Abstract
An inducible, cytosolic glutathione S-transferase (GST) was purified from Streptomyces griseus. GST isoenzymes with pI values of 6.8 and 7.9 used standard GST substrates including 1-chloro-2,4-dinitrobenzene. GST had subunit and native Mrs of 24 and 48, respectively, and the N-terminal sequence SMILXYWDIIRGLPAH.
Glutathione S-transferases (GSTs; EC 2.5.1.8) comprise a family of multifunctional proteins that catalyze the nucleophilic attack and conjugation of glutathione (GSH) with a large variety of reactive electrophiles (16). In mammals, GSTs and cytochrome P450 enzymes are induced by phenobarbital and polycyclic aromatics (27). GSTs detoxify xenobiotics as the initial step in the formation of mercapturic acids, forming hydrophilic metabolites that are readily excreted. Cytosolic human, rat, and mouse GSTs have multiple isoenzymes, all of which are composed of 2 subunits (19). GSTs are grouped into α, μ, π, σ, κ, and ζ classes based upon sequence similarity, immunological properties, and substrate and inhibitor specificities (27).
Although GSTs are known to reside in yeasts (28, 29), protozoa (24), fungi (7, 13, 14), and bacteria (3, 10, 11, 18, 20, 31), very little is known about prokaryotic GSH conjugation. GSTs have been implicated in the microbial biodegradation and detoxication of xenobiotics (31). Streptomyces griseus contains many enzymes similar to those involved in mammalian drug metabolism. These include a soluble cytochrome, P450 (30); an S-adenosylmethionine-dependent catechol O-methyltransferase (9); UDPG-pyrophosphorylase; and other glucose-activating enzymes (22). This work describes the purification and characterization of GST isoenzymes from S. griseus, the first such enzymes from Streptomycetes.
For enzyme purification, GST assays were conducted in total volumes of 1 ml of 0.1 M phosphate buffer (pH 6.5) containing 1 mM 1-chloro-2,4-dinitrobenzene (CDNB), 1 mM GSH, and 20 μg of protein. Complete reaction mixtures were incubated at 35°C for 3 min while the increase in absorption at 340 nm due to the formation of S-(2,4-dinitrophenyl)-GSH (ɛ = 9.6 M−1) by the reaction of CDNB with GSH occurred (16). A standard unit of GST activity was the amount of enzyme that catalyzed the formation of 1 μmol of S-(2,4-dinitrophenyl)-GSH per min at 35°C. The assay of GST activity with other substrates was carried out at various wavelengths according to Habig et al. (16). Protein concentrations were determined by the Bradford method with bovine serum albumin as the protein standard (4).
S. griseus strain ATCC 13273 was grown at 30°C and stored on Sabouraud dextrose agar in sealed screw-cap tubes at 4°C. The organism was cultivated in two stages in soybean meal glucose medium (9, 17). Cultures were incubated at 30°C with shaking at 250 rpm. Stage one cultures inoculated with S. griseus spore suspensions from slants provided 72-h cultures that were used to inoculate (10% inoculum) stage two cultures. Stage two cultures were harvested after 72-h growth, and cells were pelleted by centrifugation at 10,000 × g for 20 min and washed with 0.5% (wt/vol) NaCl. For cell extracts, S. griseus cell pellets (10 g) were suspended in 63 ml of 10 mM potassium phosphate buffer (pH 7.0) containing 1.0 mM EDTA, 0.2 mM GSH, and 20% (vol/vol) glycerol (buffer A) and subjected to French press homogenization. Cell homogenates were centrifuged at 20,000 × g for 30 min, and the resulting supernatants were centrifuged again at 100,000 × g for 1 h 15 min at 4°C to give cell extracts for analysis or for enzyme purification.
S. griseus cells were analyzed for thiol content, including GSH, a necessary reactant in the GST reaction (20). Cell extracts contained total thiols from 63 nmol/g of dry cells at 24 h to a maximum of 127 nmol/g of dry cells at 60 h. GST activity of the cytosolic enzyme was proportional to growth, with maximum activity at 0.0033 μmol min−1 mg of protein−1 at 48 h. GST activity was inducible in S. griseus. For induction, various chemicals were added to 24-h stage two cultures that were grown for an additional 48 h. GST levels in cell extracts from cultures containing 3MC, phenobarbital, progesterone, and genistein were three to four times higher than those in controls. CDNB and dinitrobenzene (DNB) achieved an about twofold induction of GST, while β-NF and CCl4 were not GST inducers.
All steps for the purification of S. griseus GST were performed in the cold, and results from the purification of GST are summarized in Table 1. The supernatant centrifuged at 100,000 × g was applied to a DEAE-Sephadex column (40-ml bed volume, 2 by 30 cm, 250 mg of protein) equilibrated with buffer A. After washing with the same buffer, elution was performed with a linear gradient of 0 to 0.2 M KCl in 500 ml of buffer A to give a single peak between 160 and 170 mM KCl and an eightfold increase in specific activity. Active fractions were pooled, dialyzed against 500 ml of buffer A three times, and applied to a GSH-Sepharose affinity column (1 by 10 cm), which was equilibrated with the same buffer. After loading, the column was exhaustively washed with buffer A containing 50 mM KCl to remove unbound proteins. The enzyme was eluted with 50 mM Tris-HCl (buffer B), pH 9.6, containing 5 mM GSH, giving a 100-fold increase in GST-specific activity and 50% recovery. The fractions containing GST activity were pooled, concentrated by ultrafiltration through a 10-kDa (PM 10) cutoff membrane (Amicon Division, Beverly, Mass.), and dialyzed against buffer A for 10 h. The efficient two-step purification process gave 150 μg of GST with a specific activity of 3.23 U/mg of protein in an overall recovery of 48% activity from the crude cell extract and a nearly 800-fold purification.
TABLE 1.
Purification of GST from S. griseus
| Step | Protein (mg) | GST activity (U) | Sp act (U/mg of protein)a | Yield (%) | Purification (n-fold) |
|---|---|---|---|---|---|
| 100,000 × g supernatant | 250 | 1.01 | 0.004 | 100 | |
| DEAE-Sephadex | 30 | 0.96 | 0.032 | 95.0 | 8 |
| GSH-Sepharose 4B | 0.15 | 0.48 | 3.2 | 47.5 | 800 |
Units of enzyme activity (U) are expressed as μmoles per minute.
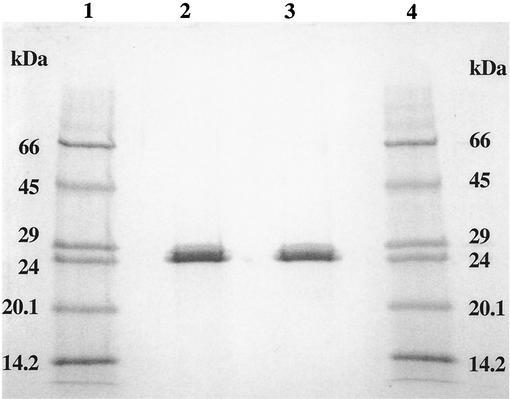
Purified GST gave a single band with an Mr of 24 by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (21). Size exclusion chromatography over a Sephadex G-150 (1 by 90 cm) column eluted with 20 mM Tris-HCl buffer (pH 7.5) containing 1 mM GSH at a flow rate of 0.5 ml/min gave an active protein with an estimated Mr of 48, indicating that GST was a homodimer. The N-terminal amino acid sequence of purified GST (20 μg) was determined to be SMILXYWDIIRGLAH (X represents ambiguity in amino acid identity) by Edman degradation on a GST band electroblotted to a polyvinylidene difluoride membrane. The UV-visible absorption at 280 nm indicated the lack of prosthetic groups such as flavin and heme.
Maximum enzyme activity occurred at 35°C and pH 6.5 in 0.1 M potassium phosphate buffer A. Activity was independent of 1 mM levels of Na+, K+, Ca2+, Mg2+, Mn2+, Zn2+, and Co2+. However, Hg2+ completely inhibited the reaction. The enzyme was stable both after lyophilization and when stored at −20°C but not at 4°C. When kept at 45°C, activity was gradually lost over a period of 2 h.
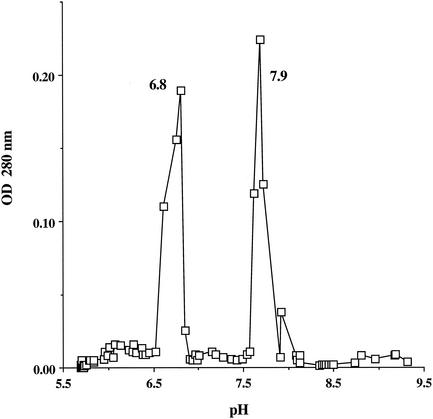
The pI of purified GST was estimated by chromatofocusing over a PBE 94 (Pharmacia) column. Samples to be examined were dialyzed in 25 mM ethanolamine-acetic acid buffer (pH 9.4). The column was washed with 30 ml of the same buffer and then eluted with polybuffer 96 acetic acid (Pharmacia) covering a pH range from 6 to 9 while 1-ml fractions were collected. The pH of each fraction was immediately measured, and fractions were assayed for enzyme activity. By this process, purified S. griseus GST was resolved into two isoenzymes, Sg-GST-6.8 and Sg-GST-7.9, with pI values of 6.8 and 7.9, respectively (Fig. 1). Both isoenzymes exhibited molecular masses of approximately 24 kDa by SDS-PAGE (Fig. 2). The isoenzymes displayed similar specific activities towards chloro-2,4-DNB (3.2 U min−1 mg of protein−1), p-nitrobenzyl chloride (1.1 U min−1 mg of protein−1), and ethacrynic acid (0.9 U min−1 mg of protein−1). However, specific activities of Sg-GST-6.8 and Sg-GST-7.9 versus CDNB were different at 2.3 and 3.0 U min−1 mg of protein−1, respectively. 1,2-Epoxy-3-(p-nitrophenoxy)-propane was not a substrate for either isoenzyme. Neither of the isoenzymes showed the GSH peroxidase activity previously found with a number of GST isoenzymes (27).
FIG. 1.
PBE column chromatographic resolution of S. griseus GST isoenzymes by isoelectric focusing at an optical density (OD) at 280 nm.
FIG. 2.
SDS-PAGE of the purified GST isoenzymes (5 μg) (lanes 2 and 3) and protein markers (3.6 μg per band) (lanes 1 and 4) stained with Coomassie blue. Markers from top to bottom are bovine albumin (66 kDa), egg albumin (45 kDa), glyceraldehyde-3-phosphate dehydrogenase (36 kDa), carbonic anhydrase (29 kDa), bovine pancreatic trypsinogen (24 kDa), soybean trypsin inhibitor (20.1 kDa), and bovine milk α-lactalbumin (14.2 kDa).
Kinetic studies with purified GST isoenzymes were conducted by measuring initial reaction rates with fixed concentrations of 1 mM CDNB and GSH varied between 0 and 1.0 mM. Alternatively, GSH concentrations were kept constant at 1.0 mM and CDNB was varied between 0.1 and 1.0 mM. Kinetic constants were determined by using Lineweaver-Burk plots and the EZ-FIT program developed by Perrella (25). Duplicate samples were incubated at 35°C by using two different protein concentrations. The apparent Km values for GSH and CDNB of Sg-GST-6.8 were 0.25 ± 0.01 and 1.32 ± 0.05 mM, respectively, and the Km values of Sg-GST-7.9 for GSH and CDNB were 0.20 ± 0.01 and 1.29 ± 0.06, respectively.
GSH has been implicated in the maintenance of an intracellular reductive environment that protects cells from the damaging effects of electrophiles (31). Cohen et al. reported that the GSH contents of Fusarium oxysporum and Rhizoctoria solani mycelia were initially decreased in the presence of electrophilic compounds such as pentachloronitrobenzene and CDNB and then increased gradually to original levels (6).
The reaction used to assay for GST in S. griseus takes advantage of the coupling of the reactive aromatic electrophile CDNB with GSH by the displacement of chlorine to produce a readily measured conjugate. In mammals, hepatic GSH and GSTs often accompany cytochrome P450 enzymes where they quench highly reactive electrophilic species formed during the oxidative metabolism of aromatics, drugs, and other xenobiotics. These phase I (oxidizing) and phase II (conjugating) enzymes together appear to be necessary for the maintenance of normal metabolic and liver function (19). Bacterial GSTs have often been implicated in aromatic compound metabolism, but no physiological roles have been assigned to the proteins coded for by putative GST gene sequences (31).
In the 100,000 × g supernatants from untreated S. griseus cells, GST activity could be reproducibly demonstrated at a level of approximately 0.004 μmol min−1 mg of protein−1. Common mammalian and microbial GST inducers clearly increased levels of S. griseus GST. Genistein, a known inducer of cytochrome P450soy induced GST activity threefold in S. griseus, thus linking the coinduction of both phase I and phase II metabolizing enzymes in this organism (30). GSTs are inducible in other organisms, such as Aspergillus ochraceus TS (7) (7.9-fold with 3MC) and the yeast Issatechaenkia orientalis GST (37-fold with o-DNB) (28, 29).
The specific activity for CDNB of the purified S. griseus enzyme was similar to that reported for Escherichia coli B (18), Proteus mirabilis (10), and A. ochraceus (8). The S. griseus enzyme was similar in functional mass to mammalian, plant, and microbial GSTs. GSTs from Pseudomonas sp. (20), E. coli B (18), and Phanerochaete chrysosporium (13) are homodimers with typical masses of 42, 45, and 58 kDa respectively. On the other hand, Tetrahymena thermophila GST is a monomer of 33,000 to 35,000 kDa (24) and the native GST of A. ochraceus TS is a 56-kDa homotetramer (8). The N-terminal amino acid sequence for S. griseus GST was different than the GST sequences reported for other bacteria and fungi (1, 5, 11, 14). Interestingly, the S. griseus GST N-terminal sequence showed nearly 65% homology to a sequence reported for rat mu GST (14), indicating a similarity with at least one mammalian microsomal GST.
Two major S. griseus GST isoenzymes represented about 0.06% of the total cytosolic proteins in cell extracts. When the purified isoenzymes were rechromatographed on chromatofocusing columns, they emerged as single peaks at the same points and had the same pI values as before. Thus, the isoenzymes were not artifacts of purification. Three isoenzymes, Sg-GST-6.8, Sg-GST-7.4, and Sg-SGT-7.9, were observed in cell extracts when S. griseus was challenged with 3MC (results not shown). This indicated the responsiveness of S. griseus to xenobiotics in terms of the range of GSTs formed. Microbial GST isoenzymes are known in P. mirabilis (10), Serratia marcescens (11), Xanthomonas campestris (12), and Mucor circinelloides (14). While acidic, neutral, and basic GST isoenzymes have been reported in mammals, only neutral and basic GST isoenzymes have been observed in microorganisms (7). Km values for GSH and CDNB for S. griseus GST isoenzymes were very similar to those reported for E. coli (3) but different from the values for Pseudomonas sp. GST (20). While the Km for GSH from A. ochraceus (8) is twice that for S. griseus, the Km for CDNB is similar in both organisms. Comparison data for inhibition studies with bacterial GSTs are unavailable. Cha et al. recently reported that GST activity in Cunninghamella elegans was inhibited by quercetin, cibacronblue, hematin, and alzarin (5).
Physiological roles of GSTs from bacteria purified by affinity chromatography remain elusive (31). It is interesting that the presence of CDNB-active GST enzymes has been associated with increased bacterial resistance to several antibiotics (26). In S. griseus, two putative roles for GST can be suggested. Our strain of S. griseus produces the cytotoxic antibiotic chromomycin A3 (23), a polyketide that undergoes one-electron oxidation reactions to form highly reactive intermediates capable of interacting with nucleophiles like GSH (2). The S. griseus GST enzyme system could play a role in this organism's resistance to the toxic effects of the chromomycin A3 that it produces. Sariaslani demonstrated that S. griseus contains a genistein-inducible cytochrome P450 enzyme system (29). It was recently reported that genistein was hydroxylated by S. griseus to catechol products (17). Catechols are readily oxidized to electrophilic quinoid products. Since genistein induces both cytochrome P450 and GST in the same strain of S. griseus, it is likely that, as in mammalian liver, both activities are simultaneously induced.
Acknowledgments
We gratefully acknowledge funding from the U.S. Department of Agriculture through the Iowa Biotechnology By-products Consortium.
REFERENCES
- 1.Altchul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PST-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anyanwutaku, I. O., R. J. Petroski, and J. P. N. Rosazza. 1994. Oxidative coupling of aureolic acids and hydroquinone catalyzed by copper oxidases. Implications for the molecular mechanism of action of Aureolic acids. Bioorg. Med. Chem. 2:543-551. [DOI] [PubMed] [Google Scholar]
- 3.Arca, P., P. Garcia, C. Hardisson, and J. E. Suarez. 1990. Purification and study of a bacterial glutathione S-transferase. FEBS Lett. 263:77-79. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the purification of microgram quatities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Cha, C. J., B. F. Coles, and C. E. Cerniglia. 2001. Purification and characterization of a glutathione S-transferase from the fungus Cunninghamella elegans. FEMS Microbiol. Lett. 203:257-261. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, E., A. Gamliel, and J. Katan. 1986. Glutamate glutathione-S-transferase in fungi. Effect of p-nitrobenzene and 1-chloro-2,4-dinitrobenzene: purification and characterization of the transferase from Fusarium. Chem. Physiol. 26:1-9. [Google Scholar]
- 7.Datta, J., T. K. Datta, and T. B. Samanta. 1994. Microsomal glutathione S-transferase (GST) isoenzymes in Aspergillus ochraceus TS: induction by 3-methylcholanthrene. Biochem. Biophys. Res. Commun. 203:1508-1514. [DOI] [PubMed] [Google Scholar]
- 8.Datta, J., and T. B. Samanta. 1992. Characterization of a novel microsomal glutathione S-transferase produced by Aspergillus ochraceus TS. Mol. Cell. Biochem. 118:31-38. [DOI] [PubMed] [Google Scholar]
- 9.Dhar, K., and J. P. N. Rosazza. 2000. Purification and characterization of Streptomyces griseus catechol O-methyltransferase. Appl. Environ. Microbiol. 66:4877-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Ilio, C., A. Aceto, R. Piccolomini, N. Allocati, A. Faraone, L. Cellini, G. Ravagnan, and G. Federici. 1988. Purification and characterization of three forms of glutathione transferase from Proteus mirabilis. Biochem. J. 255:971-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Ilio, C., A. Aceto, R. Piccolomini, N. Allocati, A. Faraone, T. Bucciarelli, D. Barra, and G. Federici. 1991. Purification and characterization of a novel glutathione transferase from Serratia marcescens. Biochim. Biophys. Acta 1077:141-146. [DOI] [PubMed] [Google Scholar]
- 12.Di Ilio, C., A. Aceto, N. Allocati, R. Piccolomini, T. Bucciarelli, B. Dragani, A. Faraone, P. Sacchetta, R. Petruzzelli, and G. Federici. 1993. Characterization of glutathione transferase from Xanthomonas campestris. Arch. Biochem. Biophys. 305:110-114. [DOI] [PubMed] [Google Scholar]
- 13.Dowd, C. A., C. M. Buckley, and D. Sheehan. 1997. Glutathione S-transferases from the white rot fungus, Phanerochaete chrysosporium. Biochem. J. 324:243-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowd, C. A., and D. Sheehan. 1999. Variable expression of glutathione S-transferase isoenzymes in the fungus, Mucor circinelloides. FEMS Microbiol. Lett. 170:13-17. [DOI] [PubMed] [Google Scholar]
- 15.Ellman, G. L. 1959. Tissue sulfhydryl groups. Archiv. Biochem. Biophys. 82:70-77. [DOI] [PubMed] [Google Scholar]
- 16.Habig, W. H., M. J. Pabst, and W. B. Jakoby. 1974. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 249:7130-7139. [PubMed] [Google Scholar]
- 17.Hosny, M., and J. P. N. Rosazza. 1999. Microbial hydroxylation and methylation of genistein by Streptomycetes. J. Nat. Prod. 62:1609-1612. [Google Scholar]
- 18.Ilzuka, M., Y. Inoue, K. Murata, and A. Kimura. 1989. Purification and some properties of glutathione S-transferase from Escherichia coli B. J. Bacteriol. 171:6039-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakoby, W. B., and W. H. Habig. 1980. Enzymatic basis of detoxication, vol. 2, p. 63-94. Academic Press, New York, N.Y.
- 20.Jung, U. H., J. Y. E. Cho, H. M. Seong, S. J. Kim, Y. Kim, and A. S. Chung. 1996. Chracterization of a novel glutathione-S-transferase from Pseudomonas sp. J. Biochem. Mol. Biol. 29:111-115. [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Liu, S. Y., and J. P. N. Rosazza. 1998. Enzymatic conversion of glucose to UDP-4-keto-6-deoxyglucose in Streptomyces sp. Appl. Environ. Microbiol. 64:3972-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montanari, A., and J. P. N. Rosazza. 1990. Biogenesis of chromomycin A3 by Streptomyces griseus. J. Antibiot. 43:883-889. [DOI] [PubMed] [Google Scholar]
- 24.Overbaugh, J. M., E. P. Lau, V. A. Marino, and R. Fall. 1988. Purification and preliminary characterization of a monomeric glutathione S-transferase from Tetrahymena thermophila. Arch. Biochem. Biophys. 261:227-234. [DOI] [PubMed] [Google Scholar]
- 25.Perrella, F. W. 1988. EZ-FIT: a practical curve fitting microcomputer program for the analysis of enzyme kinetic data on IBM PC compatible computers. Anal. Biochem. 174:437-447. [DOI] [PubMed] [Google Scholar]
- 26.Piccolomini, R., C. Di Ilio, A. Aceto, N. Allocati, A. Faraone, L. Cellini, G. Ravagnan, and G. Federici. 1989. Glutathione transferase in bacteria: subunit composition and antigenic characterization J. Gen. Microbiol. 135:3119-3125. [DOI] [PubMed] [Google Scholar]
- 27.Pickett, C. B., and A. Y. Lu. 1989. Glutathione S-transferases: gene structure, regulation and biological function. Annu. Rev. Biochem. 58:743-764. [DOI] [PubMed] [Google Scholar]
- 28.Tamaki, H., H. Kumagai, and T. Tochikura. 1990. Glutathione S-transferase in yeast: induction of mRNA, cDNA cloning and expression in Escherichia coli. Biochem. Biophys. Res. Commun. 172:669-675. [DOI] [PubMed] [Google Scholar]
- 29.Tamaki, H., K. Yamamoto, and H. Kumagai. 1999. Expression of two glutathione S-transferase genes in the yeast Issatchenkia orientalis is induced by o-dinitrobenzene during cell growth arrest. J. Bacteriol. 181:2958-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trower, M. K., F. S. Sariaslani, and D. P. O'Keefe. 1989. Purification and chracterization of a soybean flour-induced cytochrome P-450 from Streptomyces griseus. J. Bacteriol. 171:1781-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vuilleumier, S. 1997. Bacterial glutathione S-transferases: what are they good for? J. Bacteriol. 179:1431-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]