Abstract
Genetic mapping showed that the rice blast avirulence gene AVR-Pita is tightly linked to a telomere on chromosome 3 in the plant pathogenic fungus Magnaporthe grisea. AVR-Pita corresponds in gene-for-gene fashion to the disease resistance (R) gene Pi-ta. Analysis of spontaneous avr-pita− mutants indicated that the gene is located in a telomeric 6.5-kb BglII restriction fragment. Cloning and DNA sequencing led to the identification of a candidate gene with features typical of metalloproteases. This gene is located entirely within the most distal 1.5 kb of the chromosome. When introduced into virulent rice pathogens, the cloned gene specifically confers avirulence toward rice cultivars that contain Pi-ta. Frequent spontaneous loss of AVR-Pita appears to be the result of its telomeric location. Diverse mutations in AVR-Pita, including point mutations, insertions, and deletions, permit the fungus to avoid triggering resistance responses mediated by Pi-ta. A point mutation in the protease consensus sequence abolishes the AVR-Pita avirulence function.
INTRODUCTION
Rice blast disease, caused by the fungus Magnaporthe grisea (Hebert) Barr (Rossman et al., 1990), remains one of the most important constraints to rice production worldwide, despite the availability of dozens of “major” disease resistance (R) genes, which are known as Pi genes (Yamada et al., 1976; Zeigler et al., 1994). Resistance mediated by Pi genes has not been durable, typically failing under field conditions soon after new cultivars are introduced to the field as new races, or pathotypes, of the pathogen appear with the ability to attack the previously resistant rice cultivars. Sasaki first distinguished differences in cultivar specificity between rice blast field isolates in 1922 (Yamada, 1985); since then, hundreds of pathogen races have been identified based on their infection spectra on differential rice cultivars. Recently, DNA fingerprinting analyses have shown that rice blast fungal populations are asexual and consist of dozens of families (discretely separable clonal lineages) having apparently predictable responses toward specific R genes (Levy et al., 1993; Zeigler et al., 1994). Lineage exclusion strategies are currently being tested as an aid for deploying R genes for genetic control of rice blast disease.
The rice blast system is a classical gene-for-gene system (Flor, 1971) in which avirulence (AVR) genes in the pathogen show a functional correspondence with particular R genes in rice (Silué et al., 1992; Zeigler et al., 1994). AVR genes may encode pathogen molecules that are themselves recognized, directly or indirectly, by the corresponding R gene product, or they may encode enzymes involved in production of small molecule ligands that serve as recognition factors. Pathogen recognition triggers host resistance responses and stops infection. Field efficacy of any R gene thus depends on the biology of its associated AVR gene. Specifically, R gene efficacy depends on the nature of any function that an AVR gene may provide for the pathogen independent of its function in triggering R gene–mediated resistance and on the effects of AVR gene mutation on the ability of the pathogen to survive and compete under field conditions.
AVR genes are predicted to encode molecules that function during normal growth and pathogenicity (Leach and White, 1996; Laugé and De Wit, 1998). Most likely, this activity is quite distinct from the role of the AVR gene product in triggering R gene–mediated resistance in a host plant. Among the >30 bacterial AVR gene products reported in the literature, only a few have presented clues as to what this function might be. The avrD gene cloned from Pseudomonas syringae pv glycinea is believed to play a role in producing low molecular weight glycolipid elicitors (Keen et al., 1990; Midland et al., 1993). The bacterial AVR gene, avrRxv, of Xanthomonas campestris pv vesicatoria encodes a predicted protein with similarity to the YopJ protein produced by an animal pathogen, Yersinia pseudotuberculosis. YopJ appears to be involved in modulating host signaling responses (Orth et al., 1999). The avrBs2 gene, also from X. campestris pv vesicatoria, encodes a predicted protein with similarity to an enzyme that catalyzes phosphodiester linkages, agrocinopine synthase of Agrobacterium tumefaciens, and to an enzyme that hydrolyzes phosphodiester linkages, UgpQ of Escherichia coli (Swords et al., 1996). Mutants lacking avrBs2 activity fail to induce Bs2-mediated resistance, and they exhibit reduced growth in planta, suggesting a role for avrBs2 in pathogen fitness. A few other cloned bacterial avirulence genes appear to be required for full pathogen fitness, thus supporting the hypothesis that these genes perform valuable functions for the pathogen, independent of their role in triggering R gene–mediated resistance (Leach and White, 1996).
The majority of fungal AVR genes cloned to date are derived from fungi that colonize intercellular spaces in plant tissues. These genes were cloned by using reverse genetic approaches initiated by identifying extracellular proteins that elicit plant cell necrosis in a race-specific manner (reviewed in Laugé and De Wit, 1998). The genes cloned include the Avr9, Avr4, and Ecp2 genes from the tomato leaf mold pathogen Cladosporium fulvum and the nip1 gene from the barley leaf scald pathogen Rhynchosporium secalis. In contrast, reverse genetic approaches have not succeeded for intracellular fungal pathogens such as M. grisea, so positional cloning approaches were used to identify the PWL genes, AVR genes that confer host species specificity (Kang et al., 1995; Sweigard et al., 1995). Map-based cloning strategies are being used to clone other M. grisea AVR genes (Mandel et al., 1997; Farman and Leong, 1998; Dioh et al., 2000).
The utility of R genes in controlling rice blast disease has been limited by the inherent variability of the pathogen. Reports on the degree of genetic instability for AVR genes in the rice blast fungus have been the focus of much debate (reviewed in Valent and Chumley, 1991). At one extreme were reports by Ou (1980) that vegetative cultures derived from single conidia of Philippine pathogens produced conidia of so many races that it was impossible to define race for a single fungal strain. In contrast, Latterell and Rossi (1986) and Bonman et al. (1987) reported that U.S. and Philippine races were genetically stable, although mutants with altered specificity were occasionally detected. Kiyosawa (1976) failed to observe the continuous variation reported by Ou but reported relatively more genetic instability in avirulence determinants corresponding to certain rice blast R genes. Detailed understanding of AVR gene variability requires the molecular cloning and characterization of these genes.
Characterization of a corresponding AVR/R gene pair has not previously been reported for rice blast disease. We set out to clone such a pair of genes to advance the molecular understanding of processes that lead to pathogen recognition and triggering of defense responses. This study and the accompanying article by Bryan et al. (2000)(this issue) present the characterization of the fungal AVR-Pita gene and the corresponding Pi-ta rice blast resistance gene from rice. Genetic mapping of AVR-Pita, along with analysis of spontaneous avr-pita− mutants, suggested that AVR-Pita was located adjacent to a telomere. A gene was cloned from the telomeric region, and when introduced into virulent rice pathogens, the cloned gene specifically conferred avirulence toward rice cultivars that contain Pi-ta. Sequencing of wild-type and mutant alleles suggested that AVR-Pita encodes a neutral zinc metalloprotease, and maintenance of the protease motif appears to be required for avirulence gene function. We show here that the pathogen can escape Pi-ta–mediated recognition by diverse mechanisms, including point mutations, insertion mutations, and deletions of AVR gene sequences. This study provides insight into a potential source of AVR gene hypervariability: location near a telomere.
RESULTS
Initial Steps toward Map-Based Cloning of AVR-Pita
The AVR gene that prevents infection of rice cultivars with the Pi-ta blast R gene was identified through genetic analysis of the Chinese field isolate O-137. We mapped AVR-Pita to one end of linkage group 2c (Sweigard et al., 1993), which corresponds to the telomere 5 (Tel 5) region on chromosome 3 of the consolidated M. grisea restriction fragment length polymorphism (RFLP) map (Nitta et al., 1997). A chromosome walk was initiated from a cosegregating cosmid marker, cos33, and this resulted in assembly of overlapping cosmids, with steps represented by cos33, A43F2, A37H1, and A21F5, as described in Methods. A21F5, the cosmid most distal to cos33, contained a single-copy 3-kb BamHI fragment that cosegregated with AVR-Pita in the mapping population. However, no new cosmids were identified using this BamHI fragment as a probe. None of the cosmids identified were active in transforming a virulent strain of the pathogen to avirulence on rice cultivar Yashiro-mochi, which contains Pi-ta. However, sequences from A21F5 proved useful in characterizing deletion events that inactivate the AVR-Pita gene, as described below.
The AVR Gene Resides at a Telomere
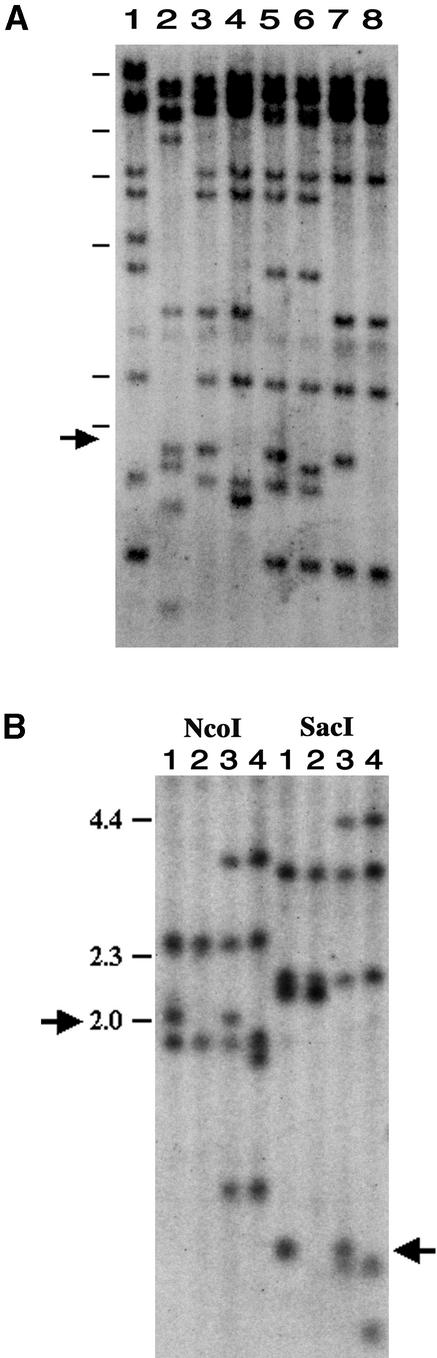
We mapped telomeric RFLPs in the mapping population to determine the distance between the cos33 cluster linked to AVR-Pita and the end of the chromosome. We used an oligonucleotide probe that recognizes the hexameric repeat sequence conserved at the ends of chromosomes in various organisms ranging from filamentous fungi to humans (Zakian, 1989; Schechtman, 1990; Farman and Leong, 1995). Discrete bands identified by probing an EcoRI digest of parental and mutant DNAs with the radiolabeled telomere repeat oligonucleotide are shown in Figure 1A. As expected, these bands mapped to the ends of the M. grisea linkage groups (Sweigard et al., 1993). One of the telomere fragments, a 1.8-kb EcoRI band, was inseparable from AVR-Pita and from the molecular markers linked to AVR-Pita among 66 progeny tested. An analysis of three spontaneous virulent mutants, CP917, CP983, and CP984, showed that all three lack the 1.8-kb EcoRI band linked to AVR-Pita. CP917 has a new EcoRI band ∼300 bp smaller and CP983 has a new band ∼100 bp smaller; for CP984, no novel bands were apparent.
Figure 1.
Identification and Mapping of the AVR-Pita Telomere by Using Virulent Mutants.
Genomic DNAs were digested with EcoRI (A) or NcoI or SacI (B), electrophoresed on 0.7% agarose gels, blotted to a Hybond-N membrane, and hybridized with the 32P-labeled telomere repeat oligonucleotide 5′-(AACCCT)4-3′.
(A) Lanes 1 and 2 contain the DNAs of 6043 (virulent on Yashiro-mochi) and 4224-7-8 (AVR-Pita), respectively, which are the parental strains of cross number 4360 (Sweigard et al., 1993). DNAs loaded in the remaining lanes are from three pairs of avirulent strains and virulent mutants derived from them: 4360-17-1/CP917 (lanes 3 and 4); 4375-R-6/CP983 (lanes 5 and 6); and 4375-R-26/CP984 (lanes 7 and 8). The arrow marks the band corresponding to Tel 5. Bars at left indicate positions of λ HindIII DNA fragments used as length standards; from the top, they are 23.1, 9.4, 6.6, 4.4, 2.3, and 2.0 kb.
(B) Distal restriction fragments of the AVR-Pita telomere were identified by restriction digestion and hybridization of genomic DNAs from two avirulent strain/mutant pairs (4375-R-26/CP984 in lanes 1 and 2, and 4360-17-1/CP917 in lanes 3 and 4). No changes were observed in telomere fragments >4.4 kb, so these fragments are not shown. The telomeric 2.0-kb NcoI fragment and the telomeric 0.8-kb SacI fragment (arrows) were altered in each of the two independent mutants. Not shown are data identifying the distal 9-kb SalI fragment, 6.9-kb BamHI fragment, 6.5-kb BglII fragment, 4-kb EcoRV fragment, and 1.3-kb HindIII fragment. Bars at left indicate positions of λ HindIII DNA length standards in kilobases.
Genomic DNAs were digested with additional restriction enzymes and probed with the telomere repeat oligonucleotide to determine which telomeric fragments were polymorphic between the mutant and parental strains (Figure 1B). This analysis identified the most distal restriction fragment linked to AVR-Pita produced by each enzyme, as summarized in Figure 2. Comparison of the telomeric restriction fragments for wild-type and mutant strains revealed that for all enzymes tested, the CP917 telomere was shortened by ∼300 bp. The deletion in CP917 occurred quite close to the end of the chromosome, as indicated by alteration of the 0.8-kb SacI telomeric fragment (Figure 1B). Although the nature of the CP984 mutation was not clear in this experiment, in a separate experiment we found it to have a larger deletion (see below).
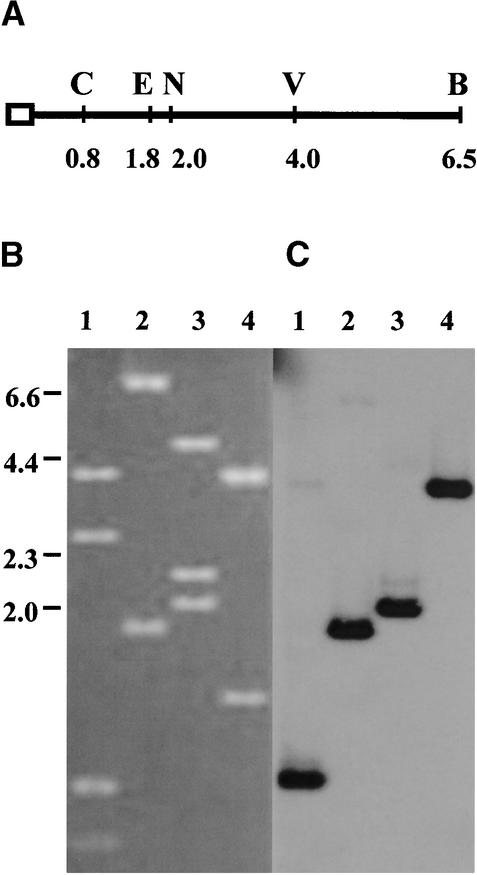
Figure 2.
Cloning of the AVR-Pita–Associated Telomere.
(A) The deduced map of distal restriction fragments of Tel 5 is summarized. The telomere repeats are represented by the open box. The numbers below each restriction site refer to the distance in kilobases from that site to the telomere end. The first SalI site, 9 kb from the chromosome tip, is not shown. B, BglII; C, SacI; E, EcoRI; N, NcoI; V, EcoV.
(B) and (C) Clone pCB780, isolated from a BglII–blunt end size-fractionated library, is shown by DNA gel blot hybridization to contain the Tel 5 BglII fragment. For double digestion of the putative AVR-Pita telomere plasmid DNA, SalI was used to cleave in the polylinker in combination with each of four enzymes: SacI (lanes 1), EcoRI (lanes 2), NcoI (lanes 3), and EcoRV (lanes 4). The SalI site of pBluescript SK+ is 20 bp from the site of insertion of the telomere repeat sequence. The SacI site of pBluescript SK+ is 35 bp from the point at which the BglII end of the telomere fragment was inserted. Restriction digests were separated by electrophoresis, blotted onto Hybond-N membrane, and probed with the radiolabeled telomere repeat oligonucleotide. The ethidium bromide–stained gel (B) and corresponding DNA gel blot (C) are shown. The cloned chromosome end fragment contains two internal SacI sites and one internal NcoI site that were not identified by using the telomere probe. Bars at left identify the positions of λ HindIII DNA length standards in kilobases.
Cloning the Telomere Associated with AVR-Pita
Changes in the sizes of telomeric restriction fragments that correlated with a loss of avirulence toward Yashiro-mochi suggested that the AVR-Pita gene is located directly adjacent to Tel 5. Because cosmid libraries were unlikely to contain chromosome ends, we decided to clone this telomere specifically. Genomic DNA from the avirulent parental strain 4224-7-8 was first treated with BAL- 31 nuclease to remove any 3′ overhang of the G-rich strand and to produce a blunt end at the telomere (Richards and Ausubel, 1988). The BAL-31–treated DNA was digested with BglII and SalI and was used to create a genomic sublibrary of 6- to 7-kb fragments. This produced an enriched fraction of genomic DNA containing the telomere fragment based on our deduction that the 6.5-kb BglII telomeric fragment containing AVR-Pita does not contain any SalI sites (Figure 2A). The sublibrary was screened for telomere-containing clones by using the telomeric oligonucleotide, and positive clones were analyzed further. A single clone, designated pCB780, was obtained with the predicted restriction fragments, shown in Figure 2B.
The selectable fungal transformation marker HPH, encoding hygromycin B resistance (Staben et al., 1989), was introduced into pCB780 to test the telomere clone for functionality. Transforming this plasmid containing the 6.5-kb BglII telomere fragment into the laboratory strain CP987, which is virulent on cultivar Yashiro-mochi, resulted in an unusually large number of unstable transformants. Only one stable transformant was obtained under conditions that typically would yield 30 to 40 stable transformants when the introduced plasmid contained the HPH gene alone. In infection assays, this stable transformant had lost the ability to infect Yashiro-mochi, although it retained full pathogenicity on rice varieties that lack Pi-ta, as illustrated in Figure 3. Thus, the only obvious difference between this transformant and the recipient strain was a gain of avirulence toward Yashiro-mochi.
Figure 3.
Functional Analysis of AVR-Pita.
Infection assay comparing specificity of virulent strain CP987 with the specificity of a transformant that contains a functional AVR-Pita gene. Pots 1 and 4 each contain rice varieties Sariceltik and Tsuyuake, which lack Pi-ta. Pots 2 and 3 contain Yashiro-mochi with Pi-ta. Pots 1 and 2 were inoculated with CP987, and pots 3 and 4 were inoculated with 839-13, a transformant of CP987 that received pCB839, containing the Tel 5 EcoRI fragment lacking the repeats. Plants are shown 7 days after inoculation.
Identification of AVR-Pita
We predicted that AVR-Pita resides very near the telomere because the virulent mutant CP917 contained a small deletion within the 800-bp telomeric SacI fragment (Figure 1B). However, we were not able to further localize the gene using pCB780 subclones retaining the telomeric repeat sequences because these subclones failed to give stable M. grisea transformants. We hypothesized that the instability of fungal transformants generated by using these subclones resulted from the integration of telomeric repeat sequences at internal sites in the recipient's chromosomes, causing chromosome breakage. At the same time, subclones lacking the telomere end produced stable transformants, but they did not confer AVR-Pita gene function. In particular, two subclones that did not confer the avirulence phenotype removed very small amounts of DNA from the telomere end: a 258-bp SspI fragment deletion and a 190-bp ApaLI fragment deletion. Apparently, therefore, the presence of the telomeric hexanucleotide repeat destabilized transformation, and sequences within 190 bp of the telomere end of the clone were required for function of the avirulence gene.
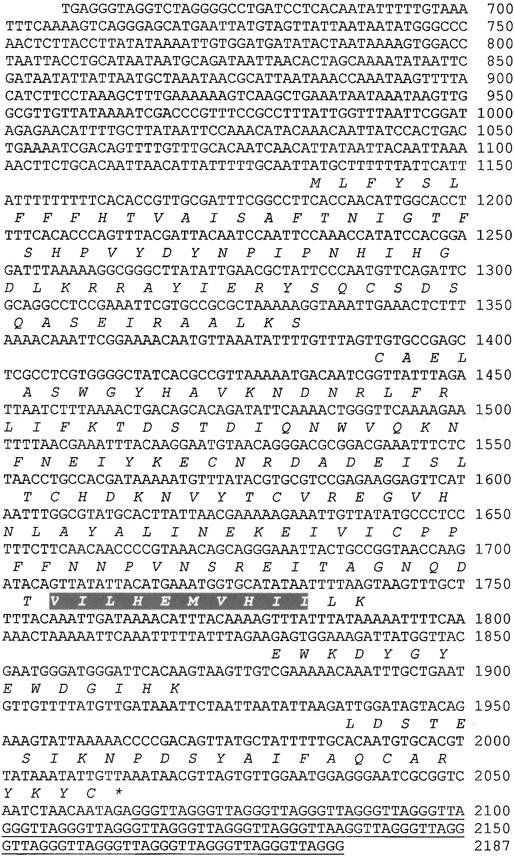
Sequencing the telomere end of the genomic DNA insert in pCB780 identified 20 complete copies and one partial copy of the telomeric repeat sequence (Figure 4). We used a digestion procedure with Exonuclease III (New England Biolabs) to generate a series of clones containing deletions extending from the telomere end into the DNA insert. One clone, pCB808, contained a 123-bp deletion that precisely removed all copies of the telomeric repeat; we incorporated this deletion into subcloned fragments subsequently tested for complementation. As predicted, derivatives that lacked the telomere repeat sequence produced stable M. grisea transformants at normal frequencies. Results presented in Figure 5 show that the activity complementing AVR-Pita was contained in the 1.8-kb EcoRI telomere fragment. However, the 1443-bp ApaI fragment and the 1273-bp HindIII fragment failed to confer avirulence. Two intermediate subclones of 1531 bp and 1595 bp were constructed, and both were active in conferring avirulence. This suggests that sequences required for AVR-Pita gene function were contained within the 89 bp between the ApaI site and the end of the 1531-bp subclone.
Figure 4.
Telomere End Sequence and Predicted Translation of AVR-Pita.
The sequence was determined for 2187 bp from the telomere end of the genomic insert in pCB780 (GenBank accession number AF207841). The distal 1531-bp sequence shown here represents the minimal sequence that confers an avirulent phenotype to strains that are virulent on Yashiro-mochi; it includes 20 complete copies and one partial copy of the telomeric hexanucleotide repeat sequence (bp 2065 to 2187). These repeats were removed (see underlined 123 nucleotides) from clones used for complementation analysis by digestion with exonuclease III. The predicted protein translation is shown below the DNA sequence in italics. Gaps in the translation correspond to introns. A Motifs search for amino acid patterns was conducted using the PROSITE Dictionary of Protein Sites and Patterns, which is available online at http://expasy.cbr.nrc.ca/prosite/ (Hofmann et al., 1999). The putative AVR-Pita protein contains PROSITE pattern PS00142, the zinc binding domain signature from neutral zinc metalloproteases (highlighted). The majority of zinc-dependent metalloproteases contain the following common primary structure: [GSTALIVN]-x-x-H-E-[LIVMFYW]-{DEHRKP}-H-x-[LIVMFYWGSPQ], in which [ ] denotes that any one of the listed residues is allowed, x denotes that any residue is allowed, and { } denotes that none of the listed residues is allowed. Residues in the AVR-Pita motif are shown in boldface.
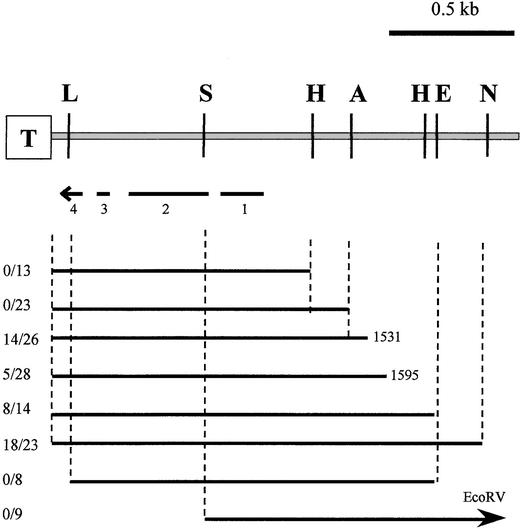
Figure 5.
Sequences Required for AVR-Pita Avirulence Activity.
The telomere repeat sequence (T) and relevant restriction sites obtained by genomic sequencing are indicated in this map of the telomere end of the cloned BglII fragment. Subclones used for complementation analyses are indicated, with the arrow labeled EcoRV indicating a subclone that extends an additional 2 kb to the EcoRV site. Inactive subclones extending from the EcoRI site to the BglII site are not shown. All subclones extending to the telomere end incorporate the 123-bp deletion of the telomere repeat sequence, shown to cause instability in transformed strains. The ratios listed at left indicate the number of avirulent transformants among the total number of hygromycin-resistant transformants tested in the infection assay. Typically, <100% of M. grisea transformants express a nonselected phenotype. The broken arrow indicates the relative sizes and locations of the four exons that make up the AVR-Pita gene. L, ApaLI; S, SacI; H, HindIII; A, ApaI; E, EcoRI; N, NcoI.
Five open reading frames (ORFs) starting with methionine and capable of encoding peptides of >50 amino acids are present between nucleotides 724 and 1770 of the complementing fragment (Figure 4). Three were oriented away from the telomere (nucleotides 879 to 724, 1120 to 926, and 1770 to 1597); the other two were oriented toward the telomere (nucleotides 1134 to 1337 and 1370 to 1762). A single-base insertion was introduced at the SacI site at nucleotide position 1401 by use of partial SacI digestion and treatment with T4 DNA polymerase. This produced a frameshift mutation in one of the ORFs oriented toward the telomere but left the other four ORFs unaffected. Subsequent transformation experiments using plasmid pCB950, which carries the 2.0-kb NcoI telomere fragment with the engineered frameshift mutation, showed that AVR-Pita complementation had been eliminated, because none of 19 independent transformants showed the avirulence phenotype. This experiment implicated the ORF between nucleotides 1370 and 1762 and eliminated further consideration of the three ORFs that would be translated in the opposite orientation.
On the basis of DNA sequence, we predicted that the two ORFs oriented toward the telomere would encode a single polypeptide, with two exons separated by a 56-bp intron that contains good matches to consensus splice sites. This prediction was supported by experiments in which modification of the putative exon 1 ATG at nucleotide 1134 eliminated activity of the AVR-Pita gene, whereas modification of the ATG at nucleotide 1370, which would be contained within the predicted intron, did not affect activity (Figure 4). Two additional smaller exons of 43 and 79 nucleotides could be joined to the exon 1/exon 2 polypeptide by removal of introns of 88 and 66 nucleotides, respectively, which would produce a 223–amino acid ORF.
AVR-Pita did not appear to be transcribed at detectable levels during axenic growth of the fungus in culture. Although the gene later was shown to be expressed in plant tissue at 24 to 32 hr after inoculation (G.T. Bryan and B. Valent, unpublished results), we were unable to synthesize a complete cDNA from RNA isolated from this tissue. We therefore subcloned the putative AVR-Pita genomic coding sequence into a constitutive expression vector, pCB963, under the control of the Aspergillus TrpC promoter and terminator (Staben et al., 1989). Strains transformed with pCB963 and grown in liquid culture expressed AVR-Pita, as determined by RNA gel blot analysis with the DraIII-EcoRI fragment from pCB780 as a probe (data not shown). A cDNA clone obtained from this transformed strain confirmed the positions of the three predicted introns. A second cDNA expression construct (pCB980) under the control of the native AVR-Pita promoter was assembled by fusing the cDNA coding sequence to a 490-bp genomic promoter fragment. When introduced into virulent strains of M. grisea, pCB980 specifically conferred avirulence toward Yashiro-mochi.
Structure of the Deduced AVR-Pita Protein
The AVR-Pita gene encodes a predicted 223–amino acid polypeptide with the stop codon located just 48 bp from the start of telomeric repeat sequences. The deduced AVR-Pita polypeptide shown in Figure 4 exhibited substantial similarity to fungal neutral metalloproteases from Aspergillus spp and Penicillium citrinum (Tatsumi et al., 1991; Matsumoto et al., 1994; Altschul et al., 1997). The fungal metalloproteases showing the greatest sequence identity to AVR-Pita fall into families M35 and M36 in this superfamily of zinc-dependent metalloproteases. AVR-Pita was most similar (27% identities and 44% conserved residues) to NpII, a neutral zinc metalloprotease from Aspergillus oryzae. Among the conserved regions are a 10–amino acid segment, between residues 173 and 182, corresponding to the consensus zinc binding domain of neutral zinc metalloproteases. Three important conserved residues in this domain are His-176 and His-180, which by analogy are predicted to function as zinc ligands, and Glu-177, which is predicted to be the active site residue for a putative metalloprotease (Tatsumi et al., 1991).
Spontaneous Mutations Associated with Loss of Avirulence Gene Function
We isolated additional spontaneous mutants and characterized a total of 11 independent mutational events associated with the gain in virulence toward Yashiro-mochi. Sizes of the telomeric DNA fragments associated with AVR-Pita in each mutant were determined by DNA gel blot analysis, with the telomere oligonucleotide as the probe. Telomeric fragments were unchanged in size for three mutants (CP918, CP1615, and CP1635), suggesting that point mutations were responsible for the change in virulence phenotype. Three other mutants (CP917, CP983, and CP1632) contained telomeric restriction fragments that appeared to have changed in size. The remaining five mutants (CP984, CP1614, CP1638, CP1641, and CP1644) had lost the telomeric fragment as well as all homology to AVR-Pita (data not shown). In the absence of AVR-Pita mutations, the associated telomere appeared to be stable, as evidenced by the sharp telomeric band seen in DNA gel blot analysis of wild-type strains (Figures 1A and 1B). Also, strains with mutations in the unlinked PWL2 host species specificity gene (Sweigard et al. 1995) showed no size changes in the AVR-Pita–associated telomere (data not shown).
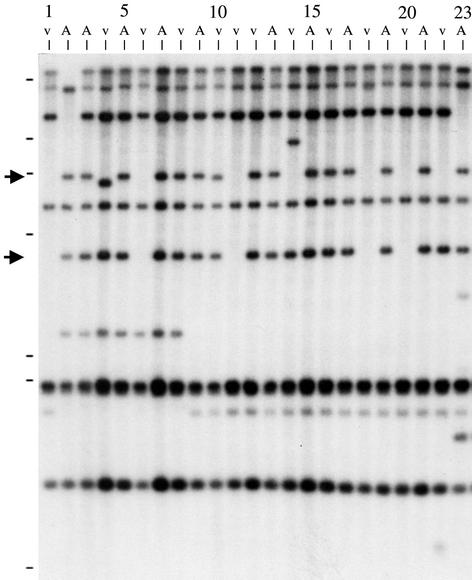
We performed DNA gel blot analyses with other sequences that were linked to AVR-Pita to further characterize the deletion mutants that appeared to have completely lost the telomeric BglII fragment. During the previous PWL2 chromosome walk, Sweigard et al. (1995) identified a 10-kb XhoI fragment from cosmid A10H8 that hybridized to seven distinct BamHI bands, one of which cosegregated with AVR-Pita and Tel 5. We sequentially probed a BglII digest of parental and mutant genomic DNAs with the radiolabeled telomeric oligonucleotide and the A10H8 XhoI fragment to determine whether the A10H8 homology occurred on the 6.5-kb telomeric BglII fragment. Interestingly, the DNA gel blot analysis shown in Figure 6 demonstrates that this probe hybridized to the telomeric 6.5-kb BglII fragment and to an additional 4-kb fragment, both of which were deleted in mutants CP984, CP1614, CP1638, and CP1641. Similarly, hybridizing the A10H8 fragment to SalI-digested parental and mutant DNAs identified the telomeric 9-kb SalI fragment and a 3.5-kb fragment, both of which were deleted in some mutants. Thus, analysis with the A10H8 low-copy repeat sequence identified the penultimate BglII and SalI restriction fragments at Tel 5 and showed that the deletions in four of the mutants extended beyond the 6.5-kb BglII fragment.
Figure 6.
A Low-Copy Repeat Sequence Identifies Restriction Fragments Distal to the AVR-Pita Telomere That Are Deleted in Some Spontaneous Mutants.
Genomic DNAs were digested with BglII for DNA gel blot analysis. Hybridization with the 10-kb XhoI fragment from cosmid A10H8 identified the 6.5-kb telomeric BglII fragment and an additional 4.0-kb fragment (arrows) that was altered in avr-pita− mutants. All avirulent laboratory strains inherited these two bands from the Chinese field isolate O-137 (lane 23) through the RFLP mapping strain 4224-7-8 (lane 2). The virulent mapping parent 6043 (lane 1) does not have these bands. Not shown are data consistent with this result but obtained by using SalI, EcoRI, and SacI. Except for lane 1, which contains DNA from the virulent parent 6043, each v lane contains DNA from a virulent mutant obtained from the first avirulent strain (A lanes) to its left. Lane 1, 6043 (v); lane 2, 4224-7-8 (A); lane 3, 4360-17-1 (A); lane 4, CP917 (v); lane 5, 4375-R-26 (A); lane 6, CP984 (v); lane 7, 4375-R-39 (A); lane 8, CP918 (v); lane 9, 4375-R-6 (A); lane 10, CP983 (v); lane 11, CP1614 (v); lane 12, CP1615 (v); lane 13, CP1631 (A); lane 14, CP1632 (v); lane 15, CP1634 (A); lane 16, CP1635 (v); lane 17, CP1637 (A); lane 18, CP1638 (v); lane 19, CP1640 (A); lane 20, CP1641 (v); lane 21, CP1643 (A); lane 22, CP1644 (v); and lane 23, O-137 (A). Markers at left identify the positions of λ HindIII DNA length standards, which are as follows (from the top): 23.1, 9.4, 6.6, 4.4, 2.3, 2.0, and 0.56 kb.
Cosmid A21F5 identified in our chromosome walking experiments contained a single-copy sequence that mapped to the AVR-Pita locus. DNA gel blot analysis with this 3-kb BamHI fragment as a probe (data not shown) indicated that this sequence did not hybridize to either the 6.5-kb telomeric BglII fragment or the 4-kb BglII fragment identified by the A10H8 sequence. This single-copy clone did hybridize to wild-type BglII fragments of 3.0 and 2.5 kb and to SalI fragments of 1.8, 1.6, and 1.0 kb. These sequences were unaltered in nine of the 11 mutants, but they had been deleted in two of the mutants (CP1614 and CP1641).
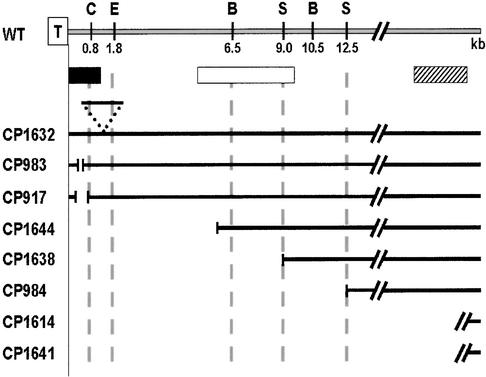
The relative sizes of the deletions in the avr-pita− mutants are summarized in Figure 7. For CP917 and CP983, containing the respective 300- and 100-bp deletions previously identified, no additional upstream deletions were detected by the A10H8 and A21F5 sequences. CP1644 lost most of the 6.5-kb BglII telomeric fragment but retained the penultimate BglII fragment, suggesting that it contained a deletion of ∼6 kb at the chromosome tip. Likewise, CP1638 and CP984 appeared to contain 9- and 12.5-kb deletions, respectively. Two mutants, CP1614 and CP1641, are missing sequences homologous to all three probes, which suggests that they contain large deletions. Finally, CP1632 appeared to have a 1.8-kb insertion between the SacI and EcoRI sites at the chromosome end.
Figure 7.
Insertion/Deletion Events Associated with Loss of Avirulence.
All DNA gel blot analyses probing with the telomeric oligonucleotide (T), the low-copy repeat 10-kb XhoI fragment from A10H8 (open bar), and the single-copy 3.0-kb BamHI fragment from chromosome walk cosmid A21F5 (striped bar) are consistent with the events depicted. The black bar indicates the location of the AVR-Pita coding sequence. The first two BglII (B) and SalI (S) sites are indicated, but only the first SacI (C) and EcoRI (E) sites are shown. CP1632 has an insertion of ∼1.8 kb between the EcoRI and SacI sites (broken tri-angle). CP983 and CP917 have small deletions (vertical broken lines) of ∼100 and 300 bp, respectively. Deletions of ∼6, 9, and 12.5 kb are shown for CP1644, CP1638, and CP984, respectively. CP1614 and CP1641 are missing the AVR-Pita linked fragments identified by all three probes. For these two mutants, the slanted broken lines indicate that the sizes of these deletions are unknown. For all other strains, the slanted broken lines indicate that the distance (in kilobases) between the A10H8 and A21F5 sequences is unknown. WT, wild types.
The CP983 avr-pita− allele, which contained the smallest visible deletion, was cloned and sequenced. This mutant contained a deletion that begins in intron 3 and extends toward the telomere into exon 4, resulting in the loss of 25 amino acids at the C terminus of the AVR-Pita polypeptide (described in Table 1).
Table 1.
Mutations Associated with Loss of Avirulence
| Mutant | Codon Change | Amino Acid Change |
|---|---|---|
| CP918 | TGG1487TAG | W100STOP |
| CP983 | Deletion from intron 3 (at nucleotide 1871) into exon4 |
Loss of C-terminal 25 amino acids |
| CP1615 | TTA1736TGA | L183STOP |
| CP1635 | GAA1718GGA | E177G |
Virulent Alleles without Size Changes in Telomeric Restriction Fragments
Virulent avr-pita− alleles that retain homology to AVR-Pita and lack telomeric polymorphisms were cloned and sequenced. All were found to contain point mutations that alter the predicted AVR-Pita polypeptide, as shown in Table 1. Two have nonsense mutations, resulting in termination at codons 100 and 183, and the other has a missense mutation that results in the substitution of glycine for glutamic acid at position 177 (E177G). Interestingly, this would correspond to removal of the active site glutamic acid in the protease motif. Thus, a missense mutation that abolishes avirulence activity would also be expected to eliminate the putative metalloprotease activity.
DISCUSSION
Avirulence genes have been identified on the basis of their role in triggering specific R gene–mediated resistance in a host plant, but only rarely has their sequence provided a clue as to what role, if any, the gene plays during normal growth and colonization of the host plant (Leach and White, 1996; Laugé and De Wit, 1998; Orth et al., 1999). AVR-Pita encodes a predicted protein that differs from other characterized fungal AVR gene products (small extracellular polypeptides that clearly act as elicitors of R gene–associated cell death). Instead, the AVR-Pita gene product has similarity to proteins with a known biochemical function. AVR-Pita appears to encode a neutral zinc metalloprotease, given its amino acid sequence similarity to fungal metalloproteases (Tatsumi et al., 1991; Matsumoto et al., 1994; Altschul et al., 1997) and the presence of an amino acid motif characteristic of neutral zinc metalloproteases (Figure 4). Although protease activity by this protein has not been demonstrated biochemically, it is implicated. Avirulence function is absent in a spontaneous mutant in which a glycine residue replaces a glutamic acid that is predicted to be important for protease function. G.T. Bryan and B. Valent (unpublished results) have used in vitro mutagenesis to alter other conserved residues in the putative zinc binding domain, leading to a loss of avirulence gene function and providing additional evidence that protease function may play an essential role. However, we do not rule out an alternative explanation for the effect of these missense mutations on AVR-Pita avirulence function: that these mutations destabilize the protein structure and lead to increased rates of degradation. Future studies should also focus on biochemical demonstration of protease activity.
AVR-Pita is unusual in its location extremely close to a telomere. The UAA stop codon of the AVR-Pita ORF is separated from the start of telomere repeat sequences by only 48 bp. It is intriguing to consider that the instability observed for AVR-Pita may be an effect of its telomeric location. Spontaneous avr-pita− mutants were easily isolated after inoculating rice plants that contain the Pi-ta resistance gene. If the phenomenon we see in laboratory studies also occurs in the field, then one mechanism the blast fungus might use to escape Pi-ta–mediated recognition of the AVR signal molecule would be to modify the product of the AVR gene or to prevent its expression completely. Some cases of loss in avirulence were due to point mutations, but the majority we analyzed were due to deletions ranging from 100 bp to >10 kb. One mutant, CP1632, has an insertion of a Pot3 transposon in the AVR-Pita promoter (S. Kang, M.-H. Lebrun, L. Farrall, and B. Valent, unpublished results). These avr-pita− mutants appeared to be normal pathogens in our laboratory studies.
Genes located at telomeres in other systems show unusual genetic and epigenetic regulation. In Saccharomyces cerevisiae, genes located near telomeres appear to be genetically unstable. The SUC gene family, which includes several genes that encode invertase, contains the nontelomeric SUC2 locus and five other SUC loci that are located in subtelomeric regions of different chromosomes (Carlson et al., 1985). Individual yeast strains contain a subset of the telomeric loci. The frequent absence of some of the subtelomeric SUC loci in individual yeast strains suggests that SUC genes have moved to different chromosomal locations since the recent divergence of closely related S. cerevisiae strains. Two other telomeric gene families in S. cerevisiae, the PHO genes, encoding acid phosphatases (Venter and Hörz, 1989), and the MAL loci, each including three genes required for fermentation of maltose (Charron and Michels, 1988), show similar strain-to-strain variation in gene organization. In the African trypanosomes, antigenic variation occurs by regularly changing the expressed copy of the gene that encodes its major surface coat protein, known as the variant surface glycoprotein. Of the ∼103 variant surface glycoprotein genes, only one is expressed, and it is part of a polycistronic transcription unit found at one of several possible telomeric positions (Borst and Rudenko, 1994). A particular gene is activated either by a mechanism of duplicative transposition from a silent position, which is often in a chromosome internal position, or by a switch to a different telomeric expression site.
In S. cerevisiae, another phenomenon observed for genes located at telomeres is the occurrence of gene silencing through transcriptional repression. This silencing is unstable, with genes switching between the repressed and derepressed states. Telomeric gene silencing has been studied primarily by the integration of reporter genes, such as URA3 and ADE2, near a telomere (Gottschling et al., 1990), but it has also recently been shown to occur for a gene normally found in a subtelomeric position (Vega-Palas et al., 2000). Whether telomeric silencing occurs at any stage in the growth of M. grisea is unknown. As was the case with the PWL2 gene (Sweigard et al., 1995), we were unable to detect expression of AVR-Pita in mycelia from axenically grown cultures. However, AVR-Pita expression can easily be detected during later stages of growth inside the host plant (G.T. Bryan and B. Valent, unpublished results). Thus, telomere silencing does not appear to be operative at the AVR-Pita locus during proliferation of the fungus within the plant.
AVR-Pita was not present in standard libraries of M. grisea genomic DNA because it lies so close to the telomere. If other fungal genes for pathogenicity and host specificity are also located near telomeres, some associated challenges in isolating them can be overcome by building and analyzing libraries of telomeric DNA fragments at an early stage of investigation. However, other challenges are inherent in libraries enriched for telomeric repeat sequences. In cloning AVR-Pita, we observed that plasmids carrying telomeric repeat sequences were much less efficient in producing stable transformants of M. grisea than were those plasmids lacking the repeats. We attribute this effect to the destabilization and possible loss of large chromosome segments when telomeric repeats are inserted at proximal sites. This sort of telomere-induced chromosome breakage has been exploited in fungi and mammalian cells to study chromosome structure and telomere-associated gene-silencing mechanisms (Vollrath et al. 1988; Farr et al., 1991; Kistler and Benny, 1992; Brown et al., 1994; Nimmo et al., 1994; Kistler et al., 1996).
AVR genes may be considered to limit the host range of a pathogen by preventing infection on certain host genotypes. The location of an AVR gene at a telomere may allow M. grisea to rapidly adapt to its host. Kiyosawa (1971)(1976) first reported a marked difference in mutation frequency that depended on the particular AVR genes and strains studied, and differences in AVR gene stability have since been confirmed (Sweigard et al., 1995; Mandel et al., 1997). Some AVR genes map near telomeres and others do not. For example, the AVR-TSUY gene that blocks infection of rice variety Tsuyuake maps to Tel 2 of chromosome 1 (Sweigard et al., 1993; Nitta et al., 1997) and pathogen strains containing this AVR gene give rise to virulent mutants at frequencies similar to those seen for AVR-Pita in this study. Dioh et al. (2000) have indirect evidence that the AVR1-Ku86 gene, corresponding to rice variety Ku86, is linked to AVR1-TSUY and the corresponding telomere. AVR1-MedNoï was linked to telomeric RFLPs that were not assigned to a chromosome (Dioh et al., 2000). In contrast, other AVR genes recombine with telomeric markers. The genetically stable AVR1-MARA gene, corresponding to an R gene in rice variety Maratelli, occurs internally on chromosome 2B (Mandel et al., 1997; chromosome 2 in Nitta et al., 1997), and two AVR genes, AVR1-Irat1 (Dioh et al., 2000) and AVR1-CO39 (Farman and Leong, 1998), occur internal to the AVR-TSUY telomere in the area around RFLP marker A11D9. Telomeric location would not explain all examples of genetic instability, however. The genetically unstable PWL2 gene does not appear to be located at a telomere (Sweigard et al., 1995).
Interestingly, three copies of a low-copy repeat sequence, present in the 10-kb XhoI fragment from cosmid A10H8, are linked to AVR genes (Sweigard et al., 1995). This XhoI fragment from A10H8 identified seven distinct BamHI bands by gel blot analysis of the RFLP mapping parents (Sweigard et al., 1993), and four of these were present in both mapping parents. Three bands segregated in the mapping population: one mapped to the PWL2 locus on linkage group 3; one mapped near cos72 and A11D9, markers that are linked to AVR1-CO39 and AVR1-Irat1 on LG1; and one mapped to the AVR-Pita locus at Tel 5. Indeed, we have now shown that this sequence occurs very near to AVR-Pita, within the telomeric 6.5-kb BglII fragment.
A detailed population analysis of AVR-Pita occurrence and evolution in the field is now possible. DNA gel blot analysis with the AVR-Pita gene as a probe determined that restriction fragments were conserved within single genetic lineages in Colombia (Montenegro-Chamorro, 1997) and in the Philippines (Zeigler et al., 1995) but generally varied between lineages. Particular AVR-Pita restriction fragments were correlated with an avirulence phenotype, and others were correlated with virulence. A few lineages lacked homology to AVR-Pita. Montenegro-Chamorro (1997) sequenced AVR-Pita alleles from Colombian lineages and demonstrated striking conservation of the AVR-Pita coding sequence relative to the gene from the Chinese rice pathogen O-137. Uniformly avirulent lineages contain genes encoding AVR-Pita proteins that differ by only two or three amino acids from the O-137 protein. Virulent lineages contain avr-pita− genes encoding proteins that uniformly differ by seven amino acids relative to AVR-Pita proteins that confer avirulence. Again, these avr-pita− proteins differ from each other by only one or two amino acids. This conservation of AVR-Pita structure among field isolates of the pathogen in Colombia contrasts with the genetic instability we see for the telomeric AVR-Pita locus in laboratory studies. If the AVR-Pita gene were to reside at different chromosomal locations in the different blast strains under study, this might account for the apparent differences in gene stability. Striking variations in the electrophoretic karyotypes seen among different M. grisea strains that infect rice (Talbot et al., 1993; Orbach et al., 1996) would be consistent with variable locations for single AVR genes within different field isolates. The full impact of telomeric location of AVR genes on the dynamics of rice blast disease in the field remains to be determined.
Some virulent avr-pita− genes from field isolates of the pathogen encode a single conservative amino acid substitution in the protease motif relative to the O-137 AVR-Pita protein (Montenegro-Chamorro, 1997; S. Kang, G.T. Bryan, and B. Valent, unpublished results). However, in these cases, the new amino acid also fits within the defined protease motif (Figure 4). This suggests that evolution of avirulence alleles may involve modifications of a protein such that its metabolic function is preserved but its recognition as an avirulence factor is altered. If protease activity is confirmed for AVR-Pita, it will be important to determine the relationship between this activity and the avirulence function of the protein in triggering Pi-ta–mediated resistance. Bryan et al. (2000)(this issue) report that the corresponding Pi-ta resistance gene encodes a predicted cytoplasmic receptor with a centrally located nucleotide binding site and a C-terminal leucine-rich domain. Jia et al. (2000) report evidence that a putative processed form of the AVR-Pita protein interacts directly with the Pi-ta protein to trigger resistance responses. These intriguing results present an opportunity for understanding the presumed dual functions an AVR gene plays in the lifestyle of the pathogen, in contributing to the pathogenicity process and in sometimes triggering R gene–mediated defense responses.
METHODS
Fungal Strains and Plant Infection Assays
The Magnaporthe grisea strains used in this study are described in Table 2. The O-137 AVR gene, originally named AVR2-YAMO for avirulence toward the Pi-ta–containing differential variety Yashiro-mochi (Yamada et al., 1976), was renamed AVR-Pita when correspondence with Pi-ta was demonstrated. Infection assays have been described (Valent et al., 1991).
Table 2.
Fungal Strains Used in This Study
| Name | Description/Reference |
|---|---|
| O-137 | AVR-Pita+ field isolate collected in 1985 at the China National Rice Research Institute (Hangzhou) (B. Valent and Y. Shen, unpublished results) |
| 4224-7-8 | AVR-Pita+ progeny of a cross between laboratory strain 4157-1-1 and field isolate O-137 (Sweigard et al., 1995) |
| 6043 | avr-pita− laboratory strain (Leung et al., 1988) |
| 4360-17-1 | AVR-Pita+ progeny of a cross between 4224-7-8 and 6043 |
| 4375-R-6 | AVR-Pita+ progeny of a cross between 4360-17-1 and 4360-R-12 |
| 4375-R-26 | AVR-Pita+ progeny of a cross between 4360-17-1 and 4360-R-12 |
| 4375-R-39 | AVR-Pita+ progeny of a cross between 4360-17-1 and 4360-R-12 |
| CP917 | avr-pita− mutant of 4360-17-1, spontaneous |
| CP918 | avr-pita− mutant of 4375-R-39, spontaneous |
| CP983 | avr-pita− mutant of 4375-R-6, spontaneous |
| CP984 | avr-pita− mutant of 4375-R-26, spontaneous |
| CP987 | avr-pita−pwl2− double mutant, obtained from CP917, spontaneous (Sweigard et al., 1995) |
| CP1614 | avr-pita− mutant of 4375-R-6, spontaneous |
| CP1615 | avr-pita− mutant of 4375-R-6, spontaneous |
| CP1631 | AVR-Pita+ culture of 4375-R-6 derived from a single conidium |
| CP1632 | avr-pita− mutant of CP1631, spontaneous |
| CP1634 | AVR-Pita+ culture of 4375-R-6 derived from a single conidium |
| CP1635 | avr-pita− mutant of CP1634, spontaneous |
| CP1637 | AVR-Pita+ culture of 4375-R-6 derived from a single conidium |
| CP1638 | avr-pita− mutant of CP1637, spontaneous |
| CP1640 | AVR-Pita+ culture of 4375-R-6 derived from a single conidium |
| CP1641 | avr-pita− mutant of CP1640, spontaneous |
| CP1643 | AVR-Pita+ culture of 4375-R-6 derived from a single conidium |
| CP1644 | avr-pita− mutant of CP1643, spontaneous |
Mutant Isolation
Some spontaneous virulent mutants, such as CP917, CP983, CP984, and CP918, were isolated during the course of the genetic analyses that identified AVR-Pita. A similar procedure was described for the host species specificity gene PWL2 (Sweigard et al., 1995). Additional spontaneous mutants were isolated from strain 4375-R-6 in two experiments. In the first, ∼20 Yashiro-mochi plants were inoculated with 15 mL of a spore suspension (100,000 conidia/mL) from a single culture of 4375-R-6. Single conidia were isolated from each of 17 fully susceptible lesions, and cultures derived from these conidia were retested for their ability to cause disease on Yashiro-mochi. All retests showed fully virulent interactions, with hundreds of coalescing lesions that killed the plants. These mutants fell into two independent classes when analyzed by DNA gel blot analysis using the telomeric oligonucleotide probe. Isolates from 14 of the lesions, represented by CP1614, were missing the 1.8-kb EcoRI telomere fragment, and isolates from the other three lesions, represented by CP1615, retained the 1.8-kb EcoRI fragment. In a second experiment, 10 genetically purified cultures were derived from strain 4375-R-6 and preserved by frozen storage for future analysis. For each culture, 6 mL of spore suspension (100,000 spores/mL) was inoculated onto seven Yashiro-mochi plants. Five independent virulent mutants were isolated, each from one or two individual lesions that were produced by inoculation with five of the independent cultures of 4375-R-6 (Table 2 and Figure 6).
Plasmids, DNA Libraries, and DNA and RNA Manipulations
Plasmids used in this study are described in Table 3. Recombinant plasmids were made using pBluescript SK+ (Stratagene) unless otherwise stated. Unless noted, Escherichia coli strain DH5α (Gibco BRL) was used. The “A” cosmid library was constructed with genomic DNA from strain 4392-1-6 in pMOcosX (Orbach, 1994), as described (Sweigard et al., 1995). Standard protocols were used for restriction enzyme digestions and RNA and DNA gel blot analysis (Sambrook et al., 1989; Ausubel et al., 1994), except as noted below for hybridizations with the oligonucleotide probe. The oligonucleotide 5′-(AACCCT)4-3′ was synthesized and radiolabeled by kinase treatment with γ-32P-ATP for detection of the hexameric telomere repeat sequence by DNA gel blot analysis. Genomic DNAs were prepared as described (Sweigard et al., 1995), digested with restriction enzymes, electrophoresed on 0.7% agarose gels, and blotted to Hybond-N membranes. For hybridization with the telomeric repeat oligonucleotide, membranes were prehybridized in a solution of 6 × SSPE (1 × SSPE is 0.15 M NaCl, 10 mM sodium phosphate, and 1 mM EDTA, pH 7.4), 5 × Denhardt's solution (1 × Denhardt's solution is 0.02% Ficoll, 0.2% polyvinylpyrrolidone, and 0.02% BSA), 0.5% SDS, and 100 μg/mL single-strand calf thymus DNA for 2 hr at 42°C and then hybridized with radiolabeled oligonucleotide overnight at 42°C. Membranes were washed first for 10 min in 2 × SSPE containing 0.1% SDS at room temperature and then for 15 min in the same solution at 30 to 34°C.
Table 3.
Key Plasmids Used in This Study
| Name | Description/Reference |
|---|---|
| pCB780 | 6.5-kb BglII blunt telomere fragment from 4224-7-8 in BamHI-EcoRV sites of pBluescript SK+ |
| pCB781 | pCB780 containing HPH subcloned as a 2.3-kb SaII fragment from plasmid pCSN43 (Staben et al., 1989) |
| pCB783 | 791-bp telomeric SacI fragment in pBluescript SK+ constructed by deletion of three SacI fragments (5.7 kb) in pCB780 |
| pCB806 | 6.5-kb BglII end fragment lacking telomere repeats constructed by replacing the 430-bp DraIII fragment from pCB780 with the 307- bp DraIII fragment from pCB808 |
| pCB808 | 668-bp SacI telomeric end fragment with 123-bp deletion of the telomere repeats by Exonuclease III digestion of pCB783 |
| pCB813 | 2-kb NcoI end fragment lacking telomere repeats constructed by deletion of the NcoI-BglII fragment from pCB806 |
| pCB831 | avr-pita− allele from CP918 in pBluescript KS− |
| pCB832 | avr-pita− allele from CP1615 in pBluescript KS− |
| pCB833 | avr-pita− allele from CP1635 in pBluescript KS− |
| pCB839 | 1.8-kb EcoRI end fragment lacking telomere repeats constructed by deletion of the EcoRI-NcoI fragment from pCB813 |
| pCB840 | 1.3-kb HindIII end fragment lacking repeats constructed by deletion of the HindIII and HindIII-NcoI fragments from pCB813 |
| pCB843 | avr-pita− allele from CP983 in pBluescript KS− |
| pCB859 | 1.4-kb ApaI end fragment lacking telomere repeats constructed by deletion of the ApaI-NcoI fragment from pCB813 |
| pCB950 | 2-kb NcoI fragment in pCB813 subjected to partial digestion by SacI and flushing with T4 polymerase, producing a frameshift at the nucleotide 1401 SacI site |
| pCB963 | Genomic AVR-Pita coding sequence replacing HPH in pCSN43 (Staben et al., 1989) |
| pCB979 | cDNA coding sequence with incorporated ClaI-BamHI sites cloned into the ClaI and BamHI sites of pBluescript SK+ |
| pCB980 | 490 bp of AVR-Pita promoter with incorporated ClaI site cloned into ClaI-HincII sites of pCB979 for expression of cDNA coding sequence |
Cosmid Contig Assembly
Filters containing the cosmid library were probed with the linked marker cos33, identifying two sets of overlapping cosmids represented by A43F2 and A1A4. A43F2 contained rice pathogen–specific repetitive DNA sequences as well as sequences that cosegregated with the AVR-Pita locus, but A1A4 was subsequently shown to be located elsewhere. Probing cosmid filters with radiolabeled A43F2 DNA identified new cosmids A29F12, A37H1, and A37H9. As a probe for subsequent walking steps, radiolabeled RNA fragments corresponding to the ends of the inserts were synthesized by making use of the T7 and T3 bacteriophage promoters that flank the cloning site in the pMOcosX cosmid vector (Orbach, 1994). The end-specific probe produced from the T3 promoter in cosmid A37H1 contained highly repetitive DNA. However, the end-specific probe produced from the T7 promoter, which hybridized to a few prominent bands against a background smear, was used to identify cosmid A21F5. Both end fragments from A21F5 contained highly repetitive DNA. However, A21F5 contained an internal single-copy 3-kb BamHI fragment that cosegregated with AVR-Pita. This BamHI fragment hybridized to previously identified cosmids A21F5, A29F12, A37H1, and A37H9 but failed to identify new cosmids.
Telomere Fragment Cloning
Genomic DNA from strain 4224-7-8, which is avirulent on rice cultivars that contain Pi-ta, was treated with 0.125 units/mL BAL-31 nuclease (New England Biolabs, Beverly, MA) for 50 min at 30°C, as described (Richards and Ausubel, 1988). Under these conditions, the size of the telomeric fragments underwent no visible decrease, as determined by DNA gel blot analysis. The DNA was then digested with the restriction enzymes BglII and SalI and subjected to electrophoresis on a 0.8% low-melting-point agarose gel. DNA fragments in the size range of 7 to 8 kb were eluted from the gel and ligated into the BamHI and EcoRV polylinker sites of pBluescript SK+.
Deletion of the Telomere Repeats
An Exonuclease III deletion procedure, modified from Henikoff (1984), was used to produce progressive deletions from the telomere repeat end of the putative AVR-Pita clone. Plasmid pCB783, containing the 791-bp telomeric SacI fragment, was digested first with KpnI and XhoI and then with Exonuclease III (New England Biolabs). The ExoIII-digested DNAs were treated with S1 nuclease (Sigma) followed by a fill-in reaction with the Klenow fragment of DNA polymerase I, ligation, and transformation into DH5αMCR cells (Gibco BRL). The extent of deletion in each clone was determined by sequencing. One deletion clone, pCB808, contained a 123-bp deletion that precisely removed the telomeric repeats. The 307-bp DraIII fragment from pCB808 was substituted for the 430-bp DraIII fragment from pCB780 to produce pCB806, containing the 6.5-kb BglII fragment minus the telomere repeat sequence.
Fungal Transformation and Complementation Analysis
Fungal transformation was performed as described (Sweigard et al., 1995). A selectable marker encoding hygromycin B resistance, the HPH gene, was subcloned into the SalI site of pCB780 to produce pCB781. However, most complementation tests for function in conferring avirulence were accomplished by cotransformation, in which a second HPH-containing plasmid, pCB725 (Sweigard et al., 1995), was transformed along with a fivefold excess of the plasmid lacking the selection marker. Strain CP987 (Sweigard et al., 1995) was used as a recipient.
Subclones tested for complementation (Figure 5), including the NcoI, EcoRI, ApaI, and HindIII fragments, were derived from pCB806, which had the deletion of the telomere repeat sequence. All constructs were verified by sequencing. Additional subclones were made to further localize sequences required for AVR gene function. Oligonucleotide LFO (5′-ACGTACGGGATCCGAGGGTAGGTCTAGG-3′) was constructed to subclone a 1531-bp fragment, and LFN (5′-ACGTACGGGATCCGCTTGAATCCGGAG-3′) subcloned a 1595-bp fragment. BamHI sites (underlined) were incorporated for ligation into pBlueScript SK+. These oligonucleotides were used for polymerase chain reaction (PCR) together with LFG (5′-ACGTGTCGACTCTATTGTTAGATTGACCG-3′; SalI site underlined), which amplifies sequences adjacent to the telomere repeat and incorporates a SalI site.
Site-Directed Mutagenesis
Megaprimer mutagenesis (Aiyar and Leis, 1993) was performed to replace each of the ATGs in the potential initiation codons at nucleotides 1134 and 1370 with a HindIII recognition site (AAGCTT) by using oligonucleotides LFP (5′-AAGCTTTTTTATTCATTATTTTTTTTT-CAC-3′) and LFQ (5′-AAGCTTAATATTTTGTTTAGTTGTGCCGAG-3′), respectively. Megaprimers were produced using 18 PCR cycles and the following primer combinations: LFG/LFP, LFG/LFQ, and LFE (5′-CTGTATCTTGGTTACCGGC-3′)/LF36 (5′-GACAACTACCATGGA-ACCC-3′). The products of the mutant oligonucleotide reactions were purified, mixed with purified LFE/LF36 product, and amplified for 25 cycles by using the LFG and LFO primers described above. Reaction products were cut with SalI and BamHI and ligated into pBluescript SK+. Precise incorporation of the desired mutations was verified by sequencing.
cDNA Cloning
For construction of a constitutive expression vector for the putative AVR-Pita gene, pCSN43 (Staben et al., 1989) was first modified to eliminate extra BamHI and ClaI sites by deleting the smaller MluI-SacI fragment. The AVR-Pita genomic coding sequence (nucleotides 1134 to 2067) was cloned by PCR using oligonucleotides designed to place a ClaI site at the start ATG (LF2C, 5′-GATCGAATCGATATGCTTTTTTATTCATTATTTTTTTTTC-3′) and a BamHI site at the telomere end (LF2D, 5′-GATCGAGGATCCCCCTCTATTGTTAGATTG-ACC-3′). We then replaced the coding sequence of HPH in pCSN43 with the sequence coding AVR-Pita to produce pCB963.
Transgenic fungus containing pCB963 was grown in liquid culture for purification of RNA. The Applied Biosystems (Foster City, CA) GeneAmp RNA PCR kit protocol was used to reverse-transcribe RNA by using random hexamer priming followed by PCR amplification of cDNA with oligonucleotides LF2C and LF2D. The cDNA was subcloned into pBluescript SK+ by using the incorporated ClaI and BamHI sites and was named pCB979. A native AVR-Pita promoter fragment containing nucleotides 645 to 1133 was fused to the cDNA-coding sequence in pCB979. The promoter fragment was obtained from pCB813 by using PCR primers LF2H (5′-AAGCATATCGATAAAAATAATGTTAATTGTGCAG-3′), incorporating a ClaI site to match the one introduced at the ATG by primer LF2C, and LF12 (5′-GCCGAGTCG-TTCTGAGGG-3′). A fill-in reaction with the PCR reaction product was performed with the Klenow fragment of DNA polymerase I, followed by digestion with ClaI. This DNA fragment was subcloned into pCB979 that had been digested with ClaI and HincII to produce pCB980.
Cloning Mutant Alleles
Mutant alleles from CP918, CP1615, and CP1635 were cloned into pBluescript KS− (Stratagene) by PCR with oligonucleotides LF17 (5′-TGACCGCGATTCCCTCCATT-3′) and LF36. The allele from CP983 was cloned by using LF8 (5′-CTTGTGAATCCCATCCC-3′) and LF36. Independently isolated clones were sequenced to eliminate PCR errors.
Acknowledgments
We thank Anne M. Carroll for her excellent technical support and especially for her assistance with constructing the RFLP map. We are grateful to Phil Smith and Kathy Kline-Smith for their dedication to maintaining the excellent plant growth facility used in this study. We thank Gregory T. Bryan, Yulin Jia, and Todd DeZwaan for thoughtful comments on the manuscript.
References
- Aiyar, A., and Leis, J. (1993). Modification of the megaprimer method of PCR mutagenesis: Improved amplification of the final product. BioTechniques 14, 366–368. [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1994). Current Protocols in Molecular Biology. (New York: Greene Publishing Associates/Wiley Interscience).
- Bonman, J.M., Vergel de Dios, T.I., Bandong, J.M., and Lee, E.J. (1987). Pathogenic variability of monoconidial isolates of Pyricularia oryzae in Korea and in the Philippines. Plant Dis. 71, 127–130. [Google Scholar]
- Borst, P., and Rudenko, G. (1994). Antigenic variation in African trypanosomes. Science 264, 1872–1873. [DOI] [PubMed] [Google Scholar]
- Brown, K.E., Barnett, M.A., Burgtorf, C., Shaw, P., Buckle, V.J., and Brown, W.R.A. (1994). Dissecting the centromere of the human Y-chromosome with cloned telomeric DNA. Hum. Mol. Genet. 3, 1227–1237. [DOI] [PubMed] [Google Scholar]
- Bryan, G.T., Wu, K.-S., Farrall, L., Jia, Y., Hershey, H.P., McAdams, S.A., Faulk, K.N., Donaldson, G.K., Tarchini, R., and Valent, B. (2000). A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12, 2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, M., Celenza, J.L., and Eng, F.J. (1985). Evolution of the dispersed SUC gene family of Saccharomyces by rearrangements of chromosome telomeres. Mol. Cell. Biol. 5, 2894–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron, M.J., and Michels, C.A. (1988). The naturally occurring alleles of MAL1 in Saccharomyces species evolved by various mutagenic processes including chromosomal rearrangement. Genetics 120, 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioh, W., Tharreau, D., Notteghem, J.L., Orbach, M., and Lebrun, M.-H. (2000). Mapping avirulence genes in the rice blast fungus, Magnaporthe grisea, with RFLP and RAPD markers. Mol. Plant-Microbe Interact. 13, 217–227. [DOI] [PubMed] [Google Scholar]
- Farman, M.L., and Leong, S.A. (1995). Genetic and physical mapping of telomeres in the rice blast fungus, Magnaporthe grisea. Genetics 140, 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farman, M.L., and Leong, S.A. (1998). Chromosome walking to the AVR1–CO39 avirulence gene of Magnaporthe grisea. Discrepancy between the physical and genetic maps. Genetics 150, 1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr, C., Fantes, J., Goodfellow, P., and Cooke, H. (1991). Functional reintroduction of human telomeres into mammalian cells. Proc. Natl. Acad. Sci. USA 88, 7006–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Gottschling, D.E., Aparicio, O.M., Billington, B.L., and Zakian, V.A. (1990). Position effect at S. cerevisiae telomeres: Reversible repression of pol II transcription. Cell 63, 751–762. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. (1984). Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 28, 351–359. [DOI] [PubMed] [Google Scholar]
- Hofmann, K., Bucher, P., Falquet, L., and Bairoch, A. (1999). The PROSITE database, its status in 1999. Nucleic Acids Res. 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, S., Sweigard, J.A., and Valent, B. (1995). The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 8, 939–948. [DOI] [PubMed] [Google Scholar]
- Keen, N.T., Tamaki, S., Kobayashi, D., Gerhold, D., Stayton, M., Shen, H., Gold, G., Lorang, J., Thordal-Christensen, H., Dahlbeck, D., and Staskawicz, B. (1990). Bacteria expressing avirulence gene D produce a specific elicitor of the soybean hypersensitive reaction. Mol. Plant-Microbe Interact. 3, 112–121. [Google Scholar]
- Kistler, H.C., and Benny, U. (1992). Autonomously replicating plasmids and chromosome rearrangement during transformation of Nectria haematococca. Gene 117, 81–89. [DOI] [PubMed] [Google Scholar]
- Kistler, H.C., Meinhardt, L.W., and Benny, U. (1996). Mutants of Nectria haematococca created by a site-directed chromosome breakage are greatly reduced in virulence toward pea. Mol. Plant-Microbe Interact. 9, 804–809. [Google Scholar]
- Kiyosawa, S. (1971). Genetical approach to the biochemical nature of plant disease resistance. Jpn. Agric. Res. Q. 6, 73–80. [Google Scholar]
- Kiyosawa, S. (1976). Pathogenic variations of Pyricularia oryzae and their use in genetic and breeding studies. SABRAO J. 8, 53–67. [Google Scholar]
- Latterell, F.M., and Rossi, A.E. (1986). Longevity and pathogenic stability of Pyricularia oryzae. Phytopathology 76, 231–235. [Google Scholar]
- Laugé, R., and De Wit, P.J.G.M. (1998). Fungal avirulence genes: Structure and possible functions. Fungal Gen. Biol. 24, 285–297. [DOI] [PubMed] [Google Scholar]
- Leach, J.E., and White, F.F. (1996). Bacterial avirulence genes. Annu. Rev. Phytopathol. 34, 153–179. [DOI] [PubMed] [Google Scholar]
- Leung, H., Borromeo, E.S., Bernardo, M.A., and Notteghem, J.L. (1988). Genetic analysis of virulence in the rice blast fungus Magnaporthe grisea. Phytopathology 78, 1227–1233. [Google Scholar]
- Levy, M., Correa-Victoria, F.J., Zeigler, R.S., Xu, S., and Hamer, J.E. (1993). Genetic diversity of the rice blast fungus in a disease nursery in Colombia. Phytopathology 83, 1427–1433. [Google Scholar]
- Mandel, M.A., Crouch, V.W., Gunawardena, U.P., Harper, T.M., and Orbach, M.J. (1997). Physical mapping of the Magnaporthe grisea AVR1-MARA locus reveals the virulent allele contains two deletions. Mol. Plant-Microbe Interact. 10, 1102–1105. [Google Scholar]
- Matsumoto, K., Yamaguchi, M., and Ichishima, E. (1994). Molecular cloning and nucleotide sequence of the complementary DNA for penicillolysin gene, plnC, and 18 kDa metalloendopeptidase gene from Penicillium citrinum. Biochim. Biophys. Acta 1218, 469–472. [DOI] [PubMed] [Google Scholar]
- Midland, S.L., Keen, N.T., Sims, J.J., Midland, M.M., Stayton, M.M., Burton, V., Smith, M.J., Mazzola, E.P., Graham, K.J., and Clardy, J. (1993). The structures of syringolides 1 and 2, novel C-glycosidic elicitors from Pseudomonas syringae pv. tomato. J. Org. Chem. 58, 2940–2945. [Google Scholar]
- Montenegro-Chamorro, M.V. (1997). Alleleic Diversity of an Avirulence Gene in Colombian Field Isolates of the Rice Blast Fungus. M.S. Thesis (West Lafayette, IN: Purdue University).
- Nimmo, E.R., Cranston, G., and Allshire, R.C. (1994). Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J. 13, 3801–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta, N., Farman, M.L., and Leong, S.A. (1997). Genome organization of Magnaporthe grisea: Integration of genetic maps, clustering of transposable elements and identification of genome duplications and rearrangements. Theor. Appl. Genet. 95, 20–32. [Google Scholar]
- Orbach, M.J. (1994). A cosmid with a HyR marker for fungal library construction and screening. Gene 150, 159–162. [DOI] [PubMed] [Google Scholar]
- Orbach, M.J., Chumley, F.G., and Valent, B. (1996). Electrophoretic karyotypes of Magnaporthe grisea pathogens of diverse grasses. Mol. Plant-Microbe Interact. 9, 261–271. [Google Scholar]
- Orth, K., Palmer, L.E., Bao, Z.Q., Stewart, S., Rudolph, A.E., Bliska, J.B., and Dixon, J.E. (1999). Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science 285, 1920–1923. [DOI] [PubMed] [Google Scholar]
- Ou, S.H. (1980). Pathogen variability and host resistance in rice blast disease. Annu. Rev. Phytopathol. 18, 167–187. [Google Scholar]
- Richards, E.J., and Ausubel, F.M. (1988). Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53, 127–136. [DOI] [PubMed] [Google Scholar]
- Rossman, A.Y., Howard, R.J., and Valent, B. (1990). Pyricularia grisea, the correct name for the rice blast fungus. Mycologia 82, 509–512. [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schechtman, M.G. (1990). Characterization of telomere DNA from Neurospora crassa. Gene 88, 159–165. [DOI] [PubMed] [Google Scholar]
- Silué, D., Notteghem, J.L., and Tharreau, D. (1992). Evidence for a gene-for-gene relationship in the Oryza sativa–Magnaporthe grisea pathosystem. Phytopathology 82, 577–580. [Google Scholar]
- Staben, C., Jensen, B., Singer, M., Pollock, J., Schechtman, M., Kinsey, J., and Selker, E. (1989). Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet. Newsl. 36, 79–81. [Google Scholar]
- Sweigard, J.A., Valent, B., Orbach, M.J., Walter, A.M., Rafalski, A., and Chumley, F.G. (1993). A genetic map of the rice blast fungus Magnaporthe grisea. In Genetic Maps, 6th ed, S.J. O'Brien, ed (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 3.112–3.117.
- Sweigard, J.A., Carroll, A.M., Kang, S., Farrall, L., Chumley, F.G., and Valent, B. (1995). Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 7, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swords, K.M.M., Dahlbeck, D., Kearney, B., Roy, M., and Staskawicz, B. (1996). Spontaneous and induced mutations in a single open reading frame alter both virulence and avirulence in Xanthomonas campestris pv. vesicatoria avrBs2. J. Bacteriol. 178, 4661–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot, N.J., Salch, Y.P., Ma, M., and Hamer, J.E. (1993). Karyotypic variability within clonal lineages of the rice blast fungus, Magnaporthe grisea. Appl. Environ. Microbiol. 59, 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi, H., Murakami, S., Tsuji, R.F., Ishida, Y., Murakami, K., Masaki, A., Kawabe, H., Arimura, H., Nakano, E., and Motai, H. (1991). Cloning and expression in yeast of a cDNA clone encoding Aspergillus oryzae neutral protease II, a unique metalloprotease. Mol. Gen. Genet. 228, 97–103. [DOI] [PubMed] [Google Scholar]
- Valent, B., and Chumley, F.G. (1991). Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea. Annu. Rev. Phytopathol. 29, 443–467. [DOI] [PubMed] [Google Scholar]
- Valent, B., Farrall, L., and Chumley, F.G. (1991). Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 127, 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Palas, M.A., Martin-Figueroa, E., and Florencio, F.J. (2000). Telomeric silencing of a natural subtelomeric gene. Mol. Gen. Genet. 263, 287–291. [DOI] [PubMed] [Google Scholar]
- Venter, U., and Hörz, W. (1989). The acid phosphatase genes PHO10 and PHO11 in S. cerevisiae are located at the telomeres of chromosomes VIII and I. Nucleic Acids Res. 17, 1353–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollrath, D., Davis, R.W., Connelly, C., and Hieter, P. (1988). Physical mapping of large DNA by chromosome fragmentation. Proc. Natl. Acad. Sci. USA 85, 6027–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, M. (1985). Pathogenic specialization of rice blast fungus in Japan. Jpn. Agric. Res. Q. 19, 178–183. [Google Scholar]
- Yamada, M., Kiyosawa, S., Yamaguchi, T., Hirano, T., Kobayashi, T., Kushibuchi, K., and Watanabe, S. (1976). Proposal of a new method for differentiating races of Pyricularia oryzae Cavara in Japan. Ann. Phytopathol. Soc. Jpn. 42, 216–219. [Google Scholar]
- Zakian, V.A. (1989). Structure and function of telomeres. Annu. Rev. Genet. 23, 579–604. [DOI] [PubMed] [Google Scholar]
- Zeigler, R.S., Leong, S.A., and Teng, P.S., eds (1994). Rice Blast Disease. (Wallingford, UK: Commonwealth Agricultural Bureau International).
- Zeigler, R.S., Cuoc, L.X., Scott, R.P., Bernardo, M.A., Chen, D.H., Valent, B., and Nelson, R.J. (1995). The relationship between lineage and virulence in Pyricularia grisea in the Philippines. Phytopathology 85, 443–451. [Google Scholar]