Abstract
The Caenorhabditis elegans hermaphrodite nervous system is composed of 302 neurons that fall into at least 118 diverse classes. Here we describe cfi-1, a gene that contributes to the development of neuronal diversity. cfi-1 promotes appropriate differentiation of the URA sensory neurons and inhibits URA from expressing the male-specific CEM neuronal fate. The UNC-86 POU homeodomain protein is present in CEM and URA neurons, and can promote expression of CEM-specific genes in both CEM and URA, but CFI-1 inhibits expression of these genes in the URA cells. cfi-1 also promotes appropriate differentiation and glutamate receptor expression in the AVD and PVC interneurons. cfi-1 encodes a conserved neuron- and muscle-restricted DNA-binding protein containing an A/T rich interaction domain (ARID). ARID proteins regulate early patterning and muscle fate in Drosophila, but they have not previously been implicated in the control of neuronal subtype identity.
Keywords: cfi-1, cell fate, ARID, CEM neurons, programmed cell death, C. elegans
In the mammalian nervous system thousands of cell types can be distinguished based on position, gene expression, connectivity, and function. Even in the nematode Caenorhabditis elegans, where the nervous system is composed of only 302 neurons, these can be divided into at least 118 different classes (Sulston and Horvitz 1977; Sulston et al. 1983; White et al. 1986). How is this multitude of neuron types generated? Studies in mammals and in C. elegans suggest that lineage-intrinsic mechanisms are important for the generation of this diversity. Transcriptional regulators that are induced during early development play key roles in determining neuronal subtype identity (Sengupta and Bargmann 1996; Tanabe and Jessell 1996), and are expressed in complex, overlapping patterns.
The expression of a particular repertoire of neuron-specific proteins results from the integration of functions of multiple transcription factors. In one well-characterized example, the hermaphrodite-specific neuron (HSN) of C. elegans, which regulates egg-laying, integrates inputs from the position of the cell, the sex of the animal, and its developmental state to achieve its terminally differentiated fate. These cues control HSN differentiation by regulating the expression of an assortment of specific transcriptional regulators. The HSN is born in the tail region of the animal (Sulston et al. 1983), and during embryogenesis its identity is specified, in part, by the tail-restricted homeodomain protein EGL-5 (Desai et al. 1988; Chisholm 1991). Appropriate expression of EGL-5 then allows the HSN to migrate from its early posterior position to its final medial position in the animal. At roughly the same time, the sex-determination pathway, in the form of the zinc-finger transcription factor TRA-1, regulates expression of the programmed cell death gene egl-1 to promote HSN survival in hermaphrodites and death in males (Conradt and Horvitz 1999). In the fourth larval stage (L4), inputs from genes that control developmental timing, such as the period-related gene lin-42, promote HSN terminal differentiation. These genes control the onset of synthesis of the neurotransmitter serotonin (Slack and Ruvkun 1997; Jeon et al. 1999) and acquisition of a unique nuclear morphology (Desai et al. 1988). Establishment of HSN fate is also controlled by the neurogenic proteins UNC-86 (a POU homeodomain protein; Finney et al. 1988) and LIN-32 (a helix–loop–helix transcriptional regulator; Zhao and Emmons 1995), which regulate many neural fates in C. elegans. UNC-86 and LIN-32 act in concert with other transcription factors, such as the EGL-43 and EGL-46 zinc-finger proteins and the EGL-44 transcription enhancer factor (TEF) protein (Garriga et al. 1993; Wu et al. 2001).
The four C. elegans CEM neurons are male-specific head neurons with morphological features suggesting a chemosensory function, perhaps in regulating male-specific behavior or attraction to hermaphrodites (Ward et al. 1975; Sulston and Horvitz 1977). The development of these cells shares some features with the development of the HSNs. For example, the CEMs migrate: they are born in the anterior of the animal and receive cues instructing them to migrate to their final, more posterior positions (Sulston et al. 1983). CEMs also undergo programmed cell death in a sex-specific manner. However, in contrast to the HSN neurons, CEMs survive in males and die in hermaphrodites (Sulston and Horvitz 1977). In addition, during the transition from the L4 stage to the adult, the CEMs acquire a prominent nucleolus (Sulston and Horvitz 1977) and begin to express the ion-channel proteins PKD-2 and LOV-1 (Barr and Sternberg 1999; Barr et al. 2001).
We have taken a genetic approach to ask how developmental cues are integrated to generate CEM-specific sensory fates. We show that the unc-86 and lin-32 genes are important for the expression of CEM-specific genes, whereas a transcription factor, CFI-1 (CEM fate inhibitor), represses the expression of CEM markers in some hermaphrodite neurons. CFI-1 acts in several neurons to affect their fates, both by promoting correct neuronal differentiation and by repressing expression of inappropriate markers. Thus, cfi-1 acts as an important element in the integration and consolidation of specific cell fates.
Results
The cfi-1 gene inhibits expression of CEM features in other C. elegans neurons
To determine how the CEMs acquire their unique characteristics, we performed a genetic screen for genes that affect CEM development and survival in hermaphrodites. In wild-type animals, a pkd-2::GFP (green fluorescent protein) transgene is not expressed in hermaphrodites but is expressed in the CEM neurons and in sensory tail neurons of males (Barr and Sternberg 1999). pkd-2::GFP hermaphrodites were mutagenized and allowed to self-fertilize for two generations. Rare F2 hermaphrodites containing GFP-positive cells in the head were then isolated and used to establish candidate mutant lines. From a screen of 9500 genomes we obtained 25 mutants. Such mutants could harbor mutations in genes regulating programmed cell death or sex determination; at least 18 mutants fell into this category and will not be discussed further. Alternatively, mutants expressing GFP could have defects in cell fate genes required to prevent the expression of CEM characteristics in other cells. Two mutants, ky650 and ky651, fell into this category and define a gene we named cfi-1.
ky650 and ky651 are recessive alleles of the same gene. Whereas wild-type hermaphrodites never expressed pkd-2::GFP, ky650 and ky651 animals contained 1.9 ± 0.6 and 3.1 ± 1.0 pkd-2::GFP-expressing cells, respectively (Table 1), and ky651/+ or ky650/+ heterozygotes contained none. ky650/ky651 heterozygous animals contained 2.5 ± 0.9 GFP-expressing cells (n = 20), thus failing to complement. ky651 defects were not enhanced in trans to the genetic deficiency qDf5, suggesting that ky651 is a strong loss-of-function or null allele.
Table 1.
Ectopic neurons expressing pkd-2::gfp in cfi-1 mutants are not surviving CEMs
| Genotype
|
Sex
|
No. cells expressing pkd-2::gfp
|
n
|
|---|---|---|---|
| Wild type | Hermaphrodite | 0 ± 0 | 20 |
| Wild type | Male | 4.0 ± 0.2 | 20 |
| cfi-l(ky651) | Hermaphrodite | 3.1 ± 1.1 | 20 |
| cfi-l(ky651) | Male | 7.1 ± 1.1 | 20 |
| cfi-l(ky650) | Hermaphrodite | 1.9 ± 0.6 | 20 |
| cfi-l(ky650) | Male | 6.1 ± 0.9 | 20 |
| ced-3(n717) | Hermaphrodite | 3.4 ± 0.8 | 20 |
| cfi-l(ky651); ced-3(n717) | Hermaphrodite | 6.4 ± 1.1 | 20 |
All animals were scored as adults. No cells indicates number ± standard deviation. n, Number of animals observed.
In cfi-1 mutants, the neurons that expressed the pkd-2::GFP reporter were generally located anterior to where genuine CEM cell bodies reside (Fig. 1). To ask whether the cells expressing pkd-2::GFP were misplaced CEM neurons that have failed to undergo programmed cell death, or other neurons, we examined expression of the reporter construct in mutant males. Wild-type males express pkd-2::GFP in four CEM neurons. If the neurons we observed were misplaced CEMs we would expect to see exactly four CEM cells expressing pkd-2::GFP in cfi-1 males. As shown in Table 1, whereas cfi-1(ky651) hermaphrodites had 3.1 ± 1.1 cells expressing the reporter, cfi-1(ky651) males had on average 7.1 ± 1.1 GFP-containing cells, roughly the sum of four CEM neurons and 3.1 additional cells. These results suggest that in cfi-1 animals, cells that would not normally express pkd-2::GFP now do so.
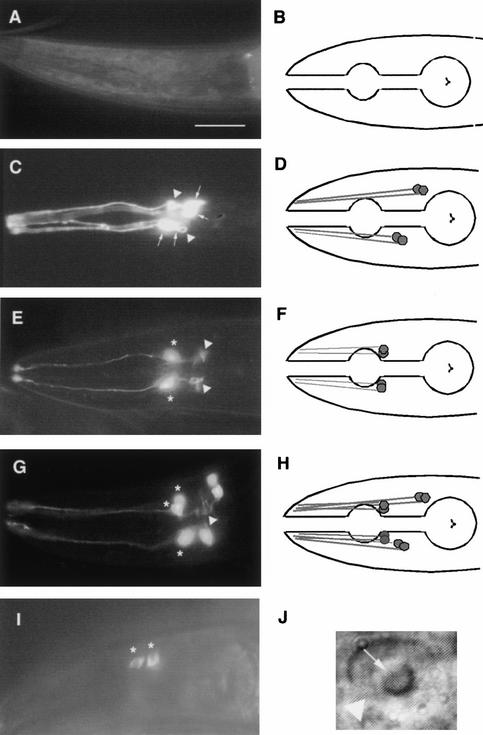
Figure 1.
cfi-1 inhibits expression of CEM features in other neurons. Bar, 50 μm. Arrowheads indicate axonal processes. Anterior is to the left, dorsal is up. (A) Head region of a wild-type hermaphrodite harboring a pkd-2::GFP integrated transgene. (B) Schematic representation of A. The outline of the pharynx is indicated. (C) Head region of a wild-type male containing the pkd-2::GFP transgene. Arrows indicate CEM cell bodies. (D) Schematic representation of C. (E) Head region of a cfi-1(ky651) hermaphrodite containing the pkd-2::GFP reporter. Asterisks indicate ectopic CEM-like cells. (F) Schematic of E. (G) Head region of cfi-1(ky651) male. Note the presence of both CEM neurons and additional neurons that express pkd-2::GFP (asterisks). (H) Schematic of G. (I) cfi-1(ky651) hermaphrodite ectopically expressing the lov-1::GFP reporter. (J) CEM-like cell in a cfi-1(ky651) L4 male animal. Note the prominent nucleolus (arrow); the nuclear membrane is indicated with an arrowhead.
To confirm this hypothesis we counted the number of GFP-expressing cells in animals containing both the cfi-1(ky651) mutation and a mutation in the programmed cell death caspase gene, ced-3 (Ellis and Horvitz 1986; Yuan et al. 1993). In ced-3 mutants CEM neurons do not undergo programmed cell death in hermaphrodites, and surviving cells acquire the neuronal characteristics of the male-specific CEM neurons (Ellis and Horvitz 1986; S. Shaham and C.I. Bargmann, unpubl.). As shown in Table 1, cfi-1(ky651); ced-3(n717) mutant hermaphrodites contained on average 6.4 ± 1.1 GFP-positive cells, roughly the sum of the number of cells present in cfi-1(ky651) and ced-3(n717) single-mutant animals. These results further support the theory that in cfi-1(ky651) animals, cells other than the CEM neurons have acquired CEM-like characteristics. The abnormal cells in cfi-1 mutants are therefore called “CEM-like cells”.
To determine the extent to which CEM-like cells have acquired CEM characteristics, we examined these cells for the expression of other features of CEM neurons. The CEM-like neurons expressed the male-specific CEM marker lov-1::GFP (Fig. 1I; Barr and Sternberg 1999) and possessed a nucleolar morphology that is unique to the CEMs among head neurons (Fig. 1J). The CEM-like neurons in cfi-1 mutants expressed the pkd-2::GFP and lov-1::GFP markers strongly in adults and not in younger animals. Normal CEM neurons express pkd-2 and lov-1 in a similar temporal pattern (Barr and Sternberg 1999). These results suggest that cfi-1 normally prevents the ectopic expression of multiple CEM-specific features in non-CEM cells.
Interestingly, not all CEM characteristics were acquired by the CEM-like cells in cfi-1 animals. The locations of the cell bodies of the CEM-like neurons usually did not resemble normal CEM positions. Normal CEMs undergo programmed cell death in hermaphrodites, but the CEM-like neurons in cfi-1 animals failed to do so. The establishment of CEM neuronal subtype identity and the decision to undergo programmed cell death may, therefore, be separate and perhaps developmentally distinct cell fate decisions.
The CEMDL/R sister cells, URADL/R, and IL2 neurons express CEM-like features in cfi-1 mutants
To determine the origins of the CEM-like cells in cfi-1 mutants, we examined the anatomical locations of these cells in the anterior ganglia. Two classes of neurons appeared to express CEM-like features: the URA neurons and the IL2 neurons. There are four radially symmetric URA-type neurons and six radially symmetric IL2-type neurons in C. elegans (White et al. 1986). The dorsal URA neurons are sister cells of the dorsal CEM neurons; URAs appear to be sensory neurons with dendritic processes that terminate in the body cavity. The IL2 neurons are unrelated to the CEMs by lineage, but both IL2s and CEMs are sensory neurons with dendritic processes that are physically exposed to the environment (Ward et al. 1975). Supporting our anatomical assignments, individual CEM-like cells appeared to have properties appropriate for either URA or IL2 neurons: 35% of cfi-1 animals contained at least one CEM-like cell with an axon resembling that of a URA neuron, and 15% of cfi-1 animals contained at least one CEM-like cell with an axon morphologically resembling an IL2 process (with the remaining animals containing unidentifiable processes, 58 animals observed).
The CEM-like cells could result from a transformation of URAs or IL2s, or from a change in cell lineage or cell death that resulted in extra cells. Although we have not examined cell lineages directly, our results favor the idea of a cell fate transformation rather than a lineage defect. The UNC-86 POU homeodomain protein and the LIN-32 helix–turn–helix transcriptional regulator are essential for many neuronal cell fates, including CEM fates (Finney and Ruvkun 1990; Zhao and Emmons 1995). As shown in Table 2, unc-86 and lin-32 are required for the expression of the CEM marker pkd-2::GFP in normal male CEM neurons, and they are also required for pkd-2::GFP expression in cfi-1 mutant CEM-like cells. Therefore, if the CEM-like cells result from lineage defects or loss of cell death, the extra cells should express unc-86. Contrary to this prediction, the expression of an unc-86::GFP reporter was normal in cfi-1(ky651) mutants (data not shown), suggesting that the CEM-like cells arose from an existing unc-86-expressing cell. UNC-86 is expressed in URA and IL2 neurons, as well as URB, URX, and URY neurons in the anterior ganglia. LIN-32 expression is less well-characterized, but lin-32 also acts in the IL2 neurons. These results are consistent with the model that the CEM-like cells arise from transformation of the URA and IL2 neurons in cfi-1 mutants.
Table 2.
unc-96 and lin-32 promote CEM neuronal identity
| Genotype
|
Sex
|
No. cells expressing pkd-2::gfp
|
n
|
|---|---|---|---|
| Wild type | Hermaphrodite | 0 ± 0 | 20 |
| Wild type | Male | 4.0 ± 0.2 | 20 |
| unc-86(n846) | Hermaphrodite | 0.05 ± 0.22 | 20 |
| unc-86(n846) | Male | 0.9 ± 0.3 | 20 |
| lin-32(u282) | Hermaphrodite | 0.12 ± 0.34 | 20 |
| lin-32(u282) | Male | 2.4 ± 0.8 | 20 |
| cfi-l(ky651) | Hermaphrodite | 3.1 ± 1.1 | 20 |
| cfi-l(ky651); unc-86(n846) | Hermaphrodite | 0.17 ± 0.38 | 18 |
| cfi-l(ky651); lin-32(u282) | Hermaphrodite | 0.13 ± 0.34 | 16 |
All animals were scored as adults. No. cells indicates number ± standard deviation. n, Number of animals observed.
If URA and IL2 cells express the CEM fate in cfi-1 mutants, we might expect an accompanying loss of expression of their normal cell-specific gene expression. We therefore examined the expression of the glutamate receptor GLR-4 in the URA neurons of cfi-1 animals. glr-4::GFP is normally expressed in URA and URY neurons in the anterior ganglia, but not in the IL2s (Brockie et al. 2001b). As shown in Figure 2, cfi-1 mutant animals failed to express glr-4::GFP in the URAs, but expressed glr-4::GFP in the URYs, supporting the notion that CEM-like neurons in cfi-1 animals are derived from transformed URA cells. A similar but less striking loss of the IL2 marker odr-2::GFP (Chou et al. 2001) was observed in cfi-1 mutants; in 9/25 animals examined, one IL2 failed to express odr-2::GFP. As described below, cfi-1 is also expressed in both URA and IL2.
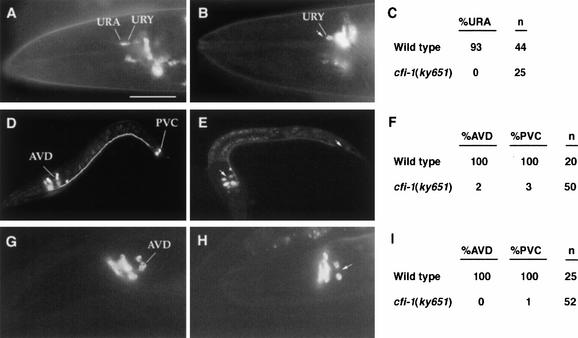
Figure 2.
The URA, AVD, and PVC neurons are defective in cfi-1 mutants. Bar, 50 μm. (A) Head region of a wild-type hermaphrodite expressing glr-4::GFP. (B) Head region of a cfi-1(ky651) hermaphrodite expressing glr-4::GFP. Arrow indicates nonexpressing URA neuron. (C) Table summarizing glr-4::GFP expression pattern in wild-type and cfi-1(ky651) animals. (%URA) The percentage of animals expressing glr-4::GFP in at least one URA neuron; (n) number of animals observed. (D) First larval stage (L1) wild-type animal expressing nmr-1::GFP. (E) cfi-1(ky651) L1 animal expressing nmr-1::GFP. Arrows indicate nonexpressing AVD and PVC neurons. (F) Table summarizing nmr-1::GFP expression pattern in wild-type and cfi-1(ky651) animals. (%AVD, %PVC) The percentage of animals expressing nmr-1::GFP in at least one AVD or PVC neuron, respectively; (n) number of animals observed. (G) Head region of a wild-type hermaphrodite expressing glr-1::GFP. (H) Head region of a cfi-1(ky651) hermaphrodite expressing glr-1::GFP. Arrow indicates nonexpressing AVD neuron. (I) Table summarizing glr-1::GFP expression in wild-type and cfi-1(ky651) animals; (%AVD, %PVC, n) as in F. PVC and AVD cell bodies are present in cfi-1 mutants and have a typical neuronal morphology by Nomarski microscopy. Weak nmr-1::GFP staining could be observed in processes of 30% of PVC neurons in cfi-1 mutants, suggesting that these cells have retained neuronal features.
As shown in Table 1, in cfi-1(ky651) animals 3.1 cells on average expressed the pkd-2::GFP marker. If all URA and IL2 neurons were consistently transformed, we would have expected 10 cells to express this reporter. Either all 10 cells have a small and equal probability of becoming CEM-like, or, alternatively, a subset of cells is more likely to acquire a CEM-like fate, with other cells transformed only occasionally. cfi-1 had a strong effect on glr-4::GFP expression in URA (Fig. 2), and a much weaker effect on odr-2::GFP expression in IL2. Therefore, we favor the possibility that the URA neurons are more likely to acquire the CEM-like fate, and the IL2 neurons only become transformed occasionally. In addition, there appeared to be a positional bias, which suggests that dorsal URAs were more likely to express CEM markers than ventral URAs. In cfi-1 hermaphrodites, of 62 CEM-like neurons observed, 38 occupied dorsal positions and 24 occupied ventral positions. Similarly in cfi-1 males, of 60 CEM-like neurons observed, 35 took dorsal positions and 25 were situated ventrally. We suggest that the probabilities of expressing the CEM fate in cfi-1 mutants follow the order: URADL/R > URAVL/R > IL2.
cfi-1 controls the fate of the AVD and PVC interneurons
The results described above suggest that cfi-1 is a regulator of neuronal subtype identity for the URA and IL2 neurons. To assess whether cfi-1 may control the fate of other C. elegans cells, we examined cfi-1 mutants for other morphological and locomotory defects. cfi-1 mutants had an unusual avoidance response to light touch. Wild-type animals stroked on the nose with an eyebrow hair move backward, whereas animals stroked on the tail move forward (Chalfie and Sulston 1981). In cfi-1 mutants the response to touch to the nose was normal, but animals stroked on the tail would often move backward instead of forward (56% moved backward when stroked on the tail, 8% stalled, and 36% moved forward; n = 100). This defect suggested that elements of the touch circuitry of C. elegans may be disrupted in cfi-1 mutants. Six touch cell neurons mediate responses to light body touch (Chalfie and Sulston 1981), and can be visualized using a mec-4::GFP reporter (Driscoll and Chalfie 1991). In cfi-1 mutants all six touch neurons expressed mec-4::GFP and had normal cell bodies and processes (data not shown). The AVD, AVA, AVB, and PVC interneurons mediate responses initiated by the touch response cells (Chalfie et al. 1985). We examined expression of the glutamate receptor NMR-1, which is normally expressed in most of these neurons (Brockie et al. 2001a,b), in cfi-1 mutants. As shown in Figure 2D–F, cfi-1 animals failed to express an nmr-1::GFP reporter in the AVD and PVC interneurons. AVD and PVC also failed to express a reporter gene for a second glutamate receptor, glr-1 (Fig. 2G–I; Hart et al. 1995; Maricq et al. 1995). The defective expression of two glutamate receptors in the AVD and PVC interneurons indicates that cfi-1 regulates AVD and PVC cell fates. Because PVC is the only interneuron that responds to tail touch, the loss of PVC interneurons in cfi-1 mutants is sufficient to account for the inappropriate response to touch to the tail (Chalfie et al. 1985; M. Chalfie, pers. comm.).
In addition to abnormal response to light touch, cfi-1 mutants also showed a slightly loopy locomotion pattern. We have not uncovered the cellular focus of this defect.
cfi-1 encodes an ARID transcription factor
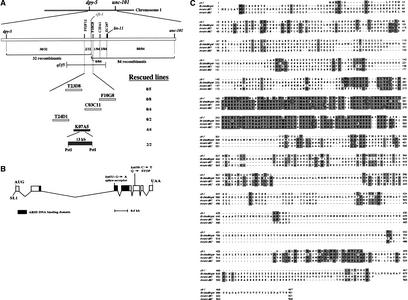
To understand the molecular mechanism by which cfi-1 exerts its effects, we cloned the gene. Using standard genetic mapping techniques we mapped cfi-1 to a genetic interval between the dpy-5 and unc-101 genes on Chromosome I, under the deficiency qDf5. Mapping of single nucleotide polymorphisms (SNPs) in the region with respect to cfi-1 identified a set of five cosmids that should contain the gene. As shown in Figure 3A, DNA corresponding to cosmid K07A5 fully rescued cfi-1(ky651) mutants. A 13-kb subclone of this cosmid rescued as efficiently as the cosmid.
Figure 3.
CFI-1 is an ARID-containing protein. (A) Mapping and rescue experiments. The numbers of recombinants isolated in each interval out of the total are indicated. Cosmids used for rescue experiments are indicated. For rescue we measured disappearance of ectopic CEM-like neurons in cfi-1(ky651) animals as assessed by expression of pkd-2::GFP. In all rescuing lines 80%–95% of animals lacked even a single ectopic CEM-like neuron. (B) Deduced mRNA structure for cfi-1. Boxes indicate exons, lines indicate introns. Dark boxes indicate exons encoding the ARID. Locations of mutations as well as the nucleotide and mRNA/protein alterations are indicated above the RNA. (C) Sequence comparison of the predicted CFI-1 protein and the related proteins Dead ringer, mouse Dril1 (Bright), and Dril2. Boxes indicate identities. Shading indicates similar amino acids.
The 13-kb subclone of K07A5 was predicted by the GeneFinder annotations of the C. elegans genome to contain a single gene (T23D8.8). We isolated a full-length cDNA of this gene (see Materials and Methods), which differed from the GeneFinder annotated prediction at the 5′ and 3′ ends. To confirm that the gene we isolated indeed corresponded to cfi-1, we determined the sequences of all exons and exon/intron boundaries of the gene in cfi-1(ky650) and cfi-1(ky651) mutants. As shown in Figure 3B, point mutations were present in both alleles. The ky650 allele contained a C → T alteration that resulted in the introduction of a TAG stop codon in place of the codon encoding glutamine 341. The ky651 allele contained a G → A mutation in the conserved G residue of the splice-acceptor sequence of the second intron. These results indicate that the gene we identified corresponds to cfi-1.
Conceptual translation of the longest open reading frame of the cfi-1 cDNA revealed a protein of 467 amino acids. A region between amino acids 174 and 296 showed high similarity to A/T rich interaction domains (ARIDs), which have been recently shown to bind DNA (Kortschak et al. 2000). ARIDs are present in many proteins, including the yeast SWI1 protein (Peterson and Herskowitz 1992), the mammalian retinoblastoma binding proteins (RBP1 and RBP2; Fattaey et al. 1993), and the mammalian MRF-1 and MRF-2 transcriptional regulators (Huang et al. 1996). CFI-1 is most similar to a subset of ARID proteins termed extended ARID (eARID; Kortschak et al. 1998) proteins and is the only eARID protein in C. elegans. eARID proteins share additional identities closely flanking the ARID domain and also share similarities in a region C-terminal to the ARID, termed REKLES (Kortschak et al. 2000). In the eARID, CFI-1 is 70%, 73%, and 72% identical to the Drosophila melanogaster Dead ringer protein (Gregory et al. 1996), and the mammalian Dril1 (Bright) and Dril2 proteins, respectively (Fig. 3C; Herrscher et al. 1995; Kortschak et al. 2000). These proteins share other regions of similarity with CFI-1 as well, suggesting that they are orthologs.
CFI-1 binds DNA in a sequence-specific manner
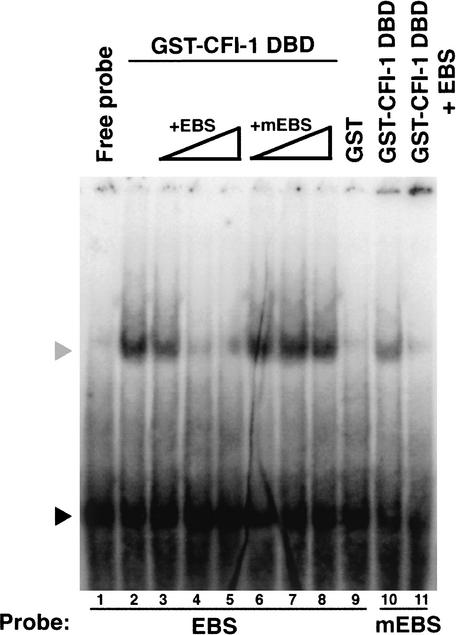
ARIDs are thought to regulate transcription by binding to DNA (Herrscher et al. 1995). Some, like the yeast SWI1 protein, do so nonspecifically (Quinn et al. 1996), whereas eARIDs bind DNA in a sequence-specific fashion (Gregory et al. 1996). To examine the DNA-binding capabilities of CFI-1, we assessed the ability of the CFI-1 ARID to bind a short oligonucleotide shown by Gregory et al. (1996) to be tightly bound by the Drosophila Dead ringer ARID (the Drosophila Engrailed homeodomain protein binding site, or EBS). As shown in Figure 4, a fusion protein between Glutathione S-transferase (GST) and the CFI-1 ARID (amino acids 163–300) bound a 32P-labeled oligonucleotide containing the EBS in a gel mobility shift assay (Fig. 4, lane 2). This binding was competed effectively with cold EBS but not with a mutant EBS containing an ATGG sequence in the ARID-binding site instead of the ATTA present in the wild-type site. These results indicate that CFI-1 is a sequence-specific DNA-binding protein.
Figure 4.
CFI-1 ARID binds DNA. Gel shift assay showing binding of CFI-1 ARID to the Engrailed binding site (EBS) or a mutant EBS (mEBS). Triangles indicate increasing concentrations of cold oligonucleotides added, either 10× (lanes 3,6), 100× (lanes 4,7), or 1000× (lanes 5,8) as much as labeled oligonucleotide. Lane 10 shows weak binding of CFI-1 ARID to the labeled mEBS that is competed well by 1× cold EBS (lane 11). Gray arrowhead indicates the position of protein–probe complexes. Black arrowhead indicates the position of free probe.
Because cfi-1 inhibited the expression of CEM-specific markers in URA and IL2 neurons, we wondered whether this inhibition was mediated by transcriptional repression complexes known to interact with other CFI-1-related proteins. We failed to detect genetic interactions between cfi-1 and either unc-37Groucho (Pflugrad et al. 1997), lin-35RB (Fattaey et al. 1993; Lu and Horvitz 1998), lin-15A repressor (Clark et al. 1994; Huang et al. 1994), or psa-1 (swi3) or psa-4 (swi2/snf2; Quinn et al. 1996; Sawa et al. 2000). These results suggest that repression of CEM fate in the URA and IL2 neurons is either directly mediated by CFI-1, or is mediated by a repression complex unrelated to Groucho, Rb, or SWI/SNF.
CFI-1 protein is expressed in neurons and muscles
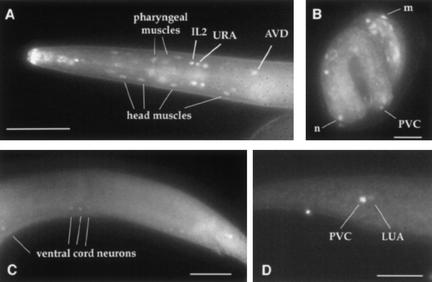
To ask where cfi-1 acts, we generated antibodies in rats against the CFI-1 ARID and examined CFI-1 expression by whole-mount immunofluorescence. As shown in Figure 5, CFI-1 protein is localized to nuclei of staining cells, supporting the idea that CFI-1 functions as a transcriptional regulator of cell identity. We detected expression of CFI-1 in mid-late embryos (twofold and later) and throughout larval development and in the adult. No staining was detected in cfi-1(ky651) animals or in animals incubated with preimmune serum. CFI-1 was expressed in a small number of cells in the head and tail regions of the animals (Fig. 5), with occasional faint staining in ventral cord neurons. In the head, only neurons and muscle cells expressed CFI-1. In the tail, only four neuronal cells expressed CFI-1. A cfi-1::GFP fusion gene was expressed in a pattern similar to that observed with anti-CFI-1 antisera.
Figure 5.
CFI-1 is a nuclear protein expressed in neurons and muscles. Expression pattern of CFI-1 visualized using anti-CFI-1 antibodies. (A) Head expression; anterior is to the left, dorsal is up. Staining in nose tip area is nonspecific staining of the anterior buccal cavity. (B) Embryonic CFI-1 expression in a 3-fold embryo. (m) Muscle nucleus in the head; (n) neuronal nucleus in the ventral cord. (C) Faint expression of CFI-1 in the ventral cord; anterior is left, dorsal is up. (D) Expression in the tail region; anterior is to the right, dorsal is up. Bars in A, C, and D, 50 μm; bar in B, 5 μm.
Using the cfi-1::GFP and anti-CFI-1 antisera we identified a number of the cells expressing cfi-1 based on their anatomical positions and neuronal morphologies. Consistent with our phenotypic analyses, CFI-1 protein was expressed in IL2 and URA neurons, and in the AVD and PVC interneurons. We also detected weaker expression in the PVC sister neuron LUA. Most, if not all, head muscle cells expressed CFI-1, and m6 and m7 muscles in the pharynx expressed the protein as well. A small number of other cells also stained occasionally.
The observation that CFI-1 protein is expressed in cells that are defective in cfi-1 mutants suggests that cfi-1 functions cell-autonomously in these cells. To ask where cfi-1 function is required, we expressed the cfi-1 cDNA under the control of the nmr-1 promoter (Brockie et al. 2001a) in the AVD and PVC neurons. This fusion gene was introduced into cfi-1(ky651) animals bearing both the nmr-1::GFP and pkd-2::GFP reporter genes. Pnmr-1::cfi-1 partly rescued expression of nmr-1::GFP in AVD and PVC neurons. For the strongest line we examined containing a Pnmr-1::cfi-1 transgene on an extrachromosomal array, expression in at least one AVD neuron of nmr-1::GFP was detected in 47% of transgenic animals, and in at least one PVC neuron in 18% of animals. The transgene did not affect the ectopic expression of pkd-2::GFP in the same line. These results support the idea that CFI-1 functions cell-autonomously to regulate cell fate.
The unc-86 POU domain homeobox gene promotes cfi-1 expression in the URA and IL2 neurons
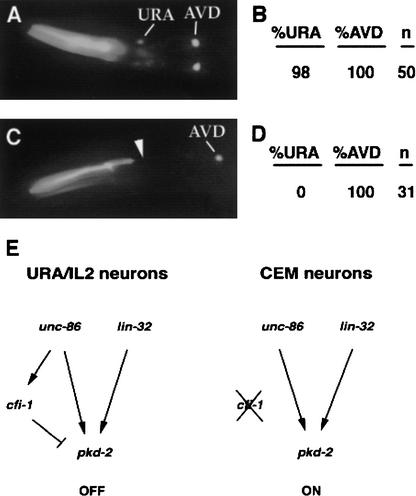
Both CFI-1 and the POU homeodomain protein UNC-86 contribute to URA and IL2 cell fates (Finney and Ruvkun 1990). As noted above, cfi-1(ky651) animals showed normal expression of unc-86::GFP. To ask whether unc-86 regulated cfi-1 expression, we performed the reverse experiment, examining expression of a cfi-1::GFP fusion gene in unc-86(n846) mutant animals that fail to express UNC-86 protein. Strikingly, although cfi-1 was still expressed in AVD, PVC, and other cells, no expression was detected in the URA and IL2 neurons (Fig. 6). These results strongly suggest that cfi-1 transcription in URA and IL2 is normally induced, directly or indirectly, by unc-86. In lin-32 mutants, 65% of animals expressed cfi-1 at reduced levels, and 35% did not express detectable cfi-1.
Figure 6.
unc-86 promotes the transcription of cfi-1 in the URA and IL2 neurons. (A) Wild-type animal expressing cfi-1::GFP. URA and AVD neurons are indicated. (B) Quantitation of cfi-1::GFP staining for wild-type animals. (%URA, %AVD) The percentage of animals expressing cfi-1::GFP in at least a single URA or AVD neuron; (n) number of animals observed. (C) cfi-1::GFP expression in an unc-86(n846) mutant animal. Note the absence of anterior neuronal staining. (D) Quantitation of cfi-1::GFP staining for unc-86(n846) mutant animals; (%URA, %AVD, n) as in B. (E) Models for regulation of CEM fate in URA/IL2 and CEM neurons. In the URA and IL2 neurons, unc-86 and lin-32 can promote CEM fate development (as indicated by expression of pkd-2), but cfi-1, which is also activated by unc-86, inhibits expression of this fate. In the CEM neurons, cfi-1 is inactive; thus, unc-86 and lin-32 can function to promote CEM differentiation.
Discussion
cfi-1 encodes a novel ARID protein regulator of neuronal subtype identity
CFI-1 regulates cell identity and promotes diversity in the C. elegans nervous system. In cfi-1 mutants, the differentiation of URA and IL2 sensory neurons and the differentiation of AVD and PVC interneurons are disrupted. In the URA and IL2 neurons, cfi-1 mutations result in the inappropriate acquisition of aspects of the CEM neuronal fate. Because the dorsal URA neurons are sister cells of the dorsal CEM neurons, it is possible that cfi-1 responds to an asymmetric cell lineage signal that is differentially distributed between the dorsal URA and the dorsal CEM neurons. Cells unrelated by lineage to the CEMs (the IL2 neurons and the ventral URAs) can also acquire CEM fate in cfi-1 mutants, suggesting that cfi-1 may also respond to other kinds of cues that regulate cell fate.
CFI-1 probably controls neuronal fate by regulating a specific set of target genes. Such a function is consistent with the sequence-specific DNA binding of CFI-1, and with the observation that the mouse Bright (Dril1) protein, a protein highly related to CFI-1, regulates expression from the immunoglobulin heavy chain enhancer (Herrscher et al. 1995). Interestingly, cfi-1 may function redundantly with other genes to regulate neuronal subtype identity, because considerable phenotypic variability is seen even in the strong cfi-1(ky651) mutant.
unc-86 both promotes and inhibits expression of CEM-specific genes in the URA and IL2 neurons
Transcriptional regulators play important roles in creating diversity in the nervous system. The most well-understood cell fate regulators are the families of homeodomain proteins. The POU homeodomain proteins, including C. elegans UNC-86, affect development of many neurons (Finney and Ruvkun 1990); the Pax homeodomain proteins subdivide the vertebrate neural tube into distinct domains (Lee and Jessell 1999); and the LIM-homeodomain proteins generate a variety of motor neurons in the vertebrate spinal cord (Tanabe and Jessell 1996) and contribute to the diversity of sensory neurons and interneurons in C. elegans (Way and Chalfie 1988; Hobert et al. 1997; Sagasti et al. 1999). Homeodomain proteins typically function in a context that is defined by other DNA-binding proteins. In the URA and IL2 neurons, CFI-1 represents an important factor that modifies the activity of UNC-86 to match a particular cell type (Fig. 6E).
unc-86 is essential for the development of the URA and IL2 neurons, where it induces expression of cfi-1. cfi-1, in turn, activates normal URA and IL2 gene expression and prevents inappropriate expression of CEM-specific genes. If cfi-1 is absent from URA and IL2 neurons, unc-86, instead, promotes expression of the CEM-specific marker pkd-2::GFP as it would normally do in CEM neurons. These results indicate that during normal development a cfi-1-dependent activity prevents unc-86-dependent induction of CEM-specific genes in URA and IL2 neurons. Such a cfi-1-dependent activity could function by qualitatively changing UNC-86 from an activator of gene expression to a repressor. Alternatively, a cfi-1-dependent activity could prevent the association of UNC-86 or an UNC-86 target protein with regulatory regions of CEM-specific genes. In either model, the absence of CFI-1 function in CEM neurons would allow UNC-86 to activate pkd-2 in these cells.
The functions of CFI-1 and its Drosophila homolog Dead ringer may be conserved
CFI-1 and the Drosophila Dead ringer protein are very similar and are likely orthologs. dead ringer is required for anterior–posterior patterning and muscle development in the Drosophila embryo (Shandala et al. 1999). dead ringer RNA is maternally contributed to embryos and is expressed in a restricted pattern during embryogenesis (Gregory et al. 1996; Shandala et al. 1999). Initially, dead ringer is expressed in three major bands in the embryo, with the anterior and posterior bands being most prominent. Following gastrulation, dead ringer expression becomes restricted to mesoderm (including muscles), pharyngeal muscles, a small group of cells in the brain, a regular array of cells in the ventral nervous system, and a small number of other tissues including the hindgut. This expression pattern is reminiscent of CFI-1 expression in C. elegans, where expression is detected primarily in head and tail cells, in pharyngeal and head muscles, in a small group of neurons near the nerve ring (the brain of the animal), and in ventral cord neurons. In contrast to Dead ringer, we did not detect early ubiquitous expression of CFI-1 in C. elegans, nor did we detect expression of CFI-1 in the gut or germ line using our antibodies, but cfi-1::GFP was expressed in posterior gut nuclei.
Their similar sequences, DNA-binding properties, and expression patterns suggest that CFI-1 and Dead ringer proteins function in similar ways to regulate cell fate. We speculate that Dead ringer will contribute to neuronal diversity in Drosophila. Although the expression patterns of the mammalian CFI-1-related proteins Bright and Dril2 have not been described in the developing nervous system, we propose that these, or other related proteins, may function during development of the mammalian nervous system as well.
Materials and methods
General methods and strains
C. elegans was cultured as described by Brenner (1974). All strains were grown at 20°C. The wild-type strains used were C. elegans variety Bristol strain N2 and the Hawaiian strain CB4856. Genetic nomenclature follows the standard C. elegans system (Horvitz et al. 1979). The mutations used are listed below: Linkage group LGI: dpy-5(e61), lin-35(n745), unc-37(s80, h763), cfi-1(ky650, ky651), unc-101(m1); LGIII: unc-86(n846), pha-1(e2123ts); LGIV: ced-3(n717), psa-4(os13); LGV: psa-1(os22); LGX: lin-32(u282), lin-15(n765).
Isolation, mapping, and cloning of cfi-1
pha-1(e2123ts) animals harboring an extrachromosomal array containing the pkd-2::GFP reporter gene, pPKD-2::GFP1 (Barr and Sternberg 1999), and the pha-1-rescuing plasmid pBX (Granato et al. 1994) were mutagenized with ethyl methanesulfonate (EMS) as described by Sulston and Hodgkin (1988). ky650 and ky651 animals isolated following mutagenesis were crossed with animals carrying an integrated pkd-2::GFP transgene, and these lines were used for all subsequent studies.
The lesion in ky651 animals was mapped to LGI using Tc1 polymorphism differences between N2 and the RW7000 strain (Korswagen et al. 1996). Phenotypic mapping placed cfi-1 between dpy-5 and unc-101. The deficiency qDf5, which uncovered cfi-1, was used to further narrow the interval containing the gene. Finally, we crossed dpy-5(e61) cfi-1(ky651) unc-101(m1) animals into CB4856 animals and mapped cfi-1(ky651) with respect to the single nucleotide polymorphic markers at position 11483 of cosmid C25A1, at position 1672 of cosmid F10G8, and at position 11546 of cosmid F52F12 (Wicks et al. 2001). The pattern of recombinants observed is shown in Figure 3.
Germ-line transformations
Germ-line transformations were carried out using standard protocols (Mello and Fire 1995). The coinjection marker used for the rescue experiments was the dominant pRF4 rol-6(su1006) plasmid at 75 ng/μL.
Plasmid constructions
cfi-1::GFP reporter genes were constructed by amplifying either the full genomic region or the region up to exon 3 from N2 genomic DNA using the polymerase chain reaction (PCR) and primers containing restriction enzyme sites, digesting the resulting products with the appropriate enzymes, and ligating to either the pPD95.75 or pPD95.69 GFP vectors (gifts from A. Fire, Carnegie Institute, Baltimore, MD), respectively. In each construct 8.17 kb of DNA 5′ to the ATG start codon was included. To make the Pnmr-1::cfi-1 fusion gene, we amplified 5 kb of the promoter region of nmr-1 (Brockie et al. 2001a) using PCR and appropriate primers and ligated the resulting product to the full-length cfi-1 cDNA in the pBluescript KS + vector (see below).
Isolation of a cfi-1 cDNA
To find a cfi-1 cDNA, we amplified a plasmid-based mixed-stage C. elegans cDNA library with two primer pairs using PCR. One pair consisted of a vector primer 5′ of the cDNA insert and an internal primer to the region encoding the CFI-1 ARID. A second pair consisted of a primer 3′ to the cDNA insert and an internal primer to the region encoding the CFI-1 ARID. Sequence analysis of the resulting products and comparison to the genomic sequence showed that a full-length cDNA contains 5′ SL1 leader sequences (Krause and Hirsh 1987) and a 3′ AATAAA transcription termination site. Once the ends of the cDNA were determined, we used primers starting at the start and stop codons to amplify the full-length cDNA.
Microscopy
Animals were examined by epifluorescence using either a fluorescence dissecting microscope or a compound microscope. Images were captured using an AxioCam CCD camera (Zeiss) and analyzed using the Axiovision software (Zeiss).
Gel mobility shift assays
Plasmid pGEX-5X-2 (Pharmacia) containing the Glutathione S-transferase gene was fused to a BamHI/SalI fragment encoding the CFI-1 ARID domain (amino acids 163–300). The resulting plasmid was used to express GST–CFI-1 (ARID) protein in Escherichia coli BL21(DE3). Bacterial supernatant was passed over a glutathione column and fusion protein was eluted with soluble glutathione. Then 10 ng of fusion protein was incubated with 0.1 ng of double-stranded oligonucleotides containing three tandem copies of the Engrailed binding site (TCAATTAAATGA; Gregory et al. 1996) or three copies of a mutant binding site (TCAATGGAATGA) in 20 μL of a solution containing 10 mM Tris (pH 7.5), 1 mM EDTA, 100 mM KCl, 0.1 mM DTT, 5% glycerol, 50 μg/mL bovine serum albumin, and 100 ng of salmon sperm DNA. Oligonucleotides were labeled with 32P by incubation with T4 polynucleotide kinase. Complexes were run on a 5% acrylamide gel in 0.5× TBE (Tris-Borate EDTA) buffer.
Generation of CFI-1 antibodies and immunofluorescence
The GST fusion protein used for the gel mobility assays (see above) was used to immunize two rats (Covance, PA). After three boosts, antiserum was tested on fixed animals for activity using an FITC-labeled mouse anti-rat IgG secondary antibody. We found that staining using either antiserum was robust at a 1:200 dilution of the antiserum and that purification of the antisera was unnecessary. Animals were fixed for whole-mount immunofluorescence using the protocols of Finney and Ruvkun (1990) for fixing either larvae and adults or embryos.
Acknowledgments
We thank Maureen Barr and Paul Sternberg for the pkd-2 and lov-1 reagents, Andres Villu Maricq for the glr-4 and nmr-1 reagents, and Scott Clark for the mec-4::GFP strain. We also thank Eli Arama, Bertrand Mollereau, Hyung Don Ryoo, and Mark Schroeder for helpful comments on the manuscript. Many of the strains used in this study were obtained from the Caenorhabditis Genetics Center (CGC), which is funded by the NIH National Center for Research Resources (NCRR). S.S. is a Brookdale National Fellow. C.I.B. is an Investigator of the Howard Hughes Medical Institute. This work was funded by the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL shaham@mail.rockefeller.edu; FAX (212) 327-7076.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.976002.
References
- Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- Barr MM, DeModena J, Braun D, Nguyen CQ, Hall DH, Sternberg PW. The Caenorhabditis elegans autosomal dominant polycystic kidney disease gene homologs lov-1 and pkd-2 act in the same pathway. Curr Biol. 2001;11:1341–1346. doi: 10.1016/s0960-9822(01)00423-7. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockie PJ, Mellem JE, Hills T, Madsen DM, Maricq AV. The C. elegans glutamate receptor subunit NMR-1 is required for slow NMDA-activated currents that regulate reversal frequency during locomotion. Neuron. 2001a;31:617–630. doi: 10.1016/s0896-6273(01)00394-4. [DOI] [PubMed] [Google Scholar]
- Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J Neurosci. 2001b;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm A. Control of cell fate in the tail region of C. elegans by the gene egl-5. Development. 1991;111:921–932. doi: 10.1242/dev.111.4.921. [DOI] [PubMed] [Google Scholar]
- Chou JH, Bargmann CI, Sengupta P. The Caenorhabditis elegans odr-2 gene encodes a novel Ly-6-related protein required for olfaction. Genetics. 2001;157:211–224. doi: 10.1093/genetics/157.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SG, Lu X, Horvitz HR. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics. 1994;137:987–997. doi: 10.1093/genetics/137.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell. 1999;98:317–327. doi: 10.1016/s0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- Desai C, Garriga G, McIntire SL, Horvitz HR. A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature. 1988;336:638–646. doi: 10.1038/336638a0. [DOI] [PubMed] [Google Scholar]
- Driscoll M, Chalfie M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature. 1991;349:588–593. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- Fattaey AR, Helin K, Dembski MS, Dyson N, Harlow E, Vuocolo GA, Hanobik MG, Haskell KM, Oliff A, Defeo-Jones D, et al. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene. 1993;8:3149–3156. [PubMed] [Google Scholar]
- Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Finney M, Ruvkun G, Horvitz HR. The C. elegans cell lineage and differentiation gene unc-86 encodes a protein with a homeodomain and extended similarity to transcription factors. Cell. 1988;55:757–769. doi: 10.1016/0092-8674(88)90132-8. [DOI] [PubMed] [Google Scholar]
- Garriga G, Guenther C, Horvitz HR. Migrations of the Caenorhabditis elegans HSNs are regulated by egl-43, a gene encoding two zinc finger proteins. Genes & Dev. 1993;7:2097–2109. doi: 10.1101/gad.7.11.2097. [DOI] [PubMed] [Google Scholar]
- Granato M, Schnabel H, Schnabel R. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 1994;22:1762–1763. doi: 10.1093/nar/22.9.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SL, Kortschak RD, Kalionis B, Saint R. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol Cell Biol. 1996;16:792–799. doi: 10.1128/mcb.16.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- Herrscher RF, Kaplan MH, Lelsz DL, Das C, Scheuermann R, Tucker PW. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: A B cell-specific trans-activator that describes a new DNA-binding protein family. Genes & Dev. 1995;9:3067–3082. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- Hobert O, Mori I, Yamashita Y, Honda H, Ohshima Y, Liu Y, Ruvkun G. Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron. 1997;19:345–357. doi: 10.1016/s0896-6273(00)80944-7. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Brenner S, Hodgkin J, Herman RK. A uniform genetic nomenclature for the nematode Caenorhabditis elegans. Mol Gen Genet. 1979;175:129–133. doi: 10.1007/BF00425528. [DOI] [PubMed] [Google Scholar]
- Huang LS, Tzou P, Sternberg PW. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol Biol Cell. 1994;5:395–411. doi: 10.1091/mbc.5.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TH, Oka T, Asai T, Okada T, Merrills BW, Gertson PN, Whitson RH, Itakura K. Repression by a differentiation-specific factor of the human cytomegalovirus enhancer. Nucleic Acids Res. 1996;24:1695–1701. doi: 10.1093/nar/24.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon M, Gardner HF, Miller EA, Deshler J, Rougvie AE. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science. 1999;286:1141–1146. doi: 10.1126/science.286.5442.1141. [DOI] [PubMed] [Google Scholar]
- Korswagen HC, Durbin RM, Smits MT, Plasterk RH. Transposon Tc1-derived, sequence-tagged sites in Caenorhabditis elegans as markers for gene mapping. Proc Natl Acad Sci. 1996;93:14680–14685. doi: 10.1073/pnas.93.25.14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortschak RD, Reimann H, Zimmer M, Eyre HJ, Saint R, Jenne DE. The human dead ringer/bright homolog, DRIL1: cDNA cloning, gene structure, and mapping to D19S886, a marker on 19p13.3 that is strictly linked to the Peutz-Jeghers syndrome. Genomics. 1998;51:288–292. doi: 10.1006/geno.1998.5259. [DOI] [PubMed] [Google Scholar]
- Kortschak RD, Tucker PW, Saint R. ARID proteins come in from the desert. Trends Biochem Sci. 2000;25:294–299. doi: 10.1016/s0968-0004(00)01597-8. [DOI] [PubMed] [Google Scholar]
- Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJ, Jessell TM. The specification of dorsal cell fates in the vertebrate central nervous system. Annu Rev Neurosci. 1999;22:261–294. doi: 10.1146/annurev.neuro.22.1.261. [DOI] [PubMed] [Google Scholar]
- Lu X, Horvitz HR. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95:981–991. doi: 10.1016/s0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- Maricq AV, Peckol E, Driscoll M, Bargmann CI. Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Pflugrad A, Meir JY, Barnes TM, Miller DM., III The Groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4 to govern motor neuron identity in C. elegans. Development. 1997;124:1699–1709. doi: 10.1242/dev.124.9.1699. [DOI] [PubMed] [Google Scholar]
- Quinn J, Fyrberg AM, Ganster RW, Schmidt MC, Peterson CL. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Hobert O, Troemel ER, Ruvkun G, Bargmann CI. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes & Dev. 1999;13:1794–1806. doi: 10.1101/gad.13.14.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa H, Kouike H, Okano H. Components of the SWI/SNF complex are required for asymmetric cell division in C. elegans. Mol Cell. 2000;6:617–624. doi: 10.1016/s1097-2765(00)00060-5. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Bargmann CI. Cell fate specification and differentiation in the nervous system of Caenorhabditis elegans. Dev Genet. 1996;18:73–80. doi: 10.1002/(SICI)1520-6408(1996)18:1<73::AID-DVG8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Shandala T, Kortschak RD, Gregory S, Saint R. The Drosophila dead ringer gene is required for early embryonic patterning through regulation of argos and buttonhead expression. Development. 1999;126:4341–4349. doi: 10.1242/dev.126.19.4341. [DOI] [PubMed] [Google Scholar]
- Slack F, Ruvkun G. Temporal pattern formation by heterochronic genes. Annu Rev Genet. 1997;31:611–634. doi: 10.1146/annurev.genet.31.1.611. [DOI] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. pp. 587–606. [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tanabe Y, Jessell TM. Diversity and pattern in the developing spinal cord. Science. 1996;274:1115–1123. doi: 10.1126/science.274.5290.1115. [DOI] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J Comp Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- Way JC, Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988;54:5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Trans R Soc Lond B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Wu J, Duggan A, Chalfie M. Inhibition of touch cell fate by egl-44 and egl-46 in C. elegans. Genes & Dev. 2001;15:789–802. doi: 10.1101/gad.857401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 β-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- Zhao C, Emmons SW. A transcription factor controlling development of peripheral sense organs in C. elegans. Nature. 1995;373:74–78. doi: 10.1038/373074a0. [DOI] [PubMed] [Google Scholar]