Abstract
Phosphorylation of clock proteins represents an important mechanism regulating circadian clocks. In Neurospora, clock protein FREQUENCY (FRQ) is progressively phosphorylated over time, and its level decreases when it is extensively phosphorylated. To identify the kinase phosphorylating FRQ and to understand the function of FRQ phosphorylation, a FRQ-phosphorylating kinase was purified and identified as casein kinase II (CKII). Disruption of the catalytic subunit gene of CKII in Neurospora resulted in hypophosphorylation and increased levels of FRQ protein. In addition, the circadian rhythms of frq RNA, FRQ protein, and clock-controlled genes are abolished in the CKII mutant. Our data suggest that the phosphorylation of FRQ by CKII may have at least three functions; it decreases the stability of FRQ, reduces the protein complex formation between FRQ and the WHITE COLLAR proteins, and is important for the closing of the Neurospora circadian negative feedback loop. Taken together, our results suggest that CKII is an important component of the Neurospora circadian clock.
Keywords: Circadian, frequency, white collar-1, Neurospora, phosphorylation
Circadian clocks control a wide variety of physiological, behavioral, cellular, and biochemical activities in most eukaryotic and certain prokaryotic organisms. At the molecular level, circadian oscillators consist of autoregulatory negative feedback loops, in which the positive elements of the loop activate the transcription of the negative elements, whereas the negative elements feedback to block their own transcriptional activation by interacting with the positive elements (Aronson et al. 1994a; Shearman et al. 1997; Darlington et al. 1998; Ishiura et al. 1998; Dunlap 1999; Kume et al. 1999; Lee et al. 1999; Young 1999; Loros and Dunlap 2001; Young and Kay 2001).
In the Neurospora frq-wc-based circadian feedback loops, two PAS domain-containing transcription factors, WHITE COLLAR-1 (WC-1) and WC-2, form heterodimeric complexes to activate the transcription of frequency (frq) (Crosthwaite et al. 1997; Talora et al. 1999; Cheng et al. 2001b, 2002), whereas two forms of FRQ proteins (large [LFRQ] and small [SFRQ]) form homodimeric complexes that feedback to repress the transcription of frq, probably by interacting with the WC-1/WC-2 complexes (Aronson et al. 1994a; Garceau et al. 1997; Liu et al. 1997; Cheng et al. 2001a; Denault et al. 2001; Merrow et al. 2001). In addition to its role in repressing frq transcription, FRQ protein positively regulates protein levels of both WC-1 and WC-2, thereby forming positive feedback loops interlocked with the negative feedback loop (Lee et al. 2000; Cheng et al. 2001b; Merrow et al. 2001). These positive feedback loops appear to be important for maintaining the robustness and stability of the clock.
Protein phosphorylation is one of the most important post-transcriptional mechanisms that regulate circadian clocks (Edery et al. 1994; Garceau et al. 1997; Kloss et al. 1998; Price et al. 1998; Sugano et al. 1998; Naidoo et al. 1999; Talora et al. 1999; Iwasaki et al. 2000; Liu et al. 2000; Lowrey et al. 2000; Schwerdtfeger and Linden 2000; Gorl et al. 2001; Lee et al. 2001; Martinek et al. 2001; Toh et al. 2001). In Drosophila, Young and colleagues have shown that two kinases, DOUBLE-TIME (DBT) and SHAGGY (SGG), play two different roles in Drosphila clock by phosphorylating two different clock components. DBT, a casein kinase I, phosphorylates PERIOD (PER) and may lead to PER degradation (Kloss et al. 1998; Price et al. 1998). In a dbt null mutant, PER is hypophosphorylated, its level is high, and the clock is not functional. SGG, the Drosophila glycogen synthase kinase-3 (GSK-3) ortholog, appears to promote the nuclear transfer of PER/TIM complex by phosphorylating TIMELESS (TIM), the heterodimeric partner of PER. The increased expression of sgg resulted in period shortening of the clock (Martinek et al. 2001).
In Neurospora, FRQ proteins are phosphorylated immediately after their synthesis and become progressively phosphorylated over time (Garceau et al. 1997). The extensive phosphorylation of FRQ results in its eventual degradation. A previous study suggested that one of the functions of FRQ phosphorylation is to trigger its degradation (Liu et al. 2000). This conclusion was supported by two lines of evidence. First, a kinase inhibitor that blocks FRQ phosphorylation in vivo reduces the degradation rate of FRQ and lengthens the period of the clock. Second, mutation of one phosphorylation site at Ser 513 of FRQ leads to reduction of the rate of FRQ degradation and a very long period (>30 h) of the clock. Thus, phosphorylation of FRQ is an important determining factor for FRQ stability and for the period length of the clock. In addition, this study also suggested that FRQ phosphorylation may have other functions as well, because the mutation of another phosphorylation site (Ser 519) resulted in a slightly shorter period and low amplitude of the overt rhythm (Liu et al. 2000).
To identify the kinases that phosphorylate FRQ, we previously purified a Neurospora calcium/calmodulin (Ca/CaM)-dependent kinase (CAMK-1) protein from Neurospora extract (Yang et al. 2001). Disruption of the camk-1 gene suggests that camk-1 plays an important role in the normal growth and development of Neurospora. However, the camk-1 null phenotype is transient and quickly reverts to the wild-type phenotype, revealing redundancy in the system. Although modest clock phenotypes were found in the camk-1 null strain, we failed to detect significant change of the FRQ phosphorylation profile in the mutant strains. Together, those data suggest that FRQ may be phosphorylated by multiple kinases in vivo. In this study, we purified another FRQ-phosphorylating kinase from the protein extracts of a camk-1 null strain. This kinase was identified as the Neurospora casein kinase II (CKII). The disruption of the gene encoding for the catalytic subunit of the kinase suggested that phosphorylation of FRQ by CKII is important for the normal function of the Neurospora circadian clock.
Results
CKII, a kinase that phosphorylates FRQ, was biochemically purified
During the purification of CAMK-1, we noted that there were several FRQ-phosphorylating Ca/CaM-independent kinase activities. One kinase activity peak eluted from the Q-Sepharose column at 0.45 M NaCl. To purify this kinase, we used the protein extracts from a camk-1ko strain to eliminate the kinase activity of CAMK-1. With the use of an in vitro kinase assay to monitor its activity to phosphorylate GST–FRQ (Yang et al. 2001), this kinase was purified from the Neurospora cell extracts after a four-step purification protocol (Fig. 1A). The elution profile in Superdex 200 suggested that this kinase was either quite large or in a protein complex (∼150–200 kD; data not shown). The results of the final step of the purification are shown in Figure 1, B and C. The peak of kinase activity was in fraction 18, eluting at 0.45 M NaCl. Silver staining of the SDS–polyacrylamide gel of the same protein fractions revealed five visible protein bands in fraction 18, with molecular weights ranging from 25 to 42 kD (Fig. 1C). Of the five protein bands, the levels of three correlated well with the activity profile.
Figure 1.
Biochemical purification of CKII, a FRQ phosphorylating kinase from Neurospora. (A) A diagram of the purification scheme for CKII. (B) Aliquots of the indicated final Mono Q column fractions were used for the in vitro kinase assay. B and F are the protein extracts loaded and the flow-through of the Mono Q column, respectively. The NaCl gradient is indicated at top. (C) Aliquots of the indicated Mono Q column fractions were subjected to 4%–15% SDS-PAGE, and the gel was subsequently silver stained. The three arrows denote the protein bands identified as containing the indicated CKII subunits. The two protein bands copurified with CKII were denoted by asterisks.
To obtain the sequence identity of these three proteins (∼40, 36, and 25 kD), these protein bands were excised from the gel and subjected to tryptic digestion followed by mass spectral analysis. The resulting mass fingerprints of the digested peptides were used to search against the NCBI database. Two independent Neurospora ESTs, c3d10np and NCM3A11, were found to match the mass fingerprints of the digested peptides of the 40-kD protein, whereas no match was found for the mass fingerprints of the 36-kD and 25-kD proteins. BLAST searches revealed that these ESTs have strong homology to the eukaryotic CKII catalytic (α) or regulatory (β) subunits.
The complete genome of Neurospora was recently determined (http://wolfram.wi.mit.edu/annotation/fungi/neurospora/). On the basis of the EST sequences and cDNA sequencing results, the corresponding full-length Neurospora CKII α and β subunit genes were identified and the entire ORF of the proteins determined. Eukaryotic CKII holoenzyme is a α2β2 heterotetramer, and most eukaryotic organisms have at least two distinct α subunits and two different regulatory β subunits. Only one α subunit and two β subunit genes were identified in the Neurospora genome, named cka, ckb1, and ckb2. When the entire ORFs of CKA, CKB1, and CKB2 were used to compare with the mass fingerprints of the digested peptides of the three proteins, the 36-kD protein was found to match CKB2 and more matches were found between the mass fingerprints of the 42-kD protein and CKA and CKB1 (Fig. 1C). The 25-kD protein band was found to be the protein degradation products of the CKA and CKB1. The fact that these proteins were found to match all three CKII subunits indicated that CKII was the kinase we were after.
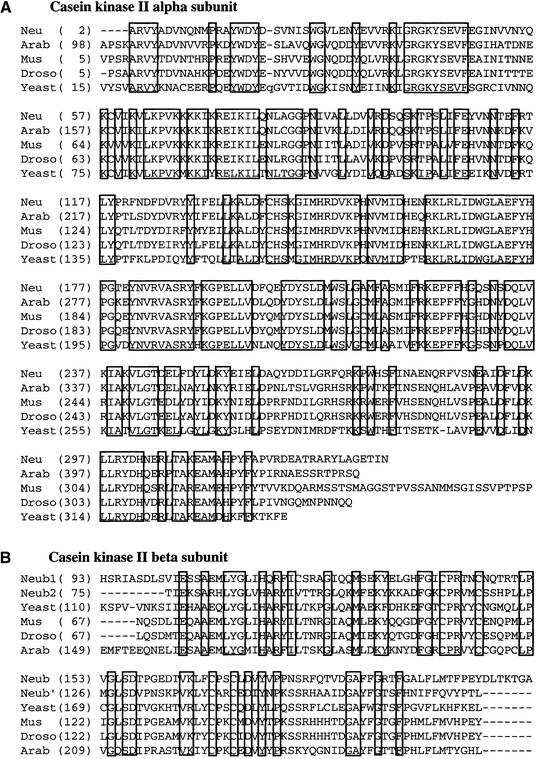
The predicted molecular weights for CKA, CKB1, and CKB2 are 39.6 kD (336 amino acids), 37.1 kD (333 amino acids), and 32.6 kD (285 amino acids), respectively, similar to their sizes estimated by SDS-PAGE (Fig. 1C). Therefore, the size of the CKII heterotetramer should be ∼150 kD, similar to the size estimated by the Superdex 200 elution profile. BLAST searches conducted with the amino acid sequences of the Neurospora CKII α and β subunits revealed that they were closely related to other eukaryotic CKII α or β subunits (Fig. 2). Interestingly, the closest homolog of the Neurospora CKA is a CKII α subunit from Arabidopsis thaliana, with 76% identity at the amino acid level spanning the entire ORF. The two Neurospora CKII β subunits are larger than most other CKII β subunits (23–30 kD), with an extended carboxyl terminus. CKB1 and CKB2 are most similar to two distinct CKII β subunits from fission yeast, with 45% and 59% amino acid sequence identity, respectively.
Figure 2.
Amino acid sequence alignment of the Neurospora CKII and other eukaryotic CKII. (A) Sequence alignment of the catalytic (α) subunits of CKII proteins. (Neu) Neurospora CKA; (Arab) Arabidopsis thaliana (AAC17823); (Mus) Mus musculus (AAA96795); (Droso) Drosophila melanogaster (P08181); (Yeast) Saccharomyces cerevisiae (NP_014704). The amino acids conserved in all proteins are boxed. (B) Sequence alignment of the regulatory (β) subunits of CKII proteins. (Neub1) Neurospora CKB1; (Neub2) Neurospora CKB2; (Yeast) Saccharomyces cerevisiae (NP_011496); (Mus) Mus musculus (NP_034105); (Droso) Drosophila melanogaster (P08182); (Arab) Arabidopsis thaliana (P40229).
Disruption of the cka gene revealed its important role in growth and development of Neurospora
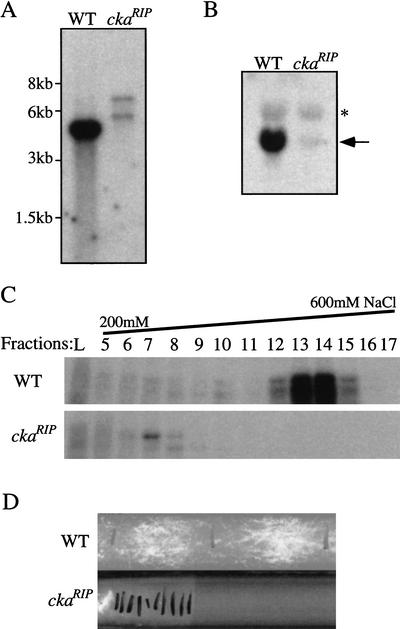
To investigate the function of the Neurospora CKII in phosphorylating FRQ in vivo, cka, the gene encoding for the catalytic subunit of the enzyme, was disrupted in Neurospora by repeat-induced point mutation (RIP) (Selker and Garrett 1988; Cambareri et al. 1989). To introduce random mutations to a target gene by RIP, the gene of interest is first duplicated in Neurospora by introducing an additional copy of this gene into a wild-type Neurospora strain. The resulting strain is then crossed with another wild-type strain. During sexual cycle, random but exclusively G-C to A-T point mutations will be introduced to the duplicated gene through an unknown gene-silencing mechanism. To mutate cka, a 2.5-kb genomic DNA containing the entire cka ORF and part of its promoter region was introduced into a wild-type strain. Afterward, a positive transformant was crossed with a wild-type strain, and individual sexual spores were picked and germinated. After the spores were germinated, we noticed that some of the progenies showed a slow growth rate. To determine whether mutations were introduced into cka in these strains, Southern blot analysis was performed. As shown in Figure 3A, the endogenous cka gene was disrupted in a RIP strain. Northern blot analysis further showed that cka expression was also mostly abolished in this mutant (Fig. 3B). DNA sequencing of the mutant cka gene found many G-C to A-T point mutations in the gene, including multiple premature stop codons after amino acid 9 (data not shown). Finally, the in vitro kinase assay results showed that the CKII kinase activity was also abolished in this mutant, confirming that CKA is the only CKII α subunit in Neurospora (Fig. 3C). Together, these results indicate that there are no transcription and translation of cka and no CKII kinase activity in the ckaRIP mutant.
Figure 3.
Disruption of the cka gene in Neurospora. (A) Southern blot analysis showing that the cka gene was disrupted in the ckaRIP strain. SacI was used to digest the genomic DNA. A 2.5-kb genomic DNA fragment containing the entire cka ORF was used as the probe. Because of the point mutations created by RIP, there is an additional SacI site in the mutated cka gene. The weaker signal in the mutatant was due to less DNA loaded. (B) Northern blot analysis showing that the expression of cka was mostly abolished in the ckaRIP strain. The position of the cka mRNA is indicated by the arrow. The asterisk denotes the large ribosomal RNA bands nonspecifically recognized by the probe. (C) In vitro kinase assay showing that the CKII activity was abolished in the ckaRIP strain. Protein extracts (5 mg) were loaded onto a Q-Sepharose column and eluted by the NaCl gradient indicated above. A wild-type strain was used as the control. (D) Race tube assay showing the slow growth phenotype of the ckaRIP strain. The cultures were first grown in LL for 2 d before being transferred into DD. The growth front of the culture was marked by a black line every 24 h.
The ckaRIP strains grew much more slowly than the wild-type strains, and they produced short aerial hyphae and little conidia. These phenotypes indicate that, although CKII is not essential for survival, it is important for the normal growth and development of Neurospora. To show the slow growth phenotype of the ckaRIP strains and to examine whether they exhibit circadian conidiation rhythm, they were analyzed by race tube assay in constant darkness. As shown in Figure 3D, the growth rate of the ckaRIP strain was <10% of the wild-type strain, 2–3 mm/day instead of 3–4 cm/day (note that each black line on the race tube represents one day's growth). Due to the slow growth and the meager production of conidia, the circadian phenotype of the ckaRIP strains could not be analyzed by the race tube assay. Our effort to identify partially functional cka allele was not successful, because all of the mutants obtained showed similar phenotype as described above and DNA sequencing revealed that they contained many mutations in the cka ORF, and some resulted in premature stop codons.
FRQ is hypophosphorylated and more stable in the ckaRIP strain
If CKII phosphorylates FRQ in vivo, we expected the phosphorylation profile of FRQ to be altered in the ckaRIP strains. In addition, if CKII phosphorylation of FRQ plays a role in triggering FRQ degradation, the steady-state level of FRQ should be high in the mutants. To test these hypotheses, Western blot analyses were performed to examine the FRQ phosphorylation profile for Neurospora cultures grown in constant light (LL) or in constant darkness (DD). Our data in Figure 4 show that the FRQ phosphorylation pattern was significantly altered in the ckaRIP strains under both conditions, and the levels of FRQ were also higher in the mutants than those of the wild-type strains. As shown in Figure 4A (DD24) and Figure 4B and C (containing LL cultures), the LFRQ and SFRQ proteins in the mutants were mostly hypophosphorylated, as indicated by their significantly faster mobilities in SDS–polyacrylamide gels than those of the wild-type FRQ. In fact, the fastest mobility species of both forms of FRQ were not found in the wild-type samples (Garceau et al. 1997), and the hyperphosphorylated LFRQ forms (indicated by the top arrow) seen in the wild-type strain were missing in the mutants (Fig. 4A,B,D). However, although FRQ was hypophosphorylated in the ckaRIP strains compared with the wild type, several phosphorylated FRQ species could be identified (Fig. 4A,B,D), indicating that at least one additional kinase phosphorylates FRQ in vivo.
Figure 4.
Western blot analyses showing that in the ckaRIP strain, the FRQ phosphorylation pattern was significantly altered, the FRQ level was higher and more stable, and FRQ protein oscillation was abolished. (A) Western blot analysis showing that in the ckaRIP strain, FRQ was mostly hypophosphorylated and its level was higher than that of the wild type. Cultures harvested in DD24 were used. (Left) The mobility range of the two alternatively translated FRQ forms is indicated at left. The top arrow indicates that the hyperphosphorylated LFRQ species found in the wild type was missing in the ckaRIP mutants. The bottom arrow indicates that the hypophosphorylated SFRQ species was found only in the ckaRIP strain. (Right) Densitometric analysis of the Western blot shown at left. (B,C) Western blot analysis showing that FRQ was more stable in the ckaRIP strain after a LD transition. Cultures were first grown in LL for 2 d before they were transferred into constant darkness and harvested at the indicated time. The top and bottom arrows at left of B indicate the hyperphosphorylated LFRQ bands in the wild-type strain and the hypophosphorylated SFRQ band in the ckaRIP strain, respectively. Densitometric analysis of the Western blot results of B is shown in C. (○) Wild type; (●) ckaRIP. (D,E) Western blot analysis showing that FRQ protein oscillation was abolished in the ckaRIP strain. Cultures were harvested in constant darkness at the indicated time. Densitometric analysis of the Western blot results in D is shown in E. Note that there was a circadian oscillation of the FRQ phosphorylation patterns in the wild type, whereas the FRQ phosphorylation pattern stayed unchanged in the ckaRIP strain at different times of the day after DD16. The representative results of three independent experiments are shown here. (○) Wild type; (●) ckaRIP.
In addition to the changes in FRQ phosphorylation profile, the amount of FRQ protein was greater in the ckaRIP strains than in the wild type under LL and DD conditions (Fig. 4A–E). At DD24, FRQ levels in the mutants were about twice as high as in the wild type (Fig. 4A). In LL (samples at 0 h in Fig. 4B–E were harvested in LL), the levels of FRQ were ∼50% higher in the mutants than in the wild type, despite the fact that similar amounts of frq mRNA were present in both strains (Fig. 5A,B, samples at 0 h). The increased amount of FRQ in the mutants is consistent with the notion that phosphorylation of FRQ triggers its degradation (Liu et al. 2000). To determine whether FRQ is more stable in the ckaRIP strain, the FRQ degradation rate was examined by measuring the changes of FRQ level following a light-to-dark transition, which has been shown to be a reliable method to compare FRQ stability (Liu et al. 2000; Gorl et al. 2001). LD transition leads to a rapid disappearance of frq RNA and degradation of FRQ protein. As in the wild-type strain, frq mRNA was also quickly degraded in the ckaRIP strain after the LD transition (Fig. 5A, second lane). As shown in Figure 4, B and C, FRQ in the wild-type strain was rapidly degraded after the LD transition, with ∼40% FRQ left after 8 h in the dark. In contrast, although FRQ protein level decreased in the mutant after the LD transition (also see Fig. 4D,E), its degradation rate was significantly slower than that of the wild type. After 8 h in the dark, ∼80% of FRQ was still left in the mutant. Slower FRQ degradation rate was also observed when a protein synthesis inhibitor (cycloheximide) was used (data not shown). These data suggest that phosphorylation of FRQ by CKII regulates the stability of FRQ.
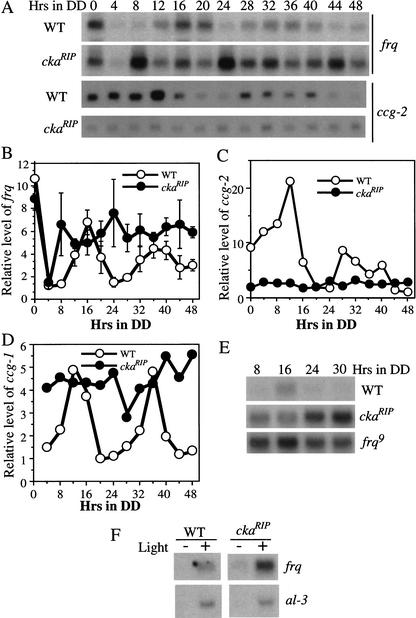
Figure 5.
Northern blot analyses showing that in the ckaRIP strain, the circadian oscillations of the levels of frq and clock-controlled genes were abolished, and the level of frq was high in constant darkness. (A) Cultures were harvested at the indicated time in constant darkness. The representative results of three independent Northern blot analyses of frq and ccg-2 mRNA in both wild-type and the ckaRIP strain were shown. (B–D) Densitometric analyses of the mRNA levels of frq, ccg-2, and ccg-1 under circadian cycles in the wild-type and the ckaRIP strains. In B, Northern blot results from three independent experiments described in A were used. Error bars, SD. (○) Wild type; (●) ckaRIP. (E) Northern blot analysis comparing the levels of frq in constant darkness in wild-type, ckaRIP, and frq9 strains. (F) Northern blot analysis showing that the light induction of frq and al-3 in the wild-type and ckaRIP strains. The cultures were harvested after 15 min of light treatment at DD24.
Circadian oscillation of FRQ is abolished in the ckaRIP strain
To examine whether the clock was still running in the ckaRIP mutant, liquid cultures of the mutant strain were grown in constant darkness and harvested at different times for two circadian cycles. Western blot analysis was performed to examine whether the level of FRQ showed circadian oscillation. In Figure 4, D and E show the results of one representative experiment. Similar to previously described results (Garceau et al. 1997), there was a robust circadian rhythm of the steady-state level of FRQ in the wild-type strain. In addition to the rhythm of FRQ amount, the phosphorylation states of FRQ also oscillated rhythmically in the wild-type strain, with FRQ being less phosphorylated at DD16–DD20 and extensively phosphorylated when its level decreased (Garceau et al. 1997). For the ckaRIP strain, although the FRQ level decreased slightly after the LD transition and then increased at DD16, the level of FRQ fluctuated very little afterwards. These data indicate that the ckaRIP strain can still respond to the LD transition, but there is no circadian oscillation of FRQ proteins. In addition, the level of FRQ protein is constantly high in the dark in the mutant, consistent with the notion that FRQ is more stable without phosphorylation by CKII. In contrast to the dramatic oscillations of FRQ phosphorylation states in the wild type, the FRQ phosphorylation pattern was the same at different time points after DD16 (Fig. 4D). These data indicate that the circadian oscillation of FRQ is abolished in the ckaRIP strain.
The levels of frq and clock-controlled genes do not change rhythmically and the amount of frq is high in constant darkness in the ckaRIP strain
Because the level of FRQ protein is constantly high in DD in the ckaRIP strain, we predicted that there should be no circadian oscillations of frq mRNA and clock-controlled genes. In addition, if the phosphorylation of FRQ by CKII affects FRQ's role to feedback to repress the transcription of frq, the amount of frq mRNA would be high in the mutant. To test these hypotheses, Northern blot analyses were performed to examine the mRNA levels of frq and the two best-studied clock-controlled genes (ccg-1 and ccg-2) (Loros et al. 1989; Bell-Pedersen et al. 1992, 1996) in the ckaRIP strain over two circadian cycles (Fig. 5). As shown in Figure 5, frq, ccg-1, and ccg-2 mRNA levels showed robust circadian rhythms in the wild-type strain. As expected, the expression of all of these genes did not cycle in the ckaRIP strain, indicating that the circadian clock was not functional in the mutant. Although the frq RNA level fluctuated, the fluctuation was random and not circadian, because the time of its peak and trough were not reproducible in other experiments (Fig. 5B). In addition, frq stayed at a high level in constant darkness (Fig. 5A,B,E). The high and randomly fluctuated frq level in constant darkness (Fig. 5A,B) is reminiscent of the frq level in the frq9 strain (Aronson et al. 1994a), a frq mutant that only produces a truncated form of FRQ protein (Aronson et al. 1994b). To compare the levels of frq directly among wild-type, ckaRIP and frq9 strains, a Northern blot experiment was performed for samples harvested in the dark (Fig. 5E). As shown in the figure, the levels of frq in the ckaRIP strain were found to be comparable with those in the frq9 strain. These data suggest that, although the FRQ protein level was high in the ckaRIP strain, it failed to inhibit its own transcription.
In contrast to the high levels of frq, ccg-2 in the ckaRIP strain stayed at a low level that was comparable with its trough level in the wild-type strain (Fig. 5A,C). This low level of ccg-2 was similar to what was found in some ccg-2 mutants in which the activating clock element (ACE) of the ccg-2 promoter was deleted (Bell-Pedersen et al. 1996), suggesting that the expression of ccg-2 has lost clock control in the ckaRIP mutant. Similar to the high levels of frq, the amounts of ccg-1 in the ckaRIP strain stayed at a high level at different time points in the dark, suggesting that the clock negatively regulates the level of ccg-1 (Fig. 5D). These data suggest that the circadian oscillator regulates ccg-1 and ccg-2 differently, even though they both are morning-specific genes. Together with the data in Figure 4, these data strongly suggest that FRQ phosphorylation by CKII is required for the function of the clock, and the hypophosphorylated FRQ proteins in the ckaRIP mutant are not fully functional. Therefore, the Neurospora CKII is an important component of the Neurospora circadian system.
To further show that the arrhythmic phenotype of the ckaRIP strain was not due to its inability to be entrained by light, we examined the light induction of frq and albino-3 (al-3) genes in the ckaRIP strain. As shown in Figure 5F, both frq and al-3 are rapidly induced by a 15-min light pulse in the mutant. This result suggests that CKII is not required for the light input of the clock.
Hypophosphorylated FRQ proteins in the ckaRIP mutant promote the complex formation between FRQ and the WC proteins
Although FRQ phosphorylation by CKII seems to destabilize FRQ, it is obvious that its phosphorylation also has additional roles in regulating the function of FRQ, because the hypophosphorylated FRQ in the ckaRIP mutant fails to close the circadian negative feedback loop, as reflected by the high levels of frq in constant darkness. It is possible that the lack of CKII-mediated phosphorylation of FRQ blocks the nuclear localization of FRQ (Komeili and O'Shea 1999), because it is an essential step for FRQ to function (Luo et al. 1998). To test this possibility, nuclei preparations from both the wild-type strain and the ckaRIP strain were examined. As seen in Figure 6A, the mutant FRQ was found in the nucleus like the wild-type FRQ. Thus, CKII is not required for the nuclear localization of FRQ. Similar to previously described results (Talora et al. 1999; Schwerdtfeger and Linden 2000; Cheng et al. 2001a), WC-1 and WC-2 were greatly enriched in the nuclear fractions in both strains.
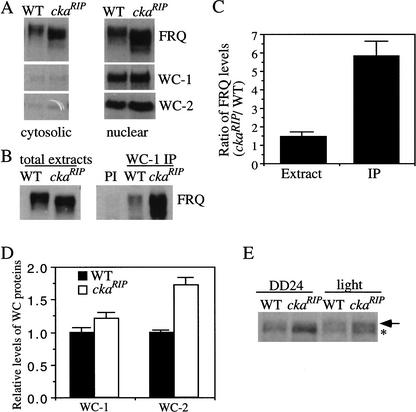
Figure 6.
The nuclear localization of FRQ was normal in the ckaRIP mutant, and significantly more of the hypophosphorylated FRQ in the ckaRIP strain were found to be associated with the WC proteins than FRQ in the wild-type strain. (A) Western blot analysis showing that FRQ in the ckaRIP mutant was found in the nucleus. Western blots were probed with FRQ, WC-1, and WC-2 antiserum. (B) Immunoprecipitation assay showing that more of the FRQ protein in the ckaRIP strain was found to coprecipitate with the WC proteins than the wild type. The Neurospora protein extracts (LL) were either immunoprecipitated by WC-1 antiserum (right) or analyzed directly (left) by Western blot analyses. (PI) Wild-type extracts were immunoprecipitated by WC-1 pre-immune serum. Note the similar FRQ phosphorylation pattern in wild-type and the ckaRIP strain after immunoprecipitation. (C) Densitometric analyses of the results from four independent immunoprecipitation experiments as described in B. Error bars, SD. (D) Comparison of the WC levels in the wild-type and the ckaRIP strains. The data is quantified from five independent experiments for cultures harvested in constant darkness (DD24 or DD28). (▪) Wild type; (□) ckaRIP. (E) Western blot analysis showing the comparison of the phosphorylation patterns of WC-1 in wild-type and ckaRIP strains. The light samples were harvested after a 15-min light pulse at DD24. To resolve the phosphorylated WC-1 bands, the electrophoresis time was twice as long as usual. The arrow indicates the hyperphosphorylated WC-1 band, whereas the asterisk marks the hypophosphorylated WC-1 band.
Another possible reason why FRQ fails to function in the ckaRIP mutant is that the phosphorylation of FRQ by CKII affects its interaction with the WC complex and therefore the negative feedback loop cannot be closed (Cheng et al. 2001a; Denault et al. 2001). To test this possibility, immunoprecipitation experiments using WC-1 or WC-2 antiserum were performed to compare the protein–protein interactions between FRQ and WC proteins in the wild-type and ckaRIP strains. The results of a representative experiment using WC-1 antiserum are shown in Figure 6, B and C. Although FRQ was found to be associated with WC-1 and WC-2 in both strains, significantly more FRQ was coprecipitated with WC-1 in the mutant, suggesting that the hypophosphorylated FRQ in the ckaRIP mutant either stabilizes or promotes the formation of the FRQ–WC complex. Similar results were obtained in four independent experiments with WC-1 antiserum and with WC-2 antiserum to perform immunoprecipitation (Fig. 6C; data not shown). In addition, we found that the wild-type FRQ that coprecipitated with the WCs lacked most of the hyperphosphorylated species (Fig. 6B, right; see also Fig. 6B in Cheng et al. 2001a). These results suggest that, even in the wild-type strain, the hypophosphorylated FRQ species are the major forms of FRQ that bind to the WC complex. Together, these data suggest that the phosphorylation of FRQ by CKII decreases FRQ's ability to interact with the WC proteins. Furthermore, these results suggest that the CKII phosphorylation of FRQ plays a role in closing the feedback loop after the binding of FRQ to the WC complex.
Besides its role in repressing its transcription, FRQ also positively regulates the levels of both WC-1 and WC-2 through different mechanisms (Lee et al. 2000; Cheng et al. 2001b; Merrow et al. 2001). To examine whether these positive feedback loops are still functional in the ckaRIP strain, the levels of WC-1 and WC-2 in the ckaRIP strain were compared with those of the wild-type strain. As shown in Figure 6D (cultures harvested in DD), the level of WC-1 in the mutant is slightly higher (∼20%) than that of the wild type, whereas the increase of WC-2 is ∼70% in the mutant. This data suggests that the hypophosphorylated FRQ is still able to positively regulate the levels of WC proteins and the high level of FRQ in the mutant is responsible for the increases of its WC levels. Because WC-1 is the limiting factor in the WC complex (Cheng et al. 2001b; Denault et al. 2001) and WC-1 is unstable without forming a complex with WC-2 (Cheng et al. 2002), the concentration of the WC complex in the ckaRIP strain should be ∼20% higher than that of wild type. Such an increase in WC complex is insufficient to explain the high levels of the frq mRNA, because there were five times more FRQ–WC complex found in the ckaRIP strain (Fig. 6C). Therefore, the high levels of frq in the ckaRIP strain, is likely due to the following: (1) FRQ cannot repress the activity of WC complex efficiently; (2) there is a higher level of WC complex in the mutant.
Because WC-1 is known to be phosphorylated, and as it becomes hyperphosphorylated after a light pulse, (Talora et al. 1999; Schwerdtfeger and Linden 2000), we examined whether the phosphorylation pattern of WC-1 was altered in the ckaRIP strain, WC-1 phosphorylation patterns of the ckaRIP strain in DD and after a light pulse were compared with those of the wild type. As shown in Figure 6E, the phosphorylation patterns of WC-1 were similar in both strains, and as in the wild type, the light treatment also resulted in the hyperphosphorylation of WC-1. Therefore, the phosphorylation of WC-1 did not appear to be altered in the CKII mutant. However, we do not know whether the activity of the WC complex is altered in the CKII mutant strain.
The RNA level of the catalytic subunit of CKII is constant under circadian conditions
Because the phosphorylation states of FRQ show a circadian rhythm in a wild-type strain, it is possible that the expression of CKII genes is controlled by a circadian clock. To test this hypothesis, Northern blot analysis was performed to examine whether the expression of cka is rhythmic in a wild-type strain over two circadian cycles. As shown in Figure 7, despite the robust circadian oscillation of frq mRNA, the level of cka mRNA appeared to be relatively constant at different times of the day. This result suggests that, like dbt in Drosophila (Kloss et al. 1998), the steady state level of cka is not clock controlled. However, we cannot rule out the possibility that the CKII activity and the cellular localization of CKII are regulated by the clock.
Figure 7.
Northern blot analysis showing that the level of cka was constant at different times of the day. (A) Wild-type cultures were harvested in DD at the indicated time. Northern blot analyses were performed to examine the expression of frq and cka genes. The representative results from two independent experiments are shown. (B) Densitometric analysis of the Northern blot results of A. (○) frq; (●) cka.
Discussion
In this study, we have biochemically purified the Neurospora CKII, a kinase that phosphorylates FRQ. Disruption of the catalytic subunit of CKII, cka, revealed that, like the previously identified Neurospora clock genes frq, wc-1, and wc-2, CKII has an essential role in the circadian system of Neurospora. The arrhythmic expressions of frq and clock-controlled genes and the levels of the mRNA of these genes in the ckaRIP mutant are consistent with lack of clock function in this mutant. To our knowledge, this is the first essential clock component identified by biochemical purification in any system.
Phosphorylation of FRQ by CKII in vivo
Previously, we purified a Ca/CaM-dependent kinase (CAMK-1) from Neurospora (Yang et al. 2001). Due to the reversion of the camk-1 knock-out strains and the redundancy of the Ca/CaM-dependent kinases in Neurospora (two additional Ca/CaM-dependent kinases were identified in the Neurospora genome database), we could not make a firm conclusion on whether CAMK-1 plays an important role in phosphorylating FRQ in vivo.
In contrast, evidence herein strongly suggest that CKII phosphorylates FRQ in vivo and such phosphorylation is important for the normal function of the circadian clock. The disruption of cka, the only catalytic subunit of Neurospora CKII, resulted in dramatic changes in the FRQ phosphorylation pattern under different conditions. FRQ was mostly hypophosphorylated in the mutant strain, and some of the hypophosphorylated FRQ species were never found in the wild-type strains. Previously, it was shown that even the least-phosphorylated FRQ species in the wild type was phosphorylated (Garceau et al. 1997), suggesting that as soon as FRQ is synthesized, it is immediately phosphorylated. We have shown that the purified endogenous CKII can directly phosphorylate FRQ in vitro. In combination with the changes of the FRQ phosphorylation profile in the ckaRIP mutant, these data suggest that CKII can also directly phosphorylate FRQ in vivo. However, as we do not have the evidence to show the direct interaction of CKII and FRQ in vivo (the interactions between the kinase and its substrates are usually weak and transient), we cannot exclude the possibility that the changes in FRQ phosphorylation pattern in the ckaRIP mutant are due to indirect regulation of FRQ phosphorylation by CKII.
Using the Motif Scanner protein phosphorylation prediction software, we found that there are at least 29 potential primary CKII phosphorylation sites on FRQ. Although the purified CKII was able to phosphorylate the small region of FRQ containing Ser 501 to Ser 519 in vitro (data not shown), the comparison of the known CKII consensus sequence, (S/T)XX(D/E/Tp/Sp), with the sequences of these three known FRQ phosphorylation sites showed that they do not resemble the typical CKII phosphorylation sites. Thus, it is unclear whether the phosphorylation of this region by CKII in vitro is physiologically relevant in vivo. Our future identification of more FRQ phosphorylation sites and the analyses of their function should inform us about the function of the specific phosphorylation sites in regulating FRQ.
Despite the dramatic differences in the FRQ phosphorylation profile between the wild-type and the mutant strains, several FRQ phosphorylation species can still be identified in the ckaRIP mutant. These data indicate that at least one additional kinase can still phosphorylate FRQ in vivo. However, it is unclear whether the FRQ phosphorylation pattern in the ckaRIP mutant is due to phosphorylation of the CKII-unrelated sites or a slower FRQ phosphorylation rate in the mutant. It is possible that CAMK-1 is one of the kinases that phosphorylates FRQ in the ckaRIP mutant. Unfortunately, our efforts to obtain the double mutant (camk-1ko, ckaRIP) failed, most likely because the disruption of both genes is lethal.
A recent study by Gorl and colleagues has suggested that casein kinase I (CKI) is involved in regulating the stability of FRQ by phosphorylating one of the PEST domains of FRQ (Gorl et al. 2001). The deletion of the PEST domain resulted in increased FRQ stability and long period rhythm (∼28 h) of the clock. However, CKI failed to phosphorylate the previously identified FRQ phosphorylation sites. Interestingly, both CKI and CKII are Ser/Thr kinases that phosphorylate Ser/Thr surrounded by acidic residues. Therefore, it is possible that both kinases work together to phosphorylate FRQ.
FRQ phosphorylation has multiple roles
We have shown that in the ckaRIP mutant, the levels of FRQ protein, and frq mRNA are high and not rhythmic. In addition, there is no clock-controlled expression of clock-controlled genes. Interestingly, in constant darkness, the levels of ccg-2 was constantly low, whereas ccg-1 stayed at a high level in the ckaRIP mutant, suggesting that the clock regulation on these two genes are different despite the fact that they are both morning-specific genes. These data suggest that the clock function is disrupted in the mutant, and that CKII-mediated phosphorylation of FRQ is essential for the function of the FRQ in the clock.
A major conclusion of this study is that FRQ phosphorylation by CKII has several important roles in regulating the function of FRQ in the circadian clock (Fig. 8). First, consistent with previous results (Liu et al. 2000), phosphorylation of FRQ by CKII is important in regulating the stability of FRQ. We have shown that the FRQ level is higher and more stable in the ckaRIP mutant than in the wild type. Thus, like phosphorylation of PER by DBT in Drosophila (Price et al. 1998; Suri et al. 2000), the phosphorylation of FRQ by CKII may be important in determining the period length and the normal function of the clock. The constitutive high levels of FRQ in constant darkness are likely the result of increased stability of FRQ and high levels of frq mRNA.
Figure 8.
Model for gene regulation within the Neurospora circadian oscillator. WC-1 and WC-2 form heterodimer to activate the transcription of frq. FRQ proteins interact with the WC-1/WC-2 complex to inhibit their transcriptional activation, forming the negative feedback loop. FRQ also positively regulate the levels of both WC-1 and WC-2, forming the positive feedback loops. The phosphorylation of FRQ have multiple roles in regulating the function of FRQ: promoting degradation of FRQ, reducing the interaction between FRQ and the WC complex, and important for the negative repression of frq transcription. WC-2 maintains the steady state of WC-1 by forming complex with WC-1 (Cheng et al. 2002).
Second, the phosphorylation of FRQ by CKII plays important roles in regulating the formation of the FRQ–WC complex and the closing the circadian negative feedback loop. We have shown that not only FRQ in the ckaRIP mutant can interact with the WC complex, but the amount of FRQ coprecipitated with the WC proteins is significantly greater than that of the wild-type FRQ. Even in a wild-type strain, the less phosphorylated FRQ forms are the major species found to be associated with the WC complex. In wild type, beside the oscillation of the level of FRQ, the phosphorylation pattern of FRQ also changes dramatically in a circadian fashion in constant darkness. Thus, the different ability for various FRQ-phosphorylated forms to interact with the WC complex is a mechanism that increases the amplitude of FRQ rhythm and results in robust oscillation of the clock, and the amplitude of the FRQ protein levels alone is not a complete reflection of the amplitude of the rhythm. Therefore, both the quantity and quality of different forms of FRQ should be taken into account when considering the amounts of FRQ protein.
Despite more complex formation between FRQ and the WCs in the ckaRIP mutant, the frq mRNA levels are high in constant darkness and are comparable with those in the frq9 strain. Although the concentration of the WC complex is ∼20% higher in ckaRIP mutant than wild type (Fig. 6D), such an increase is insufficient to explain the increase of the frq RNA level if the negative feedback is normal in the mutant, as there are about five times more of FRQ–WC complex found in the ckaRIP mutant (Fig. 6C). In fact, these results suggested that the circadian negative feedback loop was impaired in the CKII mutant and FRQ failed to inhibit the transcriptional activation activity of the WC complex effectively in the CKII mutant. Although the interaction between FRQ and the WCs is necessary for the closing of the circadian negative feedback loop (Cheng et al. 2001a), these data suggest that the interaction alone is not sufficient for FRQ to act to repress the transcriptional activation of frq by the WC proteins. In addition, these data suggest that the interaction between FRQ and the WC proteins and the transcriptional repression are two separate events, and the lack of CKII phosphorylation of FRQ at certain sites in the ckaRIP mutant prohibits FRQ from carrying out its function. However, it is also possible that the these two aspects of the FRQ phosphorylation function are related to each other.
CKII (CK2) is also involved in the function of the Arabidopsis circadian clock (Sugano et al. 1998, 1999). In vitro, CKII subunits of Arabidopsis have been shown to interact and phosphorylate the circadian clock-associated 1 (CCA1) protein. The phosphorylation of CCA1 by CKII-like activity in vitro affects the formation of a DNA–protein complex containing CCA1. In addition, overexpression of a regulatory subunit of CKII resulted in the shortening of the periods of clock-controlled genes in Arabidopsis. Because the Neurospora WC complex binds to DNA and FRQ interacts with the WCs, it is possible that the CKII phosphorylation of FRQ influences the formation of the DNA–WC complex.
In Drosophila, the phosphorylation of TIM appears to have at least two different functions. The phosphorylation of TIM by SGG promotes the nuclear transfer of the PER/TIM complex (Martinek et al. 2001), whereas a tyrosine-linked phosphorylation of TIM by an unknown kinase is implicated in the proteasome-mediated degradation of TIM (Naidoo et al. 1999). DBT of Drosophila and the casein kinase Iε in mammals phosphorylate PER and may trigger its turnover (Price et al. 1998; Lowrey et al. 2000; Toh et al. 2001). Although both CKI and CKII are Ser/Thr kinases, their structures and substrate specificities are significantly different, suggesting that they did not evolve from a common ancestral protein.
Clock effects versus growth and developmental effects
Unlike DBT and SGG in Drosophila (Price et al. 1998; Martinek et al. 2001) and CKII in Saccharomyces (Padmanabha et al. 1990), which are required for the survival of the organisms, CKII of Neurospora is not essential for the survival of Neurospora. However, the Neurospora CKII is important for the normal growth and development of the fungus. As a protein serine/threonine kinase that is ubiquitously found in eukaryotes, CKII is known to phosphorylate many substrates and regulate many processes in eukaryotic cells (Litchfield and Luscher 1993; Pinna and Meggio 1997). However, in Neurospora, we think the clock defects in the ckaRIP mutant are probably not caused by its growth and developmental defects. First, in genetic screening for clock mutants by us and others, most of Neurospora growth and developmental mutants were found to have normal functional clocks (Brody and Martin 1973; Feldman and Atkinson 1978; Feldman 1983; Lakin-Thomas et al. 1990; Bell-Pedersen et al. 1992). A partial list of the biochemical and morphological mutants screened for clock effects is reviewed by Lakin-Thomas et al. (1990). From the 85 mutants examined, only 12 mutants were found to have clock phenotypes. Of the 12 mutants, 11 had small period shortening effects on the clock (<2–3 h) and only one mutant, cel, which is deficient in fatty acid synthetase, showed fatty acid-dependent period-lengthening effects (Mattern et al. 1982). Importantly, none of the mutant examined was found to exhibit arrhythmic phenotype, and all of the morphological mutants (the ckaRIP mutant should be categorized as one), in which some of them have severe growth and developmental defects, were found to have normal period length of the clock. The 73 mutations with normal period affect a wide range of metabolic reactions, including amino acid synthesis and protein translation, lipid and vitamin synthesis, mitochrondrial metabolism, carbon and nitrogen assimilation, as well as the morphology and production of mycelial and conidia. Therefore, there is no direct link between the general cell metabolism and the function of the clock in Neurospora.
Second, it was well known that the growth rate of Neurospora is not an influencing factor in the function of the clock. For example, the growth rate of the wild-type strain can vary significantly dependent on the type and concentration of the carbon source used, however, the function of the clock has been shown to be similar under different conditions (Dharmananda 1980; Nakashima 1981; Loros et al. 1989; Aronson et al. 1994a; Cheng et al. 2001b). Third, as we have shown in this study, the molecular phenotypes of CKII is very similar to those of the frq null stain, as suggested by the high levels of frq and ccg-1 and the low levels of ccg-2 in constant darkness (Fig. 5). These results suggest that the loss of clock function in the cka mutant appears to be the result of loss of the fully functional FRQ protein. Finally, in this study, CKII was identified as a potential clock component due to its role of phosphorylating FRQ, one of the central components of the Neurospora clock, in vitro and in vivo. Based on these data, we think that the role of CKII in the Neurospora circadian oscillator may be unrelated to its function in cell growth and development. However, we cannot rule out the possibility that the growth and developmental defects indirectly contributed to the clock phenotypes we observed in the ckaRIP strains. We also do not know whether CKII can affect the clock by regulating the activity of other unknown clock genes in Neurospora.
In conclusion, we have identified Neurospora CKII as a kinase that phosphorylates FRQ. The evidence presented here suggests that CKII phosphorylation of FRQ regulates several aspects of the FRQ function, and that CKII is an important component of the Neurospora circadian clock.
Materials and methods
Strains and culture conditions
The bd, a and bd, his-3, a (wild-type clock) strains were used as the wild-type strains in this study. Protein extracts from bd, camk-1ko were used for CKII purification. The 301-6 (bd, his-3, A) strain was the host strain for the his-3-targeting construct. Liquid culture conditions were the same as described previously (Aronson et al. 1994a). Race tube assay medium contained 1× Vogel's, 0.1% glucose, 0.17% arginine, 50 ng/mL biotin, and 1.5% agar. The genotype of the ckaRIP strain was bd, ckaRIP, his-3. Therefore, histidine (0.5 mg/mL) was added to the medium in all experiments (including the medium for the wild-type strain) when the ckaRIP strain was used. To obtain similar amounts of mycelium from liquid cultures of both the wild-type and the ckaRIP strains, the ckaRIP cultures were first grown for about 2 d before the wild-type cultures were innoculated.
In vitro kinase assay
To assay kinase activity, a GST–FRQ fusion protein (Yang et al. 2001) (5 μg) containing FRQ amino acids 425–683 was incubated with Neurospora protein extracts (5–10 μg of total extracts or 40 μL of the chromatographic fractions) in a buffer containing 25 mM HEPES (pH 7.9), 10 mM MgCl, 2 mM MnCl2, 25 μM ATP, and 10 μCi/ml[γ-32P]ATP (total reaction volume of 125 μL). The reaction was incubated at room temperature for 1 h before adding 0.5 mL of PBS and 10 μL of glutathione–agarose beads. After a 30-min incubation at room temperature, the glutathione–agarose beads were washed twice in PBS before they were resuspended and boiled in 1× SDS–polyacrylamide loading buffer. The samples were then subjected to SDS-PAGE. After electrophoresis, the gel was dried and subjected to autoradiography.
Purification of CKII from Neurospora extracts
All procedures were performed on an automatic fast protein liquid chromatography (FPLC) station (Amersham-Phamacia) at 4°C. Neurospora protein extracts (1.5 g) of the camk-1ko strain in 50 mM HEPES (pH 7.4), 137 mM NaCl, and 10% glycerol were applied on a Q-Sepharose column (80 mL bed volume) equilibrated with buffer A (50 mM Tris at pH 7.5, 1 mM DTT). After washing with buffer A, the bound materials on the column were eluted with a 800-mL linear gradient from 20 mM NaCl to 500 mM NaCl in buffer A. Fractions of 9 mL were collected and assayed for kinase activity. A total of 50 mL of active fractions (∼0.45 M NaCl) were pooled together and loaded on a hydroxyapatite column (7-mL bed volume) equilibrated with 10 mM patassium phosphate buffer (KPO4). The column was washed with 10 mM KPO4, followed by 10 mM KPO4 containing 1 M NaCl. The bound proteins were eluted with a 15-mL linear gradient from 50 mM KPO4 to 350 mM KPO4 (pH 7.5). A total of 1.5 mL of active fractions (0.24–0.34 M KPO4) were pooled together and concentrated to 0.5 mL. At this time, it was loaded on a Superdex 200 (10/30) gel filtration column equilibrated with buffer A containing 150 mM NaCl and eluted with the same buffer. A total of 2 ml of active fractions were pooled and loaded on a Mono Q (5/5) column and eluted with a 10-mL linear gradient from 300 mM NaCl to 700 mM NaCl in buffer A. Fractions of 0.5 mL were collected and assayed for kinase activity. Active fractions were stored at −80°C.
Protein sequencing
Proteins in fraction 18 of the final Mono Q column were concentrated by Centricon spin column (Amicon) and loaded onto a 10% SDS–polyacrylamide gel. The gel was Coomassie stained, and the three protein bands correlated with the kinase activity were excised from the gel and subjected to tryptic digestion. A 1-μL aliquot from the digest was then used for peptide mass fingerprinting and mass values were searched against the NCBI database with the program MS-Fit.
Disruption of cka in Neurospora
A 2.5-kb PCR fragment containing the entire CKA ORF and part of the promoter was cloned into pDE3dBH (Ebbole and Sachs 1990) and introduced into the his-3 locus of a wild-type strain (301-6: bd, his-3, A) by transformation. Southern blot analysis was performed to identify transformants that carried the additional copy of cka, and a positive transformant was crossed with a wild-type strain (bd, his-3, a). Sexual spores of the cross were picked individually and germinated on slants containing histidine. Southern and Northern blot analyses were performed to identify strains in which cka was disrupted and cka was not expressed.
Protein and RNA analyses
Protein extraction, quantification, Western blot analysis, and immunoprecipitation assays were performed as described previously (Garceau et al. 1997; Cheng et al. 2001a). Neurospora nuclei were purified as described previously (Luo et al. 1998). Immunoprecipitates or equal amounts of total protein (40 μg) were loaded in each protein lane.
RNA extraction and Northern blot analysis were performed as described previously (Aronson et al. 1994a). Equal amounts of total RNA (20 μg) were loaded onto agarose gels for electrophoresis, the gels were blotted and probed with an RNA probe specific for frq, cka, and clock-controlled genes.
Acknowledgments
We thank Lixing Wang and Weiming Rong for excellent technical assistance. We thank Drs. James Stull, Stephen Hammes, Carl Johnson, and Duojia Pan for critical reading of the manuscript. Supported by a grant from the National Institute of Health (GM 62591) to Y.L. Y.L. is an Louise W. Kahn endowed scholar in Biomedical Research at The University of Texas Southwestern Medical Center.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Yi.Liu@UTsouthwestern.edu; FAX (214) 648-7891.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.965102.
References
- Aronson B, Johnson K, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: Autoregulation in the clock gene frequency. Science. 1994a;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- Aronson BD, Johnson KA, Dunlap JC. The circadian clock locus frequency: A single ORF defines period length and temperature compensation. Proc Natl Acad Sci. 1994b;91:7683–7687. doi: 10.1073/pnas.91.16.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D, Dunlap JC, Loros JJ. The Neurospora circadian clock-controlled gene, ccg-2, is allelic to eas and encodes a fungal hydrophobin required for formation of the conidial rodlet layer. Genes & Dev. 1992;6:2382–2394. doi: 10.1101/gad.6.12a.2382. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Dunlap JC, Loros JJ. Distinct cis-acting elements mediate clock, light, and developmental regulation of the Neurospora crassa eas (ccg-2) gene. Mol Cell Biol. 1996;16:513–521. doi: 10.1128/mcb.16.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody D, Martin S. Effects of morphological and auxotrophic mutations on the circadian rhythms of Neurospora crassa. Genetics. 1973;74:s31. [Google Scholar]
- Cambareri EB, Jensen BC, Schabtach E, Selker EU. Repeat-induced G-C to A-T mutations in Neurospora. Science. 1989;244:1571–1575. doi: 10.1126/science.2544994. [DOI] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Heintzen C, Liu Y. Coiled-coil domain mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. EMBO J. 2001a;20:101–108. doi: 10.1093/emboj/20.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Liu Y. Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc Natl Acad Sci. 2001b;98:7408–7413. doi: 10.1073/pnas.121170298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Gardner KH, Liu Y. PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol Cell Biol. 2002;22:517–524. doi: 10.1128/MCB.22.2.517-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite SK, Dunlap JC, Loros JJ. Neurospora wc-1 and wc-2: Transcription, photoresponses, and the origins of circadian rhythmicity. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Stankis D, Gekakis N, Steeves T, Weitz CJ, Takahashi J, Kay SA. Closing the circadian loop: CLOCK induced transcription of its own inhibitors, per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Denault DL, Loros JJ, Dunlap JC. WC-2 mediates WC-1-FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora. EMBO J. 2001;20:109–117. doi: 10.1093/emboj/20.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmananda S. PhD thesis, Studies of the circadian clock of Neurospora crassa: Light induced phase shifting. Santa Cruz: University of California; 1980. [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Ebbole D, Sachs M. Targeting chromosomal insertion to the his-3 locus. Fung Genet News. 1990;37:15–16. [Google Scholar]
- Edery I, Zweibel L, Dembinska M, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JF. Genetics of circadian clocks. BioScience. 1983;33:426–431. [Google Scholar]
- Feldman JF, Atkinson CA. Genetic and physiological characterization of a slow growing circadian clock mutant of Neurospora crassa. Genetics. 1978;88:255–265. doi: 10.1093/genetics/88.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garceau N, Liu Y, Loros JJ, Dunlap JC. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- Gorl M, Merrow M, Huttner B, Johnson J, Roenneberg T, Brunner M. A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J. 2001;20:7074–7084. doi: 10.1093/emboj/20.24.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Anderson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Williams SB, Kitayama Y, Ishiura M, Golden SS, Kondo T. A kaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Komeili A, O'Shea EK. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Lakin-Thomas P, Coté G, Brody S. Circadian Rhythms in Neurospora: biochemistry and genetics. Crit Rev Micro. 1990;17:365–416. doi: 10.3109/10408419009114762. [DOI] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: A basis for circadian transcription. Mol Cell Biol. 1999;19:5316–5325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lee K, Loros JJ, Dunlap JC. Interconnected feedback loops in the Neurospora circadian system. Science. 2000;289:107–110. doi: 10.1126/science.289.5476.107. [DOI] [PubMed] [Google Scholar]
- Litchfield DW, Luscher B. Casein kinase II in signal transduction and cell cycle regulation. Mol Cell Biochem. 1993;127–128:187–199. doi: 10.1007/BF01076770. [DOI] [PubMed] [Google Scholar]
- Liu Y, Garceau N, Loros JJ, Dunlap JC. Thermally regulated translational control mediates an aspect of temperature compensation in the Neurospora circadian clock. Cell. 1997;89:477–486. doi: 10.1016/s0092-8674(00)80228-7. [DOI] [PubMed] [Google Scholar]
- Liu Y, Loros J, Dunlap JC. Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc Natl Acad Sci. 2000;97:234–239. doi: 10.1073/pnas.97.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros JJ, Dunlap JC. Genetic and molecular analysis of circadian rhythms in NEUROSPORA. Annu Rev Physiol. 2001;63:757–794. doi: 10.1146/annurev.physiol.63.1.757. [DOI] [PubMed] [Google Scholar]
- Loros JJ, Denome SA, Dunlap JC. Molecular cloning of genes under the control of the circadian clock in Neurospora. Science. 1989;243:385–388. doi: 10.1126/science.2563175. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Loros JJ, Dunlap JC. Nuclear localization is required for function of the essential clock protein FREQUENCY. EMBO J. 1998;17:1228–1235. doi: 10.1093/emboj/17.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- Mattern DL, Forman LR, Brody S. Circadian rhythms in Neurospora crassa: A mutation affecting temperature compensation. Proc Natl Acad Sci. 1982;79:825–829. doi: 10.1073/pnas.79.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow M, Franchi L, Dragovic Z, Gorl M, Johnson J, Brunner M, Macino G, Roenneberg T. Circadian regulation of the light input pathway in Neurospora crassa. EMBO J. 2001;20:307–315. doi: 10.1093/emboj/20.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N, Song W, Hunter-Ensor M, Sehgal A. A role for the proteasome in the light response of the timeless clock protein. Science. 1999;285:1737–1741. doi: 10.1126/science.285.5434.1737. [DOI] [PubMed] [Google Scholar]
- Nakashima H. A liquid culture system for the biochemical analysis of the circadian clock of Neurospora. Plant Cell Physiol. 1981;22:231–238. [Google Scholar]
- Padmanabha R, Chen-Wu JL, Hanna DE, Glover CV. Isolation, sequencing, and disruption of the yeast CKA2 gene: Casein kinase II is essential for viability in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4089–4099. doi: 10.1128/mcb.10.8.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna LA, Meggio F. Protein kinase CK2 (“casein kinase-2”) and its implication in cell division and proliferation. Prog Cell Cycle Res. 1997;3:77–97. doi: 10.1007/978-1-4615-5371-7_7. [DOI] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Adodeely M, Kloss B, Young MW. double-time is a new Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Schwerdtfeger C, Linden H. Localization and light-dependent phosphorylation of white collar 1 and 2, the two central components of blue light signaling in Neurospora crassa. Eur J Biochem. 2000;267:414–422. doi: 10.1046/j.1432-1327.2000.01016.x. [DOI] [PubMed] [Google Scholar]
- Selker EU, Garrett PW. DNA sequence duplications trigger gene inactivation in Neurospora crassa. Proc Nat Acad Sci. 1988;85:6870–6874. doi: 10.1073/pnas.85.18.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF, Jr, Reppert SM. Two period homologs: Circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- Sugano S, Andonis C, Green R, Wang ZY, Tobin EM. Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci. 1998;95:11020–11025. doi: 10.1073/pnas.95.18.11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Ong MS, Green RM, Tobin EM. The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc Natl Acad Sci. 1999;96:12362–12366. doi: 10.1073/pnas.96.22.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri V, Hall JC, Rosbash M. Two novel doubletime mutants alter circadian properties and eliminate the delay between RNA and protein in Drosophila. J Neurosci. 2000;20:7547–7555. doi: 10.1523/JNEUROSCI.20-20-07547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talora C, Franchi L, Linden H, Ballario P, Macino G. Role of a white collar-1-white collar-2 complex in blue-light signal transduction. EMBO J. 1999;18:4961–4968. doi: 10.1093/emboj/18.18.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- Yang Y, Cheng P, Zhi G, Liu Y. Identification of a calcium/calmodulin-dependent protein kinase that phosphorylates the Neurospora circadian clock protein FREQUENCY. J Biol Chem. 2001;276:41064–41072. doi: 10.1074/jbc.M106905200. [DOI] [PubMed] [Google Scholar]
- Young MW. Molecular control of circadian behavioral rhythms. Recent Prog Horm Res. 1999;54:87–94. [PubMed] [Google Scholar]
- Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]