Abstract
We examined the bacterial aerobic nasal flora of 216 healthy volunteers to identify potential competitive interactions among different species, with special emphasis on the influence of staphylococcal agr alleles. The Staphylococcus aureus colonization rate correlated negatively with the rate of colonization by Corynebacterium spp. and non-aureus staphylococci, especially S. epidermidis, suggesting that both Corynebacterium spp. and S. epidermidis antagonize S. aureus colonization. Most of the S. aureus and S. epidermidis isolates were agr typed by a PCR method. Only one S. aureus agr (agrSa) allele was detected in each carrier. Multiple logistic regression of the two most prevalent agrSa alleles (agr-1Sa and agr-2Sa) and the three S. epidermidis agr (agrSe) alleles showed a specific influence of the agr system. The results of this model did not support conclusions drawn from previous in vitro agr-specific cross-inhibition experiments. Our findings suggest that the agr alleles, which are strongly linked to the bacterial genetic background, may simply be associated with common biological properties—including mediators of bacterial interference—in the strains that bear them.
Staphylococcus aureus is present in the nasal vestibule of at least 30% of individuals in the normal population, and this carriage is a major risk factor for infection (12). S. aureus is an important cause of community- and hospital-acquired infections (14), and strategies for eliminating nasal carriage have been developed (11).
Longitudinal studies of healthy individuals have shown three patterns of S. aureus carriage: (i) noncarriage, (ii) intermittent carriage, and (iii) persistent carriage of the same or different strains (23, 24). The differences could be due to host factors and/or to antagonism between members of the nasal flora. Indeed, a lower incidence of S. aureus colonization is observed in individuals heavily colonized by Corynebacterium spp. (22), and interaction between these two species was confirmed by in vivo experiments showing that experimental colonization by Corynebacterium spp. inhibits colonization by S. aureus (22). Inconsistent results have been obtained with other species, including non-aureus staphylococci (18, 22).
Expression of cell wall-associated and extracellular proteins in staphylococci is controlled by the agr locus, which encodes a two-component signaling pathway whose activating ligand is a bacterial-density-sensing peptide (autoinducing peptide [AIP]) which is also encoded by agr (10). A polymorphism in the AIP amino acid sequence and in that of its corresponding receptor has been described in staphylococci (4, 7, 9). S. aureus strains can be divided into four major groups (designated agr-1Sa to agr-4Sa), such that within each group, all strains produce a peptide that can activate the agr response in the other members of the same group whereas autoinducing peptides are usually mutually inhibitory between members of different groups (7, 9). Functional agr loci are present in other staphylococcal species, including S. epidermidis (agr-1Se to agr-3Se) (3, 4, 26), which are different from each other and from agrSa. The agr-1Se AIP inhibits the activity of agr-1Sa to agr-3Sa but not agr-4Sa, while among S. aureus AIPs, only type 4 (weakly) inhibits agr-1Se activity (20). It has been proposed that agr-2Sa S. aureus strains hinder umbilical stump colonization by agr-1Sa strains (19). The biological mechanism of this interference is unknown but might be caused by molecular cross-interference between agr alleles.
The aim of the present investigation was to determine the qualitative and quantitative composition of the nasal flora of healthy individuals, focusing on S. aureus, coagulase-negative staphylococci, and corynebacteria, and to identify potential interactions between these bacteria. Staphylococcal isolates were analyzed at the species and agr allele level, and a mathematical model of bacterial nasal interference was constructed.
MATERIALS AND METHODS
Subjects.
The nasal floras of 216 healthy volunteer students (defined as subjects with no history of S. aureus disease and no current antibiotic use) from four medical and nursing schools (75, 69, 22, and 50 volunteers, respectively) were sampled. The mean age of the volunteers was 21 years (range, 17 to 35 years), and there were 64 males and 152 females.
Estimation of the nasal vestibule flora.
The standard cotton swabbing technique was used to sample the nasal vestibule. Swabs were streaked on sheep blood agar and incubated at 37°C in an aerobic atmosphere for 48 h. Bacterial density was estimated by counting CFU in logarithmic graduations. The representative colonies were subcultured and identified using standard methods, as described below. Twenty randomly selected S. aureus-positive swabs were inoculated in brain heart broth (bioMérieux) and cultured for 24 h at 37°C. After centrifugation, pellets were harvested and stored at −20°C until used for DNA extraction.
Identification of isolates.
Staphylococcus species were identified on the basis of conventional phenotypic characteristics, namely, Gram staining, cell morphology and cell arrangement, colony morphology and pigmentation on P agar and Trypticase soy agar (bioMérieux) supplemented with horse blood, catalase activity, coagulase production in rabbit plasma (bioMérieux), and production of clumping factor (Pastorex Staph Plus; bioMérieux). For species identification of coagulase-negative staphylococci, we used individual tests (susceptibility to furazolindone [300 μg], bacitracin [0.02 U], desferrioxamine [250 μg], and novobiocin) and the ID32 Staph gallery (bioMérieux). Corynebacterium spp. were identified on the basis of colony morphology and pigmentation on Trypticase soy agar supplemented with horse blood and also on the basis of cell morphology and cell arrangement after Gram staining; they were not identified to the species level.
agr typing by multiplex PCR.
Genomic DNA was extracted from staphylococci grown on agar plates or in brain heart infusion broth (13) and used as an amplification template with primers (Table 1) designed from the agr-1Sa to agr-4Sa and agr-1Se to agr-3Se sequences (GenBank accession numbers X52543, AF001782, AF001783, AF288215, Z49220, AF346724, and AF346725, respectively) to amplify specific agr alleles. For multiplex PCR, two primer sets were prepared: one to amplify agrSa alleles and another to amplify agrSe alleles. Amplification was carried out under the following conditions: an initial 5-min denaturation step at 95°C followed by 25 stringent cycles (1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 1 min of extension at 72°C) and a final extension step at 72°C for 10 min. The quality of the DNA extracts and the absence of PCR inhibitors were confirmed by amplification of gyrA (S. aureus) or the 16S-23S intergenic region of the ribosomal DNA operon (S. epidermidis) (13). PCR products were analyzed by electrophoresis through 0.8% agarose gels (Sigma). The following strains were used to control the specificity of PCR amplification: (i) S. aureus RN6390 (agr-1Sa), RN6923 (agr-2Sa), RN8462 (agr-3Sa), and A880740 (agr-4Sa) (7); (ii) S. epidermidis CCM2124 (agr-1Se), N910160 (agr-2Se), and N910191 (agr-3Se) (4).
TABLE 1.
Nucleotide sequences of agr type-specific oligonucleotide primers used in this study, and anticipated sizes of PCR products
| Gene | Primer | Oligonucleotide sequence (5′-3′)a | Size of amplified product (bp)a | Multiplex PCR set |
|---|---|---|---|---|
| agrSa | agr1-4Sa-1 | ATGCACATGG TGCACATGC | 1 | |
| agr-1Sa | agr1Sa-2 | GTCACAAGTA CTATAAGCTG CGAT | 439 | 1 |
| agr-2Sa | agr2Sa-2 | TATTACTAAT TGAAAAGTGC CATAGC | 572 | 1 |
| agr-3Sa | agr3Sa-2 | GTAATGTAAT AGCTTGTATA ATAATACCCA G | 321 | 1 |
| agr-4Sa | agr4Sa-2 | CGATAATGCC GTAATACCCG | 657 | 1 |
| agr-1Se | agr1Se-1 | GGCATTAGTC GGATTAATTA TTACG | 438 | 2 |
| agr1Se-2 | TGTAGGCCTG CAAACGG | 2 | ||
| agr-2Se | agr2Se-1 | TTTACCATTT GCAGCTATAC AAGTG | 575 | 2 |
| agr2Se-2 | ATAACAATAA TATAACCAAA CTCAAAAGTA CAG | 2 | ||
| agr-3Se | agr3Se-1 | GAAAGAGTGT ATTCAATGGA TGAGC | 338 | 2 |
| agr3Se-2 | TAAATATTAT GTATTATATC TTCAGTATAT AAAGAGATGA | 2 |
Statistical methods.
Colony counts were log10 transformed for analysis. Interspecies relationships were first described on a two-by-two basis, looking at the presence or absence of S. aureus (of each of the four agrSa alleles) with respect to the number of colonies (CFU) of one group of bacteria including Corynebacterium spp., non-aureus staphylococci (including S. epidermidis), S. epidermidis (of each of the three agrSe alleles), and S. aureus (of each of agrSa alleles), and validated by the χ2 test. To explain the probability that S. aureus is present in terms of the number of colonies of the other species, standard linear regression does not apply. Hence, a multiple logistic regression model was used to analyze simultaneously the influence of Corynebacterium spp. and S. epidermidis agr alleles on the probability of the presence of S. aureus agr-1Sa or agr-2Sa. Age, sex, and school of origin were used as candidate covariates for adjustment. Those with a P value below 0.05 were finally retained in the final model. In this model, the colony counts were categorized into four groups for corynebacteria (<102 [reference group], 102 to 103, 103 to 104, and ≥104) and into two groups for agrSe-1 to agrSe-3 (<102 [reference group] and ≥102). Statistical analyses were done with software from SAS Inc.
RESULTS
Aerobic flora of the nasal vestibule.
Colony counts in the 216 volunteers ranged from 102 to 106 CFU/swab (median 104). Sixty-five volunteers were positive for S. aureus (30%), 209 were positive for non-aureus staphylococci (97%), 150 were positive for Corynebacterium spp. (69%), 1 was positive for Streptococcus (<1%), 2 were positive for Micrococcus (1%), and 18 were positive for gram-negative species (8%). Of the 209 non-aureus Staphylococcus carriers, 198 were positive for S. epidermidis (93%), 12 were positive for S. capitis (6%), 9 were positive for S. haemolyticus (4%), 9 were positive for S. warneri (4%), 8 were positive for S. hominis (4%), 4 were positive for S. lugdunensis (2%), 3 were positive for S. cohnii subsp. cohnii (1%), and 1 was positive for S. auricularis (<1%); three isolates were not confidently identified to the species level (1%). The observed composition of the aerobic nasal flora was qualitatively and quantitatively similar to that in previous studies (5, 11).
Determination of the agr type of staphylococcal isolates.
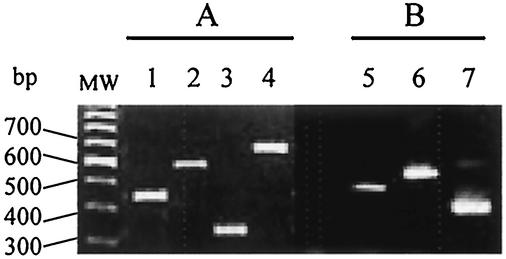
The agr type of all isolates belonging to the two most prevalent staphylococcal species, S. aureus and S. epidermidis, was determined by using specific multiplex PCR. To reduce possible bias of strain cloning, a minimum of 10 colonies of each morphological type were randomly selected for multiplex PCR. As shown in Fig. 1, strong specific signals of the expected sizes were obtained with the reference strains. All 65 S. aureus isolates fell into one of the four previously described agrSa groups (alleles). Only one agrSa allele was detected in each carrier: 34 isolates belonged to agr-1Sa, 19 belonged to agr-2Sa, 7 belonged to agr-3Sa, and 5 belonged to agr-4Sa. To verify that each carrier harbored only one agrSa allele, PCR was also performed on total DNA extracted from brain heart infusion broth cultures of samples from 20 of the S. aureus carriers. In all cases, only one agrSa allele (identical to one of the four previously identified alleles) was detected in each culture by multiplex PCR.
FIG. 1.
Multiplex PCR detection of S. aureus (A) and S. epidermidis (B) agr alleles. Lanes 1 to 4, PCR amplicons from primer set agrSa alleles using DNA from S. aureus RN6390 (agr-1Sa), RN6923 (agr-2Sa), RN8462 (agr-3Sa), and A880740 (agr-4Sa), respectively; lanes 5 to 7, PCR amplicons from set agrSe alleles using DNA from S. epidermidis CCM2124 (agr-1Se), N910160 (agr-2Se), and N910191 (agr-3Se), respectively.
Of the 198 S. epidermidis isolates, 194 were positive by PCR for at least one of the previously described agrSe alleles: 96 isolates belonged to agr-1Se, 63 belonged to agr-2Se, and 97 belonged to agr-3Se. Most carriers (n = 140) harbored only one agrSe allele (agr-1Se in 53 carriers, agr-2Se in 34 carriers, and agr-3Se in 53 carriers), while two agrSe alleles were detected in 50 carriers (agr-1Se and agr-2Se in 11 carriers, agr-1Se and agr-3Se in 27 carriers, and agr-2Se and agr-3Se in 12 carriers) and three were detected in four carriers.
Interactions between staphylococci and corynebacteria.
To detect possible interference between Corynebacterium and staphylococcal species in carriers, we examined the number of culture-positive samples for each staphylococcal species as a function of the number of Corynebacterium CFU. As shown in Table 2, within the range of 0 to 103 Corynebacterium CFU, the proportion of samples positive for S. aureus was quite similar (34 to 46%), whereas it fell significantly at higher Corynebacterium CFU levels (16 and 11% of samples were S. aureus positive at 104 and >104 Corynebacterium CFU, respectively; P = 0.08 and 0.001, respectively). The multiple-regression model (Table 3) confirmed that the probability of S. aureus isolation was reduced by about 3-fold when the Corynebacterium CFU level rose from <102 to 104 (odds ratio = 0.36; P = 0.16) and by more than 10-fold when it rose to ≥105 (odds ratio = 0.08; P = 0.0001). In contrast, the rate of non-aureus staphylococcal colonization (including S. epidermidis) was not affected by the degree of Corynebacterium colonization (Table 2). These results confirm those of previous studies indicating that Corynebacterium spp. specifically inhibit colonization by S. aureus but not by non-aureus staphylococci (22).
TABLE 2.
Incidence of nasal carriage of S. aureus, non-aureus staphylococci, and S. epidermidis according to the degree of colonization by Corynebacterium spp.
| No. of Corynebacterium sp. colonies (CFU) | No. of carriers of Corynebacterium spp. | No. (%) of healthy volunteers colonized witha:
|
|
|---|---|---|---|
| S. aureus | Non-aureus staphylococci | ||
| 0-103 | 142 | 56 (39)b | 136 (96)b |
| 103-104 | 19 | 3 (16), P < 0.08 | 19 (100), NSc |
| 104-106 | 55 | 6 (11), P < 0.001 | 54 (98), NS |
A χ2 test was used to analyze the effect of Corynebacterium spp. on the presence of S. aureus, S. epidermidis, and non-aureus staphylococci. The number of colonies was grouped into three categories.
Reference groups.
NS, not significant.
TABLE 3.
Influence of the number of Corynebacterium or S. epidermidis colonies on the probability of S. aureus isolation
| No. of colonies (CFU) |
S. aureus odds ratio (CI95%), P valuea for:
|
|
|---|---|---|
| Corynebacterium spp. | S. epidermidis | |
| <102 | 1 | 1 |
| 102-103 | 1.30 (0.57-2.89), 0.52 | 0.09 (0.03-0.30), 0.0001 |
| 103-104 | 0.36 (0.09-1.50), 0.16 | 0.16 (0.05-0.59), 0.005 |
| ≥104 | 0.08 (0.03-0.27), 0.0001 | 0.11 (0.04-0.41), 0.0008 |
A multiple logistic regression model was used to analyze simultaneously the effect of Corynebacterium spp. and S. epidermidis on the presence of S. aureus. The number of colonies was grouped into four categories, as follows: <102 (reference group, for which only the odds ratio is given), 102 to 103, 103 to 104, and ≥104 CFU. Corynebacteria and S. epidermidis were independently associated with the presence of S. aureus (P = 0.001). CI95%, 95% confidence interval.
To determine whether S. aureus, non-aureus staphylococci, or S. epidermidis sensu stricto inhibited Corynebacterium colonization, we determined the number of Corynebacterium culture-positive samples as a function of the staphylococcal CFU level. The percentage of Corynebacterium isolates fell from 73 to 42% (not a significant change) when the S. aureus CFU value increased from 0 to >105 but was unaffected by the non-aureus staphylococci and S. epidermidis CFU values (not shown). Thus, Corynebacterium specifically inhibited S. aureus colonization but not vice versa.
Relationships between staphylococcal species.
To determine if non-aureus staphylococci, especially S. epidermidis, inhibited S. aureus nasal colonization, we examined the number of S. aureus culture-positive samples as a function of the non-aureus staphylococci (including S. epidermidis) and S. epidermidis sensu stricto CFU values. The number of S. aureus-positive samples fell markedly as the non-aureus staphylococcal (not shown) and S. epidermidis CFU values increased (from 78 to 23%, P = 0.001; and from 66 to 30%, P = 0.004, respectively) (Table 4), in particular in the volunter with <103 UFC of Corynebacterium spp. (from 88 to 31%, P = 0.001). Multiple-regression analysis (Table 5) showed that the probability of S. aureus isolation fell by about 6- to 10-fold when the S. epidermidis CFU value rose from <102-103 to 104 and ≥105 (odds ratios, 0.1 to 0.16; P, 0.0001 to 0.005). This model also showed that the presence of S. epidermidis and Corynebacterium spp. was independently associated with the presence or absence of S. aureus (P = 0.0001). Colonization by >103 non-aureus staphylococci, and especially S. epidermidis, was clearly protective against S. aureus colonization. The rate of non-aureus staphylococcal and S. epidermidis colonization fell from 100 to 81% and from 99 to 81% as the number of S. aureus CFU increased (not shown); however, these results were not statistically significant, probably owing to the small number of volunteers strongly colonized by S. aureus. Lastly, some rarely detected staphylococcal species such as S. warneri, S. lugdunensis, S. haemolyticus, and S. cohnii (but not S. capitis or S. hominis) were never isolated simultaneously with S. aureus.
TABLE 4.
Incidence of nasal carriage of S. aureus according to the degree of colonization by S. epidermidis
| No. of S. epidermidis colonies (CFU) | No. of healthy volunteers colonized with S. aureus [no. of cases/total no. (%), P value)a
|
||
|---|---|---|---|
| Without adjustment with Corynebacterium spp. | In presence of <103 CFU of Corynebacterium spp. | In presence of ≥103 CFU of Corynebacterium spp. | |
| 0-10 | 12/18 (66)b | 10/11 (91)b | 2/7 (26)b |
| 10-102 | 6/13 (46)b | 6/8 (75)b | 0/5 (0)b |
| 102-103 | 25/78 (32), 0.004 | 23/73 (31), 0.001 | 2/25 (8), NSc |
| 103-104 | 11/40 (27), 0.004 | 7/22 (32), 0.001 | 4/18 (22), NS |
| 104-106 | 11/37 (30), 0.004 | 10/28 (36), 0.001 | 1/19 (5), NS |
A χ2 test was used to analyze the effect of S. epidermidis in the presence of S. aureus with and without adjustment with the number of Corynebacterium CFU acording to the results of Table 3. The number of S. epidermidis colonies was grouped into five categories.
Reference groups.
NS, not significant.
TABLE 5.
Influence of the number of colonies of corynebacteria on the probability of isolating S. aureus strains bearing agr-1Sa and agr-2Sa
| No. of Corynebacterium colonies (CFU) |
S. aureus odds ratio (CI95%), P valuea for:
|
|
|---|---|---|
| agr-1Sa | agr-2Sa | |
| <102 | 1 | 1 |
| 102-103 | 0.93 (0.38-2.19), 0.84 | 1.69 (0.55-5.26), 0.35 |
| 103-104 | NIb | 0.77 (0.08-7.20), 0.81 |
| ≥104 | 0.10 (0.02-0.48), 0.004 | 0.33 (0.06-1.61), 0.17 |
In this model, the colony counts were categorized into four groups for corynebacteria as follows: <102 (reference group, for which only the odds ratio is given), 102 to 103, 103 to 104, and ≥ 104 CFU. CI95% 95% confidence interval.
NI, not informative because of the small number of subjects.
Relationships between staphylococci bearing different agr alleles.
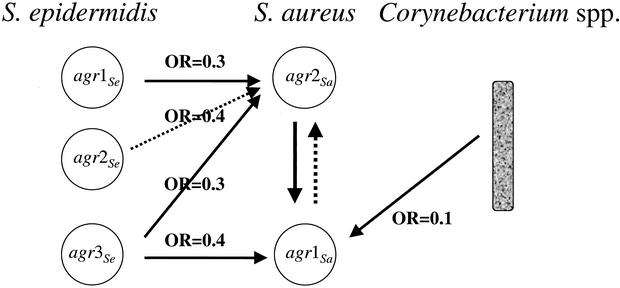
As stated above, even though multiple colonies of S. aureus from each swab were typed by PCR, we detected only one agrSa allele per sample. The presence of agr-2Sa was significantly associated with the absence of agr-1Sa (P = 0.02; Fig. 2). Other agrSa allele associations were not statistically significant, owing to the small number of subjects colonized by the relevant strains. To determine whether the interaction of S. epidermidis with S. aureus was linked to agr alleles, we used a multiple logistic regression model to analyze the influence of S. epidermidis agr alleles (agr-1Se, agr-2Se, and agr-3Se) and Corynebacterium spp. (as control) on the presence of the two most frequent S. aureus agr alleles (agr-1Sa and agr-2Sa) (Tables 5 and 6). The probability of isolating agr-1Sa S. aureus was reduced by about 2.7-fold when the CFU count of agr-3Se S. epidermidis was ≥102 versus <102 (odds ratio, 0.39; P = 0.004), while no statistical differences were observed with other agrSe alleles (P > 0.35 or higher). The probability of isolating agr-1Sa S. aureus was reduced by a factor of about 10 when the number of Corynebacterium CFU was ≥104 versus <102 (odds ratio, 0.10; P = 0.004), as observed above. In contrast, the probability of isolating agr-2Sa S. aureus was reduced by a factor of about 3 when the CFU count of agr-1Se, agr-2Se, or agr-3Se S. epidermidis was ≥102 versus <102. The results were statistically significant for agr-1Se and agr-3Se but not for agr-2Se, probably because of the small number of isolates (odds ratio, 0.31, P = 0.06; odds ratio, 0.37, P = 0.12; and odds ratio, 0.33; P = 0.05, respectively). Surprisingly, the probability that the agr-2Sa S. aureus colonization rate fell as the Corynebacterium colonization rate rose was not statistically significant, despite the large number of isolates (odds ratio minimal 0.33, P = 0.17) (Table 5; Fig. 2). agr-3Se S. epidermidis and Corynebacterium isolation was independently associated with the presence of agr-1Sa S. aureus (P = 0.027 and P = 0.0002, respectively), while agr-1Se and agr-2Se S. epidermidis isolation was independently associated with the presence of agr-2Sa S. aureus (P = 0.046 and P = 0.044, respectively).
FIG. 2.
Model of agr-dependent staphylococcal interference. Based on semiquantitative analysis of the aerobic nasal flora, a multiple logistic regression model was used to analyze simultaneously the effect of S. aureus agrSa alleles, Corynebacterium spp., and S. epidermidis agrSe alleles on S. aureus colonization. OR, odds ratio; threshold of 102 CFU for S. epidermidis and 104 CFU for Corynebacterium spp.
TABLE 6.
Influence of the number of colonies of agr-1Se to agr-3Se allele-bearing S. epidermidis on the probability of isolating S. aureus strains bearing agr-1Sa and agr-2Sa
| No. of colonies (CFU) |
S. aureus agr-1Sa odds ratio (CI95%), P value, for S. epidermidisa:
|
S. aureus agr-2Sa odds ratio (CI95%), P value, for S. epidermidisa:
|
||||
|---|---|---|---|---|---|---|
| agr-1Se | agr-2Se | agr-3Se | agr-1Se | agr-2Se | agr-3Se | |
| <102 | 1 | 1 | 1 | 1 | 1 | 1 |
| ≥102 | 0.96 (0.41-2.26), 0.92 | 0.64 (0.15-2.76), 0.35 | 0.39 (0.16-0.93), 0.004 | 0.31 (0.09-1.07), 0.06 | 0.37 (0.10-1.32), 0.12 | 0.33 (0.10-1.03), 0.05 |
In this model, the colony counts were categorized into two groups for S. epidermidis agr-1Se to agr-3Se (<102 [reference group, for which only the odds ratio is given] and ≥102 CFU). CI95%, 95% confidence interval.
DISCUSSION
We analyzed the composition of the aerobic nasal flora of 216 healthy volunteers to identify potential competitive species interactions, with special emphasis on the influence of staphylococcal agr alleles. We found that the S. aureus colonization rate in subjects colonized by Corynebacterium spp. and/or non-aureus staphylococci, especially S. epidermidis, was significantly lower than in subjects not colonized by these species, suggesting that both Corynebacterium spp. and S. epidermidis antagonize nasal colonization by S. aureus.
Most of the S. aureus and S. epidermidis isolates were agr typed by PCR. Only one agrSa allele was detected in each individual's nasal flora, as previously described (25). Moreover, multiple logistic regression analysis with the two most prevalent agrSa alleles (agr-1Sa and agr-2Sa) and the three agrSe alleles confidently showed agr-specific interaction in the nasal vestibule (Fig. 2). Our initial hypothesis was that staphylococcal interaction in the nose would reflect the heteroallelic inhibitory activity previously detected in vitro. In these in vitro experiments, it was shown that the autoinducing peptides produced by the different groups are usually mutually inhibitory on the expression of RNAIII, the effector of the agr system, and subsequently on the expression of exoproteins and toxins of S. aureus. Hence, agrSa alleles, with the exception of agr-1Sa and agr-4Sa, are all mutually inhibitory in vitro on the expression of RNAIII (7, 9), and agr-1Se is also inhibitory for agrSa but not for agr-4Sa (20). If these in vitro interferences were relevant in nasal colonization, we would have observed an inhibitory effect of agr-1Se on agr-1Sa and agr-2Sa and frequent simultaneous detection of agr-4Sa and agr-1Sa or agr-4Sa and agr-1Se. In fact, we found no correlation between in vitro and in vivo data on agr alleles. Our results were not in support of a predominant role of the agr system in staphylococcal interaction in the human nasal vestibule, as previously observed in several staphylococcal diseases (8). Jarraud et al. (8) examined the possible relationship between agr groups and human S. aureus disease by studying 198 S. aureus strains isolated from patients with suppurative infections and acute toxemia. A relationship between the genetic background, agr group, and disease type was observed in most cases of toxin-mediated disease and in several suppurative infections such as infective endocarditis. Jarraud et al concluded that the agr type had no direct responsibility for disease initiation and speculated that the preferential association between certain agr alleles, certain toxin genes, and a particular genetic background may reflect an ancient evolutionary division of S. aureus in terms of this fundamental biology of the species (8).
Hence, the apparent agr-dependent in vivo interactions observed in the present study may have been due to other mechanisms. One possibility is the synthesis of antagonists such as bacteriocins, bacteriolytic enzymes, hydrogen peroxide, lactic acids or fatty acids, and ammonia (2). Bactericidal exoproteins have been already detected in staphylococcal and Corynebacterium spp. Among the staphylococci, previous studies have identified S. aureus bactericiocin against some Corynebacterium species (17, 21) and S. epidermidis bacteriocin against S. aureus (6, 16). However, no bacteriocin-like activity produced by corynebacteria against S. aureus or S. epidermidis or by one S. aureus strain against another has been described. Another possible mechanism of bacterial interference is competition among Corynebacterium spp., S. aureus, and S. epidermidis for specific attachment to epithelial cells (1). Uehara et al. suggested that binding competition might involve the carbohydrate portion of the human nasal mucin support and showed that Corynebacterium spp. had higher affinity for mucus than did S. aureus and that S. aureus had higher affinity than did S. epidermidis (22). In our study, Corynebacterium spp. inhibited S. aureus colonization, but only strains harboring the agr-1Sa allele (Table 5). We did not find other concordant results between those reported by Uehara and our in vivo data. The physiological role of mucus is to bind and remove bacteria, not to promote bacterial adhesion to the epithelium. In adhesion experiments with human airway epithelial cells, Mongodin et al. recently showed that S. aureus did not adhere in vivo to intact mucus-producing airway epithelium but did adhere to the basolateral plasma membrane of columnar cells, to basal cells, and to the basement membrane (15).
Finally, while we did not identify the precise mechanism of the observed bacterial interference in nasal colonization, our mathematical analysis of ecological data produced a working model of bacterial interactions in the nasal vestibule. Our subsequent experiments aimed at determining these mechanisms will focus on specific interactions identified by the model. Importantly, our results show that the likelihood of nasal colonization by S. aureus in healthy subjects varies with the composition of the local flora. The relevance of our model to patients with underlying diseases remains to be tested, but it is noteworthy that most methicillin-resistant S. aureus (MRSA) strains harbor agr-1Sa (reference 25 and unpublished personal data) and that colonization by agr-1Sa strains was specifically associated with a low rate of colonization by Corynebacterium spp. and agr-3Se S. epidermidis. Indeed, our model predicted that the probability of agr-1Sa S. aureus (probably MRSA) colonization in such cases would be increased by a factor of 33. Larger cross-sectional and longitudinal studies are required to understand how one S. aureus strain can displace another, especially in the case of MRSA colonization.
Acknowledgments
We are grateful to N. Violland, A. Meyret, C. Courtier, and C. Gardon for technical assistance and to D. Young for editing the manuscript.
This work was supported by a grant from Recherche en Microbiologie et Maladies Infectieuses et Parasitaires of the Ministère de l'Education Nationale.
REFERENCES
- 1.Bibel, D. J., R. Aly, C. Bayles, W. G. Strauss, H. R. Shinefield, and H. I. Maibach. 1983. Competitive adherence as a mechanism of bacterial interference. J. Can. Microbiol. 29:700-703. [DOI] [PubMed] [Google Scholar]
- 2.Brook, I. 1999. Bacterial interference in upper respiratory tract infections. Rev. Med. Microbiol. 10:225-233. [Google Scholar]
- 3.Donvito, B., J. Etienne, T. Greenland, C. Mouren, V. Delorme, and F. Vandenesch. 1997. Distribution of the synergistic haemolysin genes hld and slush with respect to agr in human staphylococci. FEMS Microbiol. Lett. 151:139-144. [DOI] [PubMed] [Google Scholar]
- 4.Dufour, P., S. Jarraud, F. Vandenesch, T. Greenland, R. P. Novick, M. Bes, J. Etienne, and G. Lina. 2002. High genetic variability of the agr locus in Staphylococcus species. J. Bacteriol. 184:1180-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleurette, J. 1995. Les flores microbiennes commensales de la peau et des muqueuses, p. 362-403. In J. Fleurette, J. Freney, and M.-E. Reverdy (ed.), Antiseptie et désinfection. ESKA, Paris, France.
- 6.Heilmann, C., and G. Peters. 2000. Biology and pathogenicity of Staphylococcus epidermidis, p. 442-449. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 7.Jarraud, S., G. J. Lyon, A. M. Figueiredo, G. Lina, F. Vandenesch, F. Etienne, T. W. Muir, and R. P. Novick. 2000. Exfoliatin-producing strains define a four agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meunier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr type (alleles), and human disease type. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 10.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kluytmans, J., A. van Belkum, and H. Verbrugh. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lecomte, F., M. Nouvellon, and H. Levesque. 2001. Nasal carriage of Staphylococcus aureus. N. Engl. J. Med. 344:1399-1400. [DOI] [PubMed] [Google Scholar]
- 13.Lina, G., A. Quaglia, M.-E. Reverdy, R. Leclercq, F. Vandenesch, and J. Etienne. 1999. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 43:1062-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 15.Mongodin, E., O. Bajolet, J. Hinnrasky, E. Puchelle, and S. de Bentzmann. 2000. Cell wall-associated protein A as a tool for immunolocalization of Staphylococcus aureus in infected humain aiway epithelium. J. Histochem. Cytochem. 48:523-533. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura, T., K. Hirai, Y. Shibata, and S. Fujimura. 1998. Purification and properties of a bacteriocin of Staphylococcus epidermidis isolated from dental plaque. Oral Microbiol. Immunol. 13:387-389. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura, T., N. Yamazaki, H. Taniguchi, and S. Fujimurar. 1983. Production, purification, and properties of a bacteriocin from Staphylococcus aureus isolated from saliva. Infect. Immun. 39:609-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicoll, T. R., and M. M. Jensen. 1987. Preliminary studies on bacterial interference of staphylococcosis of chickens. Avian Dis. 31:140-144. [PubMed] [Google Scholar]
- 19.Novick, R. P. 1999. Regulation of pathogenicity in Staphylococcus aureus by a peptide-based density-sensing system, p. 129-146. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, D.C.
- 20.Otto, M., H. Echner, W. Voelter, and F. Gotz. 2001. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:1957-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skalaka, B., J. Pillich, and L. Pospisil. 1983. Further observations of Corynebacterium renale as an indicator organism in the detection of the exfoliative-positive strains of Staphylococcus aureus. Zentralbl. Bakteriol. Mikrobiol. Hyg. Ser. A 256:168-174. [PubMed] [Google Scholar]
- 22.Uehara, Y., H. Nakama, K. Agematsu, M. Uchida, Y. Kawakami, A. S. Abdul Fattah, and N. Maruchi. 2000. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium spp. J. Hosp. Infect. 44:127-133. [DOI] [PubMed] [Google Scholar]
- 23.van Belkum, A., N. H. Riewarts Eriksen, M. Sijmons, W. van Leeuwen, M. van den Bergh, J. Kluytmans, F. Espersen, and H. Verbrugh. 1997. Coagulase and protein A polymorphisms do not contribute to persistence of nasal colonisation by Staphylococcus aureus. J. Med. Microbiol. 46:222-232. [DOI] [PubMed] [Google Scholar]
- 24.VandenBergh, M. F., E. P. Yzerman, A. van Belkum, H. A. Boelens, M. Sijmons, and H. A. Verbrugh. 1999. Follow-up of Staphylococcus aureus nasal carriage after 8 years: redefining the persistent carrier state. J. Clin. Microbiol. 37:3133-3140. [DOI] [PMC free article] [PubMed]
- 25.van Leeuwen, W., W. van Nieuwenhuizen, C. Gijzen, H. Verbrugh, and A. van Belkum. 2000. Population studies of methicillin-resistant and -sensitive Staphylococcus aureus strains reveal a lack of variability in the agrD gene, encoding a staphylococcal autoinducer peptide. J. Bacteriol. 182:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Wamel, W. J., G. van Rossum, J. Verhoef, C. M. Vandenbroucke-Grauls, and A. C. Fluit. 1998. Cloning and characterization of an accessory gene regulator (agr)-like locus from Staphylococcus epidermidis. FEMS Microbiol. Lett. 163:1-9. [DOI] [PubMed] [Google Scholar]