Abstract
Molecular identification techniques based on total DNA extraction provide a unique tool for identification of mycelium in soil. Using molecular identification techniques, the ectomycorrhizal (EM) fungal community under coniferous vegetation was analyzed. Soil samples were taken at different depths from four horizons of a podzol profile. A basidiomycete-specific primer pair (ITS1F-ITS4B) was used to amplify fungal internal transcribed spacer (ITS) sequences from total DNA extracts of the soil horizons. Amplified basidiomycete DNA was cloned and sequenced, and a selection of the obtained clones was analyzed phylogenetically. Based on sequence similarity, the fungal clone sequences were sorted into 25 different fungal groups, or operational taxonomic units (OTUs). Out of 25 basidiomycete OTUs, 7 OTUs showed high nucleotide homology (≥99%) with known EM fungal sequences and 16 were found exclusively in the mineral soil. The taxonomic positions of six OTUs remained unclear. OTU sequences were compared to sequences from morphotyped EM root tips collected from the same sites. Of the 25 OTUs, 10 OTUs had ≥98% sequence similarity with these EM root tip sequences. The present study demonstrates the use of molecular techniques to identify EM hyphae in various soil types. This approach differs from the conventional method of EM root tip identification and provides a novel approach to examine EM fungal communities in soil.
Ectomycorrhizal (EM) fungi are major components of the soil fungal community in most boreal and temperate forests and several (sub)tropical forests. The EM fungi efficiently take up water and organic as well as inorganic nutrients from the soil and translocate these to colonized tree roots, receiving host carbohydrates in return. The extraradical mycelium is the part of the EM association actively involved in uptake of nutrients and water. Species-specific variations in EM extraradical mycelial structure and activity enable single EM fungal species to exploit specific resources (30). The concept of ecological specialization among EM fungi is now generally accepted (14) and underlines the proposed importance of EM fungal species diversity in soil ecosystems (2, 7, 21). To gain more insights into the functional roles of different EM fungal species and understand their functional diversity, it is essential to obtain information about ecologically relevant features of the symbiosis, in particular the spatial distribution of EM extraradical mycelium.
Until the advent of molecular techniques, it had been impossible to acquire data on the mycelial distribution of individual fungal species in soil. Most EM diversity studies are currently based on EM root tip inventories. Morphotype techniques combined with molecular techniques enable identification of EM fungi on root tips extracted from soil and provide information on EM fungal species diversity on a fraction of the tree root system. When using molecular techniques for identification of fungal material on roots, the distribution of extraradical mycelium in the bulk soil remains unknown. Yet, molecular identification techniques can also be used to detect fungal hyphae in substrates other than roots and therefore enable identification of mycelium in soil.
Molecular identification based on the extraction of total environmental DNA enables identification of fungi directly from the environment, irrespective of morphology or growth stage. Fungal DNA can be selectively amplified from total environmental DNA extracts, using PCR and a pair of primers that allow specific amplification of the fungal ribosomal DNA (rDNA) gene cluster. With temperature gradient gel electrophoresis (35), denaturing gradient gel electrophoresis (25, 29, 36), or single-strand conformation polymorphism (23), fungal communities can subsequently be analyzed. The 18S rDNA and internal transcribed spacer (ITS) regions have been used for construction of fungal clone libraries, enabling identification of nonmycorrhizal fungal species from church window glass (33), soil (3), and decaying plant material (6) through selective clone sequencing.
These molecular approaches have recently also been applied to the investigation of EM fungal communities in soil, using restriction fragment length polymorphism (9) and terminal restriction fragment length polymorphism (11). EM fungi are not a monophyletic group, and EM-specific primers therefore do not exist. Primers generally used for identification of EM fungi have enhanced specificity for basidiomycetes (13) and target the ITS regions of the rDNA gene cluster, including the 5.8S rDNA gene. The rDNA gene cluster is represented by one to several hundred copies per genome within the fungal nucleus and therefore provides a good target region (4, 37). Until now, the basidiomycete-specific ITS1F-ITS4B primer pair (13) has been used to amplify DNA extracted from fruit bodies (7, 8, 31), fungal cultures (13, 32), infected plant material (13), and EM root tips (7).
This study presents the use of the ITS1F-ITS4B primer pair to amplify basidiomycete DNA from soil DNA extracts, followed by a cloning and sequencing procedure, in order to identify the EM fungi present. Through identification of basidiomycete mycelium in soil, the EM fungal community structure can be analyzed in a novel way, excluding EM root tips from the analysis. The soil samples that were analyzed originated from four distinct horizons of a podzol profile, under coniferous vegetation, situated in the northern part of Sweden. The clone sequences obtained were grouped and compared to sequences from a selection of morphotyped EM root tips collected concomitantly from the same podzol profiles.
The aim of the underlying study was threefold: (i) to use molecular methods for identification of EM mycelium in soil, (ii) to investigate the distribution of EM mycelium in a vertical soil profile, and (iii) to compare the clone sequence data obtained to EM root tip sequence data.
MATERIALS AND METHODS
Soil samples.
The investigated site is located in the northern part of Sweden within the area of the Svartberget Forest Research station (Nyänget; 64°15′N, 19°45′E). The soil is a basal glacial till, and the soil type is classified as a typic haplocryod or a haplic arenosol (19), characterized by four distinct podzolic horizons. The organic mor layer (O layer, 0 to 3 cm) is underlain by a strongly weathered eluvial horizon (E horizon, 3 to 18 cm) and an enriched illuvial horizon (B horizon, 18 to 35 cm). The transition of the B horizon into parent material (C, >40 cm) at the bottom of the soil profile is gradual. The coniferous forest is dominated by Norway spruce (Picea abies) and Scots pine (Pinus sylvestris), and the main undergrowth consists of Blueberry (Vaccinium myrtillus), Lingonberry (Vaccinium vitis-idaea), and Wavy hair-grass (Deschampsia flexuosa).
In August 1999 three pits were dug about 6 m apart from each other. Pits 1 and 3 were situated 1 m from a Norway spruce tree, while pit 2 was closest (1 m) to a Scots pine tree. From one side of each pit, the soil was sampled along three vertical columns. Working downward from the O layer, a small block of soil (5 by 5 cm) was carefully removed from the center of each horizon (E, B, and C horizons) by using fine stopping knives. The soil of each horizon was sealed separately in a plastic bag, transported back to the lab, and frozen at −70°C. The E horizon was divided in two parts when sampled, an upper E1 horizon and a lower E2 horizon. In so doing, a distinction could be made between fungi found in the upper E1 horizon, close to the O layer, and fungi found closer to the B horizon. Table 1 gives average pH values and average total C contents (grams per kilogram of soil) of the pooled soil samples from each horizon of the three pits. The total C content of the soil samples was determined by a wet oxidation procedure (KurmiesC procedure) (15). There are large variations in carbon percentages (30 to 62%) of organic matter with different soil types, and therefore total C contents cannot be converted to organic matter contents of the different soil horizons.
TABLE 1.
pH and total C contents of soil samples taken from five horizons of three pits
| Horizon | pH (CaCl)a | Total-C, g/kg (KurmiesC)a |
|---|---|---|
| E1 | 3.7 ± 0.1 | 6.5 ± 1.7 |
| E2 | 3.8 ± 0.1 | 5.6 ± 0.3 |
| B1 | 4.9 ± 0.6 | 21.7 ± 8.1 |
| B2 | 5.7 ± 0.2 | 7.6 ± 1.1 |
| C | 5.3 ± 0.1 | 3.9 ± 0.5 |
Results are averages and standard deviations for three pits.
DNA extractions from soil samples.
From the three pits, DNA was extracted from three soil samples taken from each horizon. Mineral soil was gently sieved (1 mm) to discard EM root tips. Visual checks were performed to make sure no root tips had passed the sieve. O horizon samples that could not be sieved were checked visually under the dissecting microscope, and root tips were removed manually. DNA was extracted from 5 g (wet weight) of mineral soil and 1 g (wet weight) of the organic samples, using a Bead-Beater as described by Smalla et al. (34). DNA was purified twice with the Wizard DNA Clean-Up System (Promega) according to the manufacturer's purification protocol before amplification.
PCR amplification.
For each pit separately, the three DNA extracts from each horizon were pooled prior to PCR to reduce sample numbers for the cloning procedure. DNA extracts from the B1 and B2 horizons were also pooled. DNA extracts from the C horizons could not be amplified due to low template concentrations and were left out of any further analyses. The remaining 12 samples (four soil horizons [O, E1, E2, and B] from three different pits) were used for PCR and for the cloning procedure. DNA amplification of the ITS regions was performed on a Hybaid PCR Express thermocycler with the primers ITS1F (5′-CTT GGT CAT TTA GAG GAA GTA A-3′) and ITS4B (5′-CAG GAG ACT TGT ACA CGG TCC AG-3′) (13). The PCR mix consisted of 5 μl of 10× PCR buffer 2 (Roche), a 200 μM concentration of each deoxynucleoside triphosphate, a 200 nM concentration of each primer, 0.5 μl of Expand enzyme mix (Roche), 38 μl of sterile Ultrapure water, and 1 μl of 1:50-diluted template DNA (see Table 2 for template DNA concentrations). The following thermocycling pattern was used: 94°C for 3 min (1 cycle); 94°C for 1 min, 50°C for 1 min, and 72°C for 3 min (30 cycles); and 72°C for 10 min (1 cycle). Amplified DNA fragments (0.7 kb) were visualized on 1% agarose gels.
TABLE 2.
Amounts of total double-stranded DNA extracted from soil samples of five horizons of three pits
| Horizon | Double-stranded DNA (μg/g soil)a |
|---|---|
| O | 23.7 ± 7.5 |
| E1 | 7.2 ± 2.9 |
| E2 | 5.8 ± 1.1 |
| B | 4.4 ± 3.2 |
| C | 0.42 ± 0.2 |
Results are averages and standard deviations for three pits.
Cloning of ITS1F-ITS4B PCR products.
The successfully amplified PCR products (0.7 kb) were ligated into the pGem-T vector, which has 3′-T overhangs to facilitate cloning of PCR products (Promega, Madison, Wis.). Ligation mixtures were transformed into Ultracompetent E. coli XL1-Blue (Stratagene, Cambridge, United Kingdom) according to the manufacturer's instructions. The bacteria were plated out in three dilutions. From each sample 100 white colonies were picked, cultured overnight in 2 ml of Luria broth, and then frozen at −70°C.
Identification of the cloned fungal ITS fragments.
Thirty clones from each sample were amplified and analyzed on a 1% agarose gel. Clones with bands of the right size (0.7 kb) were purified with the QIAquick purification kit (Qiagen) according to the manufacturer's protocol. The 0.7-kb fragments were sequenced in one direction on an ABI377 DNA sequencer by cycle sequencing with the dye terminator system (Eurogentec, Seraing, Belgium) and the ITS1F primer. In order to identify the obtained clone sequences, all clone sequences were subjected to multiple alignments with ClustalW (DNA Star Program) and used to construct phylogenetic trees by the neighbor-joining method with the Treecon program (version 1.36; Yves van der Peer). Based on the clusters that were formed, clones were sorted into groups, or operational taxonomic units (OTUs). A representative of each OTU was compared phylogenetically to sequences of known species in the GenBank database of the National Center for Biotechnology Information (NCBI) by using the Blast program. A phylogenetic tree was then constructed with all clones from each OTU and the most homologous sequences from the database. Database sequences yielding the greatest percent similarity to the clone sequences were chosen as the best match for each OTU. The purpose of this study was to assign a taxon name to each OTU rather than to provide a phylogenetic analysis of each obtained OTU. Assignment to taxon categories was done as follows: sequence similarity of ≥99%, identification to species level; sequence similarity of 95 to 99%, identification to genus level; sequence similarity of ≤95%, identification to family or ordinal level. Neighbor-joining trees were then constructed with Amanita muscaria as an outgroup and 100 bootstrap replicates.
Statistical analysis of clone distribution.
A permutation test was performed to determine whether the occurrence of OTUs was related to soil horizons. A statistic (summore) was calculated on the actual data, measuring the association between OTU and soil horizons. This summore was compared to the null permutation distribution (i.e., under the hypothesis of no association) of a statistic calculated from random permutations of the data. The following three assumptions have been used: (i) the numbers of OTUs per pit and per horizon were held fixed, (ii) the quantitative aspect of the number of clones representing each OTU was ignored (in each soil horizon an OTU was considered to be either present or not), and (iii) the presence of an OTU in a horizon was considered to be dependent on the presence of the same OTU in adjoining, contiguous layers, while the presence of an OTU was considered to be independent of the presence of other OTUs.
The data sets for the three separate soil pits were combined for the permutation test. The test was performed exclusively with 11 OTUs represented by clones originating from more than one soil pit (OTUs 4, 5, 7, 8, 9, 10, 12, 15, 18, 21, and 24). The data set shows that over all three pits, 18 out of 44 fields in the 11-by-4 matrix (11 OTUs × 4 horizons) are empty and contain no OTU. This variable 18 is called the summore. Subsequently, a test was performed to determine the probability under the null permutation distribution of finding a summore value of 18 or more, i.e., to test whether the summore value of 18 is coincidental. A statistical program designed for this purpose was used to take random samples from all possible OTU-horizon distributions having total values equal to the actual total values found. In total, 24,000 permutations were performed (12). For 24,000 permutations, 1,200 summore values of ≥18 are required to reject the null hypothesis of a random OTU distribution at a P value of ≤0.05.
Comparison of clone and root tip sequences.
Using the Blast program (NCBI), clone sequence data were compared to sequence data obtained from root tips extracted from soil samples taken concomitantly with the soil samples used in the present study (A. Rosling, R. Landeweert, B. Lindahl, K.-H. Larsson, T. W. Kuyper, A. F. S. Taylor, and R. Finlay, submitted for publication). The nucleotide sequences from the root tip data are accessible under NCBI accession numbers AF476965 to 477005, AF481369 to 481389, and AF494441 to 494447.
Nucleotide sequence accession numbers.
The nucleotide sequences of the clones representing each OTU have been submitted to NCBI and are accessible under accession numbers AY097035 to 097059.
RESULTS
DNA extractions and PCRs.
Total DNA was successfully extracted from all soil samples (Table 2). Due to too-low template concentrations, however, DNA extracts from the C horizons could not be amplified at template dilutions equal to those for the other samples, and therefore the C samples were left out of any further analyses. DNA extracts from the O and B horizons had a dark brown color, probably due to coextracted organic matter. Double purification of extracted DNA allowed successful PCR amplification of 1:50-diluted template DNA, using the ITS1F-ITS4B primer pair.
Phylogenetic grouping of clone sequences.
Sequencing of 30 clones from all 12 soil samples resulted in 318 successful sequencing results. To determine whether analysis of 30 clones from each sample was sufficient to detect the most common fungal species, 20 extra clones from one sample (E1 horizon sample from pit 1) were sequenced and rarefaction analysis was applied. Rarefaction is a statistical method to estimate the number of species expected in a random sample of individuals taken from a collection (20). From the collection of 50 sequences, representing six fungal species (OTUs), rarefaction analysis shows that the expected number of species in a subsample of 30 clones is 5.1 ± 0.7. Therefore, detection of the most common species seems likely when 30 clones/sample are analyzed.
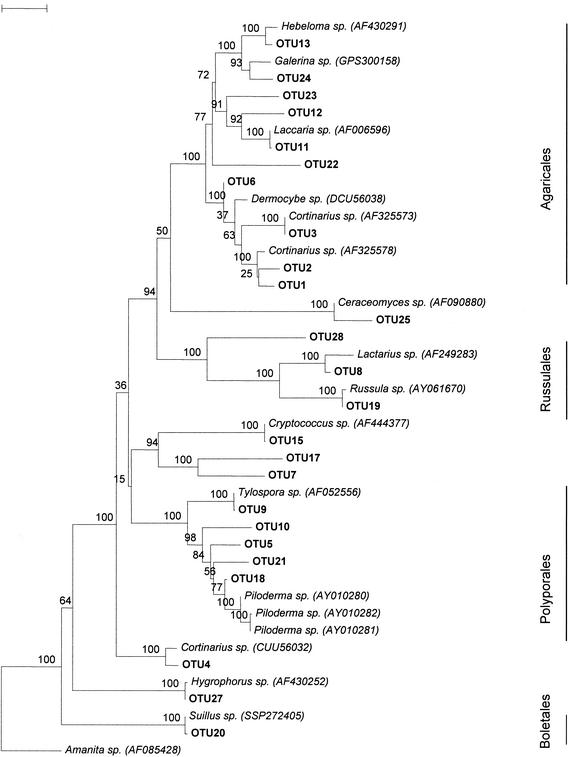
After multiple alignments and constructions of phylogenetic trees, the obtained clone sequences were sorted in 29 different OTUs. Four OTUs that were represented by one single clone sequence (OTUs 14, 16, 26, and 29) were not used further, leaving 25 different OTUs. Figure 1 presents a phylogenetic tree with the 25 OTUs and sequences from the GenBank database with highest sequence similarity to clone sequences.
FIG. 1.
Neighbor-joining tree representing the phylogenetic relationships of 25 OTU sequences obtained from three soil pits to the most closely related sequences obtained from Blast searches. The scale bar indicates 0.1 substitutions per site.
Four main fungal orders were distinguished, based on the classification of Hibbett and Thorn (17). Within these four orders, most terminal clades are well supported with high bootstrap values. The Boletales order includes OTU 20 (Suillus-like sp.). The Polyporales order includes the Piloderma-like OTUs 5, 10, 18, and 21 and the Tylospora-like OTU 9 (Fig. 1). Strong bootstrap support is found for the Russulales order, including OTU 8 (Lactarius-like sp.), OTU 19 (Russula-like sp.), and OTU 28 (unknown). The order Agaricales includes OTUs 1 to 3, 6, 11 to 13, and 22 to 24. Within the Agaricales order, the Cortinariaceae clade is strongly supported. However, another presumed member of this clade (OTU 4) does not group within this well-supported Cortinariaceae clade.
For 14 out of the 25 OTUs, a Blast search sequence similarity of ≥95% was found (Table 3). Out of the total 25 OTUs, 19 OTUs represented basidiomycetes with known affinity, while the taxonomic position of 6 other basidiomycete OTUs remained unclear. Four unidentified OTUs were represented by clones from the mineral E and B horizons only. Of the 19 basidiomycete OTUs, 7 OTUs showed a high (≥99%) nucleotide similarity to known EM fungal sequences and could be assigned species names. Five basidiomycete OTUs showed 95 to 99% sequence similarity to known EM fungal species and could be assigned to the genus level. Four basidiomycete OTUs with a low sequence similarity (≤95%) to known EM fungal sequences were assigned to the family level. For three OTUs the highest sequence similarity was found with nonmycorrhizal fungal species (Ceraceomyces eludens, Galerina pseudomycenopsis, and Cryptococcus terricola).
TABLE 3.
Fungal species from the GenBank database with highest sequence similarity to each OTU and distribution of OTUs over four horizonsa
| OTU | Blast matchb | Accession no. | Similarity (%) | No. of clones in soil horizon:
|
Total no. of clones | |||
|---|---|---|---|---|---|---|---|---|
| O | E1 | E2 | B | |||||
| 22 | None | 3 | 3 | |||||
| 23 | None | 3 | 3 | |||||
| 25 | Ceraceomyces eludens | AF090880 | 99.0 | 9 | 9 | |||
| 24 | Galerina pseudomycenopsis | GPS300158 | 92.1 | 3 | 1 | 4 | ||
| 1 | Cortinarius acutus | AF325578 | 95.3 | 27 | 2 | 1 | 30 | |
| 18 | Piloderma fallax | AY010281 | 94.9 | 5 | 4 | 4 | 13 | |
| 19 | Russula decolorans | AY061670 | 99.2 | 23 | 20 | 2 | 45 | |
| 21 | Piloderma fallax | AY010281 | 97.6 | 8 | 1 | 2 | 11 | |
| 15 | Cryptococcus terricola | AF444377 | 99.5 | 7 | 1 | 1 | 1 | 10 |
| 3 | Cortinarius collinitus | AF325573 | 99.8 | 19 | 3 | 22 | ||
| 6 | Dermocybe crocea | DCU56038 | 99.6 | 5 | 2 | 7 | ||
| 11 | Laccaria laccata | AF006596 | 99.6 | 1 | 5 | 6 | ||
| 27 | Hygrophorus olivaceoalbus | AF430252 | 99.3 | 8 | 7 | 15 | ||
| 10 | Piloderma fallax | AY010280 | 93.2 | 7 | 6 | 19 | 32 | |
| 4 | Cortinarius umbilicatus | CUU56032 | 96.0 | 1 | 14 | 8 | 23 | |
| 2 | Cortinarius acutus | AF325578 | 92.7 | 6 | 6 | |||
| 8 | Lactarius deliciosus | AF249283 | 93.5 | 4 | 4 | |||
| 9 | Tylospora asterophora | AF052556 | 99.8 | 2 | 2 | |||
| 5 | Piloderma fallax | AY010282 | 92.0 | 9 | 25 | 34 | ||
| 7 | None | 2 | 13 | 15 | ||||
| 12 | None | 1 | 2 | 3 | ||||
| 13 | Hebeloma incarnatulum | AF430291 | 96.1 | 4 | 4 | |||
| 17 | None | 2 | 2 | |||||
| 20 | Suillussp. | SSP272405 | 99.5 | 5 | 5 | |||
| 28 | None | 6 | 6 | |||||
Data for the three soil pits are pooled.
Boldface indicates EM fungal species.
Pattern of distribution of OTUs in soil profile.
The 25 OTUs consisted of variable numbers of clone sequences, often originating from different soil horizons. Ten OTUs were represented by clones from just one horizon, while the other 15 OTUs were represented by clones from two or more adjoining horizons. Table 3 gives the combined results for three soil pits. Three OTUs were found exclusively in the organic horizon, while 16 OTUs were found exclusively in the mineral soil. Six OTUs (OTUs 1, 15, 18, 19, 21, and 24) were represented by clones from the organic as well as the mineral E soil horizons. Seven OTUs were found exclusively in the E horizons, and of these, three (OTUs 2, 8, and 9) were found exclusively in the deeper E2 horizon. OTUs 4 and 10 were found in the E1, E2, and B horizons and were not seen in the O layer. Three OTUs (5, 7, and 12) were found in both the E2 and B horizons, and four OTUs occurred in the B horizon only. Clones representing OTU 15 were found in all four soil horizons.
Seven OTUs with a high sequence similarity (≥99%) to EM fungal sequences from the GenBank database were distributed as follows: in the O and E horizons, OTU 19 (Russula decolorans) was found; in the E horizons, OTUs 3 (Cortinarius collinitus), 6 (Dermocybe crocea), 9 (Tylospora asterophora), 11 (Laccaria laccata), and 27 (Hygrophorus olivaceoalbus) were found; and in the B horizon, OTU 20 (Suillus sp.) was found.
Statistical analysis of OTU distribution.
A permutation test was performed to determine whether the OTU distribution is related to soil horizons. This test was performed with 11 OTUs represented by clones from more than one soil pit. The fact that one pit was dominated by a different tree species probably increased the variation in fungal species composition between the three pits. As a result, 14 OTUs did not appear in more than one soil pit and were not included in the statistical analysis. In total, 24,000 permutations were carried out, resulting in 1,489 summore values of ≥18, leading to a calculated statistic of 0.06. At a probability level of 6%, the test indicates that the distribution of the OTUs over the four horizons is not random.
Clone sequences versus root tip sequences.
An EM root tip inventory of the same soil pits carried out concomitantly with the present study revealed the presence of 25 different EM fungal types on a selection of the root tips sampled (Rosling et al., submitted). Of the 19 basidiomycete OTUs, 2 OTUs had 100% sequence similarity to root tip sequences (OTUs 6 and 19). Similarity percentages of 98 to 99% were found for OTUs 3, 5, 8, 9, 10, 18, 21, and 22. While OTUs 5, 10, 18, and 21 matched poorly (92 to 97%) with three Piloderma fallax sequences from the GenBank database, they matched well (≥98%) with four different root tip sequences representing four potentially different Piloderma species. Although the identity of OTU 22 remained unresolved after the Blast search, a good match (98.6%) was found with a root tip sequence. This strongly indicates that this OTU groups within an EM fungal genus. Of 13 OTUs with ≥92% sequence similarity to root tip sequences, 10 OTUs occur in the same horizons as from where the matching root tips were extracted.
DISCUSSION
This paper demonstrates the use of molecular identification techniques to study the occurrence of EM hyphal material in various soil horizons.
For this study the use of ITS sequences was chosen to enable comparison with ITS sequences available from EM root tips sampled concomitantly from the same pits. In most cases, the resolving power of the ITS proved to be enough to retrieve closely related taxa from the GenBank database. Rarefaction analysis showed that the amount of clones analyzed was sufficient to detect the most common fungal species. Construction of neighbor-joining phylogenetic trees with 25 OTUs and related GenBank sequences placed most OTUs in four main fungal orders. For 10 OTUs a Blast search sequence similarity of ≥99% was found with GenBank basidiomycete sequences, enabling identification to the species level. Four OTUs with 95 to 99% sequence similarity were assigned to the genus level, while five OTUs with ≤95% sequence similarity were assigned to the family level (Table 3). Other studies considered nucleotide homologies of 87% (8) and 94% (24) sufficient for the placing of unknown sequences in known fungal taxa.
In some cases, however, the resolving power of the ITS sequence proved to be too weak for correct taxonomic placement of an OTU, as illustrated in the Cortinariaceae clade (Fig. 1). Within the Cortinariaceae clade, up to 30% base pair variation within the ITS regions and 5.8S gene sequences is possible (22), likely affecting the taxonomic placement of the Cortinarius-like OTU 4 in this study. In addition, the taxonomic positions of six basidiomycete OTUs remained unresolved, as ITS sequence data from closely related species were not available in the database. While the resolving power of the fungal ITS regions would often enable identification of distantly related species (7), unknown ITS sequences remain uninformative if they belong to groups that are underrepresented in sequence databases (18). Sequence databases tend to be biased towards sequences from fungal species that either produce distinctive sporocarps or grow well in culture, and the continuing addition of sequences from more fungal species is needed to get beyond the present “unknowns” (18). Four unresolved OTUs came from the mineral E1, E2, and B horizons. These OTUs might represent fungal species that have been missed in earlier studies, as they might occur only in deeper soil layers or not form sporocarps.
Most present inventories concerning EM fungal communities are based on EM root samples taken from organic horizons. The present study reveals that 16 out of 25 OTUs were exclusively detected in the deeper mineral soil. This demonstrates that the general concept of fungal species from deeper soil profiles not accounting for much of the fungal species diversity (5) might no longer be valid. In a podzol profile, apart from being situated at different depths, each soil horizon has distinct chemical and physical properties. Several factors could influence fungal species distribution along a vertical soil profile (27), as species differ in sensitivity to environmental factors such as O2 and CO2 (5), pH (10, 27), soluble Al3+ (26), soil moisture and organic matter content (1), or predation (28). This study was not set up to determine factors influencing fungal species distribution along a vertical gradient, yet the results indicate that the detected species are not equally distributed over four podzol horizons. Higher pH values and total C amounts (Table 1) in the B horizon compared to the E horizon could influence OTU distribution. In this study, EM fungal species such as Hebeloma or Suillus spp. were found in the mineral B soil, which is high in total C contents, whereas Cortinarius, Dermocybe, Tylospora, and Laccaria spp. were found in the strongly weathered mineral E soil, which has low total C contents. Organic and mineral nutrient availability vary in each horizon, and the presence of a fungal species in a certain horizon could depend on its ability to take up nutrients from organic or inorganic sources. Considerable amounts of extramatrical mycelium of Suillus spp. in the mineral E and B horizons have been observed in microcosm systems and have been related to the suggested role of these species in mineral weathering processes (16).
The permutation test performed indicated a nonrandom vertical distribution of OTUs along the podzol profile at a probability level of 6%. The distribution of the 11 analyzed OTUs appears to have some relation to the soil horizons, taking into account that the test performed relies on some important assumptions that influence its outcome. Based on the fact that OTUs from two or more horizons always occur in bordering horizons (Table 3), it was assumed that the occurrence of an OTU in one horizon might depend on its occurrence in one or two adjoining horizons. Quantitative clone aspects were not taken into account in this study, and the finding of one clone in a horizon was considered to represent the presence of an OTU in that particular horizon. Due to the continuity of mycelium in the soil, the occurrence of (small amounts of) mycelium in more than one soil horizon is likely. These detected amounts of mycelium in bordering horizons, however ecologically insignificant, might have been the cause of the test's outcome being only marginally significant (P ≤ 0.06). This fact illustrates the difficulty of accurately detecting spatial structure when sampling in natural systems. In fact, for many spatial analysis methods it is difficult to detect significant spatial patterns for small sample sizes (n ≤ 10) (12), and analysis of 11 OTUs proved not to be ideal.
Clone sequence data were compared to sequence data obtained from root tips to find out whether the distribution of mycelium was related to root tip distribution. EM fungal identification through root tip identification presents a very dissimilar approach to EM fungal community analysis when compared to molecular identification of fungi in soil DNA extracts. In this study, soil and root samples came from the same pits and soil horizons, but DNA was extracted from soil samples other than the samples that the root tips originated from. Nevertheless, the results (Table 4) show that for seven OTUs, clone sequence data matched well (≥99%) with the root tip data, indicating that the demonstrated molecular approach is technically robust. One unidentified OTU (OTU 22) matched with a root tip sequence, indicating that the fungus belongs to an EM fungal group that is not yet present in the GenBank database. Four Piloderma-like OTUs had a high sequence similarity with four different Piloderma-like root tip sequences, indicating that several Piloderma species await identification. In addition, the OTU-root tip comparison shows that the EM mycelium primarily colonizes the horizon in which the EM root tip is found.
TABLE 4.
OTUs with high sequence similarity to sequences from EM root tips extracted from the same soil pits, ranked according to nucleotide similarity percentage
| Clone sequence data | Blast match (%) | Root tip sequence data
|
||||
|---|---|---|---|---|---|---|
| OTU | Genus | GenBank accession no. | Horizona | GenBank accession no. | Horizona | |
| 6 | Dermocybe | AY097040 | E1, E2 | 100 | AF494447 | O |
| 19 | Russula | AY097051 | O, E1, E2 | 100 | AF476996 | O |
| 3 | Cortinarius | AY097037 | E1, E2 | 99.8 | AF476991 | E1 |
| 8 | Lactarius | AY097042 | E2 | 99.8 | AF476975 | B1 |
| 10 | Piloderma | AY097044 | E1, E2, B | 99.7 | AF476986 | B1 |
| 18 | Piloderma | AY097050 | O, E1, E2 | 99.5 | AF481388 | E1 |
| 5 | Piloderma | AY097039 | E2, B | 99.4 | AF494446 | E2 |
| 21 | Piloderma | AY097053 | O, E1, E2 | 98.8 | AF494442 | O |
| 22 | Unknown | AY097054 | O | 98.6 | AF476965 | O |
| 9 | Tylospora | AY097043 | E2 | 98.3 | AF476970 | B2 |
| 1 | Cortinarius | AY097035 | O, E1, E2 | 97.1 | AF476989 | O |
| 4 | Cortinarius | AY097038 | E1, E2, B | 95.9 | AF481373 | B1 |
| 20 | Suillus | AY097052 | B | 92.1 | AF476994 | B1 |
Horizon(s) from which the clone or root tip was extracted.
This study shows that the use of molecular methods enables identification of EM mycelium in soil and provides a complementary approach to conventional EM root tip identification in EM community studies. Both approaches, however, are subject to bias. While the sampling of clustered root tips often inadequately reflects the full species richness below ground (18), each step involved in the molecular analyses of an environment could be a source of bias, which will lead to a distorted view of the real world (38). How these molecular uncertainties affect the outcome of molecular fungal community analyses needs to be studied in greater detail.
In conclusion, the present study demonstrates that molecular identification methods provide a novel tool for future studies on EM fungal community dynamics. These molecular methods allow identification of EM mycelium in soil and therefore permit a unique view of the EM fungal community below ground, independent of EM root tip occurrence. The active soil mycelium has clear functional significance (18), and compared to data on colonized root tip numbers, the spatial distribution of EM mycelium better reflects substrate preference and potential nutrient-mobilizing and uptake capacities of EM fungal species. EM mycelial distribution data will provide significant information on potential ecological roles of different EM fungal species or functional groups in soil ecosystems. Although the accuracy of molecular methods for quantitative fungal community analysis and fungal activity measures needs to be assessed, the use of molecular identification methods will provide valuable new insights into EM fungal community dynamics.
Acknowledgments
We thank Christiaan Veenman for his help with sample preparation and sequencing. We are indebted to Nico Nagelkerke (RIVM) for performing the statistical analysis of the OTU distribution data. We thank Lijbert Brussaard for critically reading the manuscript.
Financial support to R.L and E.H. from The Netherlands Organization for Scientific Research (NWO) is gratefully acknowledged.
REFERENCES
- 1.Bååth, E., and B. Söderström. 1982. Seasonal and spatial variation in fungal biomass in a forest soil. Soil Biol. Biochem. 14:353-358. [Google Scholar]
- 2.Baxter, J. W., and J. Dighton. 2001. Ectomycorrhizal diversity alters growth and nutrient acquisition of grey birch (Betula populifolia) seedlings in host-symbiont culture conditions. New Phytol. 152:139-149. [DOI] [PubMed] [Google Scholar]
- 3.Borneman, J., and R. J. Hartin. 2000. PCR primers that amplify fungal rRNA genes from environmental samples. Appl. Environ. Microbiol. 66:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridge, P., and B. Spooner. 2001. Soil fungi: diversity and detection. Plant Soil 232:147-154. [Google Scholar]
- 5.Bruns, T. D. 1995. Thoughts on the processes that maintain local species diversity of ectomycorrhizal fungi. Plant Soil 170:63-73. [Google Scholar]
- 6.Buchan, A., S. Y. Newell, J. I. L. Moreta, and M. A. Moran. 2002. Analysis of internal transcribed spacer (ITS) regions of rRNA genes in fungal communities in a southeastern U.S. salt marsh. Microb. Ecol. 43:329-340. [DOI] [PubMed] [Google Scholar]
- 7.Buscot, F., J. C. Munch, J. Y. Charcosset, M. Gardes, U. Nehls, and R. Hampp. 2000. Recent advances in exploring physiology and biodiversity of ectomycorrhizas highlight the functioning of these symbioses in ecosystems. FEMS Microbiol. Rev. 24:601-614. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, S. M., N. A. Sawyer, and J. W. G. Cairney. 1999. Molecular identification of co-occurring Cortinarius and Dermocybe species from southeastern Australian sclerophyll forests. Mycorrhiza 9:85-90. [Google Scholar]
- 9.Chen, D. M., and J. W. G. Cairney. 2002. Investigation of the influence of prescribed burning on ITS profiles of ectomycorrhizal and other soil fungi at three Australian sclerophyll forest sites. Mycol. Res. 106:532-540. [Google Scholar]
- 10.Danielson, R. M., and S. Visser. 1989. Effects of forest soil acidification on ectomycorrhizal and vesicular-arbuscular mycorrhizal development. New Phytol. 112:41-47. [Google Scholar]
- 11.Dickie, I. A., B. Xu, and R. T. Koide. Vertical niche differentiation of ectomycorrhizal hyphae in soil as shown by T-RFLP analysis. New Phytol. 156:527-535. [DOI] [PubMed]
- 12.Fortin, M.-J., and J. Gurevitch. 1993. Mantel tests: spatial structure in field experiments, p. 342-359. In M. S. Scheiner, and J. Gurevitch (ed.), Design and analysis of ecological experiments. Chapman & Hall, New York, N.Y.
- 13.Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 14.Goodman, D. M., and J. A. Trofymow. 1998. Distribution of ectomycorrhizas in microhabitats in mature and old-growth stands of Douglas-fir on southeastern Vancouver island. Soil Biol. Biochem. 30:2127-2138. [Google Scholar]
- 15.Haenes, D. L. 1984. Determination of total organic-C in soils by an improved chromic acid digestion and spectrophotometric procedure. Commun. Soil Sci. Plant Anal. 15:1191-1213. [Google Scholar]
- 16.Heinonsalo, J., K. S. Jorgensen, and R. Sen. 2001. Microcosm-based analyses of Scots pine seedling growth, ectomycorrhizal fungal community structure and bacterial carbon utilization profiles in boreal forest humus and underlying illuvial mineral horizons. FEMS Microbiol. Ecol. 36:73-84. [DOI] [PubMed] [Google Scholar]
- 17.Hibbett, D. S., and R. G. Thorn. 2001. Basidiomycota: Homobasidiomycetes. Mycota VII Syst. Evol. B 7:121-168. [Google Scholar]
- 18.Horton, T. R., and T. D. Bruns. 2001. The molecular revolution in ectomycorrhizal ecology: peeking into the black-box. Mol. Ecol. 10:1855-1871. [DOI] [PubMed] [Google Scholar]
- 19.Ilvesniemi, H., R. Giesler, P. A. W. Van Hees, T. Magnussson, and P. A. Melkerud. 2000. General description of the sampling techniques and the sites investigated in the Fennoscandinavian podzolization project. Geoderma 94:109-123. [Google Scholar]
- 20.Krebs, C. J. 1999. Ecological methodology, 2nd ed. Cummings, Menlo Park, Calif.
- 21.Leake, J. R. 2001. Is diversity of ectomycorrhizal fungi important for ecosystem function? New Phytol. 152:1-3. [DOI] [PubMed] [Google Scholar]
- 22.Liu, Y. J., S. O. Rogers, and J. F. Ammirati. 1997. Phylogenetic relationships in Dermocybe and related Cortinarius taxa based on nuclear ribosomal DNA internal transcribed spacers. Can. J. Bot. 75:519-532. [Google Scholar]
- 23.Lowell, J. L., and D. A. Klein. 2001. Comparative single-strand conformation polymorphism (SSCP) and microscopy-based analysis of nitrogen cultivation interactive effects on the fungal community of a semiarid steppe soil. FEMS Microbiol. Ecol. 36:85-92. [DOI] [PubMed] [Google Scholar]
- 24.McKendrick, S. L., J. R. Leake, D. L. Taylor, and D. J. Read. 2000. Symbiotic germination and development of myco-heterotrophic plants in nature: ontogeny of Corallorhiza trifida and characterization of its mycorrhizal fungi. New Phytol. 145:523-537. [DOI] [PubMed] [Google Scholar]
- 25.Mohlenhoff, P., L. Muller, A. A. Gorbushina, and K. Petersen. 2001. Molecular approach to the characterisation of fungal communities: methods for DNA extraction, PCR amplification and DGGE analysis of painted art objects. FEMS Microbiol. Lett. 195:169-173. [DOI] [PubMed] [Google Scholar]
- 26.Morris, S. J., and R. E. J. Boerner. 1999. Spatial distribution of fungal and bacterial biomass in southern Ohio hardwood forest soils: scale dependency and landscape patterns. Soil Biol. Biochem. 31:887-902. [Google Scholar]
- 27.Neville, J., J. L. Tessier, I. Morrison, J. Scarrat, B. Canning, and J. N. Klironomos. 2002. Soil depth distribution of ecto- and arbuscular mycorrhizal fungi associated with Populus tremuloides within a 3-year-old boreal forest clear-cut. Appl. Soil Ecol. 19:209-216. [Google Scholar]
- 28.Newell, K. 1984. Interaction between two decomposer basidiomycetes and a collembolan under Sitka spruce: grazing and its potential effects on fungal distribution and litter decomposition. Soil Biol. Biochem. 16:235-239. [Google Scholar]
- 29.Pennanen, T., L. Paavolainen, and J. Hantula. 2001. Rapid PCR-based method for the direct analysis of fungal communities in complex environmental samples. Soil Biol. Biochem. 33:697-699. [Google Scholar]
- 30.Read, D. J. 1991. Mycorrhizas in ecosystems. Experientia 47:376-391. [Google Scholar]
- 31.Redecker, D., T. M. Szaro, R. J. Bowman, and T. D. Bruns. 2001. Small genets of Lactarius xanthogalactus, Russula cremoricolor and Amanita francheti in late-stage ectomycorrhizal successions. Mol. Ecol. 10:1025-1034. [DOI] [PubMed] [Google Scholar]
- 32.Salazar, O., M. C. Julian, and V. Rubio. 2000. Primers based on specific rDNA-ITS sequences for PCR detection of Rhizoctonia solani, R. solani AG 2 subgroups and ecological types, and binucleate Rhizoctonia. Mycol. Res. 104:281-285. [Google Scholar]
- 33.Schabereiter Gurtner, C., G. Pinar, W. Lubitz, and S. Rolleke. 2001. Analysis of fungal communities on historical church window glass by denaturing gradient gel electrophoresis and phylogenetic 18S rDNA sequence analysis. J. Microbiol. Methods 47:345-354. [DOI] [PubMed] [Google Scholar]
- 34.Smalla, K., N. Cresswell, L. C. Mendonca-Hagler, A. Wolters, and J. D. van Elsas. 1993. Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J. Appl. Bacteriol. 74:78-85. [Google Scholar]
- 35.Smit, E., P. Leeflang, B. Glandorf, J. D. van Elsas, and K. Wernars. 1999. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2614-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vainio, E. J., and J. Hantula. 2000. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol. Res. 104:927-936. [Google Scholar]
- 37.Viaud, M., A. Pasquier, and Y. Brygoo. 2000. Diversity of soil fungi studied by PCR-RFLP of ITS. Mycol. Res. 104:1027-1032. [Google Scholar]
- 38.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]