at least six expressed α-tubulin genes. Plant Cell. 1992;4:539–547. doi: 10.1105/tpc.4.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitzer G, Marmorstein A, Okamoto P, Vallee R, Rodriguez-Boulan E. Kinesin and dynamin are required for post-Golgi transport of a plasma-membrane protein. Nat Cell Biol. 2000;2:125–127. doi: 10.1038/35000081. [DOI] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G. The ArabidopsisKNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol. 1997;139:1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux MR, Hartl FU. Protein folding: Versatility of the cytosolic chaperonin TRiC/CCT. Curr Biol. 2000;10:R260–R264. doi: 10.1016/s0960-9822(00)00432-2. [DOI] [PubMed] [Google Scholar]
- Lewis SA, Tian G, Cowan NJ. The α- and β-tubulin folding pathways. Trends Cell Biol. 1997;7:479–484. doi: 10.1016/S0962-8924(97)01168-9. [DOI] [PubMed] [Google Scholar]
- Liu CM, Meinke DW. The titan mutants of Arabidopsisare disrupted in mitosis and cell cycle control during seed development. Plant J. 1998;16:21–31. doi: 10.1046/j.1365-313x.1998.00268.x. [DOI] [PubMed] [Google Scholar]
- Llorca O, Martin-Benito J, Ritco-Vonsovici M, Grantham J, Hynes GM, Willison KR, Carrascosa JL, Valpuesta JM. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J. 2000;19:5971–5979. doi: 10.1093/emboj/19.22.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M, Aloria K, Campo R, Padilla R, Avila J, Sanchez-Pulido L, Zabala JC. The β-tubulin monomer release factor (p14) has homology with a region of the DnaJ protein. FEBS Lett. 1996;397:283–289. doi: 10.1016/s0014-5793(96)01198-2. [DOI] [PubMed] [Google Scholar]
- Lloyd C, Hussey P. Microtubule-associated proteins in plants—Why we need a map. Nat Rev Mol Cell Biol. 2001;2:40–47. doi: 10.1038/35048005. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Mayer U, Herzog U, Berger F, Inzé D, Jürgens G. Mutations in the PILZ group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsisembryo and endosperm. Eur J Cell Biol. 1999;78:100–108. doi: 10.1016/S0171-9335(99)80011-9. [DOI] [PubMed] [Google Scholar]
- McElver J, Patton D, Rumbaugh M, Liu C-M, Yang LJ, Meinke D. The TITAN5 gene of Arabidopsisencodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell. 2000;12:1379–1392. doi: 10.1105/tpc.12.8.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melki R, Rommelaere H, Leguy R, Vandekerckhove J, Ampe C. Cofactor A is a molecular chaperone required for β-tubulin folding: Functional and structural characterization. Biochemistry. 1996;35:10422–10435. doi: 10.1021/bi960788r. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nogales E. Structural insights into microtubule function. Annu Rev Biochem. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- Otegui M, Staehelin LA. Cytokinesis in flowering plants: More than one way to divide a cell. Curr Opin Plant Biol. 2000;3:493–502. doi: 10.1016/s1369-5266(00)00119-9. [DOI] [PubMed] [Google Scholar]
- Poehling H-M, Neuhoff V. One and two-dimensional electrophoresis in micro-slab gels. Electrophoresis. 1980;1:90–102. [Google Scholar]
- Radcliffe PA, Toda T. Characterisation of fission yeast alp11 mutants defines three functional domains within tubulin-folding cofactor B. Mol Gen Genet. 2000;263:752–760. doi: 10.1007/s004380000252. [DOI] [PubMed] [Google Scholar]
- Radcliffe PA, Hirata D, Vardy L, Toda T. Functional dissection and hierarchy of tubulin-folding cofactor homologues in fission yeast. Mol Biol Cell. 1999;10:2987–3001. doi: 10.1091/mbc.10.9.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe PA, Garcia MA, Toda T. The cofactor-dependent pathways for α- and β-tubulins in microtubule biogenesis are functionally different in fission yeast. Genetics. 2000a;156:93–103. doi: 10.1093/genetics/156.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe PA, Vardy L, Toda T. A conserved small GTP-binding protein Alp41 is essential for the cofactor-dependent biogenesis of microtubules in fission yeast. FEBS Lett. 2000b;468:84–88. doi: 10.1016/s0014-5793(00)01202-3. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Nurse P. Regulation of cell polarity by microtubules in fission yeast. J Cell Biol. 1998;142:457–471. doi: 10.1083/jcb.142.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel J, Pierre P, Rickard JE, Diamantopoulos GS, Valetti C, van der Goot FG, Haner M, Aebi U, Kreis TE. Purification and analysis of authentic CLIP-170 and recombinant fragments. J Biol Chem. 1999;274:25883–25891. doi: 10.1074/jbc.274.36.25883. [DOI] [PubMed] [Google Scholar]
- Schwahn U, Paland N, Techritz S, Lenzner S, Berger W. Mutations in the X-linked RP2 gene cause intracellular misrouting and loss of the protein. Hum Mol Genet. 2001;10:1177–1183. doi: 10.1093/hmg/10.11.1177. [DOI] [PubMed] [Google Scholar]
- Smith LG. Plant cell division: Building walls in the right places. Nat Rev Mol Cell Biol. 2001;2:33–39. doi: 10.1038/35048050. [DOI] [PubMed] [Google Scholar]
- Snustad DP, Haas NA, Kopczak SD, Silflow CD. The small genome of Arabidopsiscontains at least nine expressed β-tubulin genes. Plant Cell. 1992;4:549–556. doi: 10.1105/tpc.4.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin LA, Hepler PK. Cytokinesis in higher plants. Cell. 1996;84:821–824. doi: 10.1016/s0092-8674(00)81060-0. [DOI] [PubMed] [Google Scholar]
- Steinbacher S. Crystal structure of the post-chaperonin β-tubulin binding cofactor Rbl2p. Nat Struct Biol. 1999;6:1029–1032. doi: 10.1038/14912. [DOI] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Gälweiler L, Palme K, Jürgens G. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;286:316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- Straight AF, Field CM. Microtubules, membranes and cytokinesis. Curr Biol. 2000;10:R760–R770. doi: 10.1016/s0960-9822(00)00746-6. [DOI] [PubMed] [Google Scholar]
- Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ. Pathway leading to correctly folded β-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- Tian G, Lewis SA, Feierbach B, Stearns T, Rommelaere H, Ampe C, Cowan NJ. Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J Cell Biol. 1997;138:821–832. doi: 10.1083/jcb.138.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Bhamidipati A, Cowan NJ, Lewis SA. Tubulin folding cofactors as GTPase-activating proteins. GTP hydrolysis and the assembly of the α/β-tubulin heterodimer. J Biol Chem. 1999;274:24054–24058. doi: 10.1074/jbc.274.34.24054. [DOI] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- Van Gestel K, Le J, Verbelen JP. A comparison of F-actin labeling methods for light microscopy in different plant specimens: Multiple techniques supplement each other. Micron. 2001;32:571–578. doi: 10.1016/s0968-4328(00)00054-8. [DOI] [PubMed] [Google Scholar]
- Vidali L, McKenna ST, Hepler PK. Actin polymerization is essential for pollen tube growth. Mol Biol Cell. 2001;12:2534–2545. doi: 10.1091/mbc.12.8.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger I, Lukowitz W, Assaad F, Schwarz H, Jürgens G, Mayer U. The Arabidopsis KNOLLE and KEULEgenes interact to promote vesicle fusion during cytokinesis. Curr Biol. 2000;10:1371–1374. doi: 10.1016/s0960-9822(00)00775-2. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO. MOR1 is essential for organizing cortical microtubules in plants. Nature. 2001;411:610–613. doi: 10.1038/35079128. [DOI] [PubMed] [Google Scholar]