Abstract
A simple and efficient delivery system was developed for making targeted gene knockouts in Mycobacterium smegmatis. This delivery system relies on the use of a pair of replicating plasmids, which are incompatible. Incompatible plasmids share elements of the same replication machinery and so compete with each other during both replication and partitioning into daughter cells. Such plasmids can be maintained together in the presence of antibiotics; however, removal of selection leads to the loss of one or both plasmids. For mutagenesis, two replicating plasmids based on pAL5000 are introduced; one of these plasmids carries a mutated allele of the targeted gene. Homologous recombination is allowed to take place, and either one or both of the vectors are lost through the pressure of incompatibility, allowing the phenotypic effects of the mutant to be studied. Several different plasmid combinations were tested to optimize loss in the absence of antibiotic selection. pAL5000 carries two replication genes (repA and repB), which act in trans, and the use of vectors that each lack one rep gene and complement each other resulted in the loss of both plasmids in M. smegmatis and Mycobacterium bovis BCG. The rate of loss was increased by the incorporation of an additional incompatibility region in one of the plasmids. To facilitate cloning when the system was used, we constructed plasmid vector pairs that allow simple addition of selection and screening genes on flexible gene cassettes. Using this system, we demonstrated that M. smegmatis pyrF mutants could be isolated at high frequency. This method should also be useful in other species in which pAL5000 replicates, including Mycobacterium tuberculosis.
Production of mutations in mycobacteria is a fundamental approach for discovering the function of genes in order to obtain knowledge concerning biology and pathogenicity. This knowledge may contribute to vaccine development by identifying virulence determinants for rational attenuation, and it may contribute to drug discovery by identifying possible targets. Mutagenesis may be achieved in a random or targeted manner. Both allelic exchange and transposon mutagenesis are rare genetic events in mycobacteria, so an efficient delivery system is required, in which the disrupted gene or transposon is introduced on a vector, homologous recombination or transposition takes place, and the vector is then lost. This has been accomplished in mycobacteria by the use of nonreplicating (suicide) plasmids (10, 14, 16), a temperature-sensitive plasmid (6), and a temperature-sensitive phage (1). In this study we took a different approach and used plasmid incompatibility. This technique has the advantage of using replicating plasmids, which results in a prolonged time in the cell for homologous recombination or transposition to occur but avoids the use of a temperature-sensitive replicon which can be difficult to lose in slowly growing species with narrow temperature ranges, such as Mycobacterium tuberculosis.
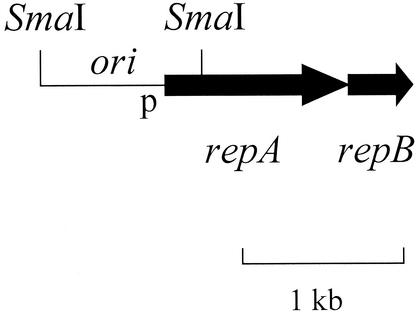
Plasmid incompatibility is defined as the inability of a pair of coresident plasmids to be stably maintained in the absence of external selection. This is due to sharing of one or more components of plasmid partitioning or replication systems. Plasmid loss due to incompatibility is commonly due to interference with the ability of the plasmid to correct stochastic fluctuations in its copy number (13). By imposing different kinds of selection on pairs of incompatible plasmids, copy number can be manipulated. We have previously mapped the replication functions of pAL5000 (23, 24) (Fig. 1). Two replication genes, repA and repB, encoding a putative primase and a DNA-binding protein, respectively, are essential for replication and can be supplied in trans. The incompatibility functions (inc) of pAL5000 were mapped to a cis-acting 120-bp fragment upstream of repA, which includes the origin of replication (oriM). The oriM/inc region is capable of conferring incompatibility to otherwise unrelated replicons.
FIG. 1.
Diagram of the pAL5000 replication genes. The repA and repB genes are both essential for replication. The origin and incompatibility functions map upstream of these genes, within the 1-kb SmaI fragment.
We show here that maintenance or loss of plasmids can be controlled in both Mycobacterium smegmatis mc2155 and Mycobacterium bovis BCG by using pairs of pAL5000-derived plasmids, and we developed an efficient delivery system for targeted gene replacement using incompatibility as a pressure to eliminate plasmids.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Plasmids used in this work are listed in Table 1. M. smegmatis strain mc2155 (22) was cultured in Lemco medium (10 g of peptone per liter, 5 g of Lemco powder [Oxoid] per liter, 5 g of NaCl per liter) containing 0.05% (wt/vol) Tween 80 or on Lemco agar plates (Lemco medium containing 15 g of agar per liter). M. bovis BCG strain NCTC 5692 was grown in Middlebrook 7H9 liquid medium (Difco) supplemented with 10% (wt/vol) OADC (oleic acid, albumin, dextrose, catalase; Becton Dickinson) and 0.05% (wt/vol) Tween 80 or on Middlebrook 7H10 agar (Difco) supplemented with 10% (wt/vol) OADC. Antibiotics were added at the following concentrations when appropriate: kanamycin, 20 μg ml−1; hygromycin, 100 μg ml−1; and gentamicin, 10 μg ml−1. When gentamicin and kanamycin were used in combination, the concentration of kanamycin was increased to 50 μg ml−1 as there is a degree of cross-resistance from gentamicin to kanamycin.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Relevant genotypea | Reference or source |
|---|---|---|---|
| pACYC184 | Cloning vector | oriE (P15A) cm | 5 |
| pGOAL18 | Cassette vector | oriE amp PAg85lacZ Phsp80sacB | 16 |
| pGOAL22 | Cassette vector | oriE amp PAg85lacZ | 16 |
| pINC9 | oriM/inc × 2 (adjacent and in same orientation) in pNUT5 | oriE amp kan repB oriM/inc.oriM/inc | This study |
| pINC52 | PAg85lacZ/Phsp60sacB (SmaI) from pGOAL18 in pINC9 (ScaI)b | oriE amp kan repB oriM/inc.oriM/inc PAg85lacZ Phsp60sacB | This study |
| pINC11 | repB PCR product and oriM/inc × 2 in pUC19 | oriE oriM/inc.oriM/inc amp repB | This study |
| pINC12 | PAg85lacZ from pGOAL22 in pINC13 | oriE oriM/inc.oriM/inc amp repB PAg85lacZ | This study |
| pINC13 | PacI-containing fragment from p2NIL in pINC11 | oriE oriM/inc.oriM/inc amp repB PacI site | This study |
| pINC15 | kan in pINC12 | oriE oriM/inc.oriM/inc amp kan repB PAg85lacZ | This study |
| p2NIL | Manipulation vector for gene replacement | oriE amp kan | 16 |
| pNUT2 | hyg in pUH36 | oriE oriM/inc amp hyg repA | This study |
| pNUT5 | kan and oriM/inc in pUH73 | oriE amp kan repB | This study |
| pNUT12 | repA and oriE/inc from pUH36 in pACYC184 | oriE (P15A) cm oriM/inc repA | This study |
| pNUT20 | pNUT12 with HindIII deletion | oriE (P15A) oriM/inc cm repA | This study |
| pNUT21 | PacI-containing fragment from p2NIL in pNUT20 | oriE (P15A) oriM/inc cm repA PacI site | This study |
| pNUT22 | PAg85lacZ from pGOAL22 in pNUT21 | oriE (P15A) oriM/inc cm repA PAg85lacZ | This study |
| pNUT23 | kan in pNUT22 | oriE (P15A) oriM/inc cm kan repA PAg85lacZ | This study |
| pUH36 | repA and oriM/inc in pUC18 | oriE oriM/inc amp repA | 23 |
| pUH73 | repB in pBluescript | oriE amp repB | Pelle Stolt |
| pUH76 | oriM/inc from pAL5000 as a 1-kb SmaI fragment in pUC19 plus kan | oriE oriM/inc amp kan | Pelle Stolt |
| pY6001 | M. smegmatis pyrF in pUC19 | oriE amp pyrF | 8 |
| pY6001G1 | gm in pY6001 | oriE amp kan pyrF::gm | This study |
| pYRANA3 | pyrF::gm in pNUT12 | oriE oriM/inc cm repA pyrF::gm | This study |
oriE is from ColE1 unless otherwise specified.
See reference 16.
Electroporation.
Competent M. smegmatis and M. bovis BCG cells were prepared as described by Parish and Stoker (15). Transformations were performed by using 400-μl aliquots of cells with 1 to 2 μg of DNA for M. smegmatis and 1 to 5 μg of DNA for M. bovis BCG. Cotransformants (cells transformed with two different plasmids) were selected on plates supplemented with antibiotics selecting for both plasmids.
Plasmid stability assay.
A single M. smegmatis or M. bovis BCG cotransformant colony was used to inoculate 5 to 10 ml of medium (see above) with antibiotics selecting for both plasmids and incubated at 37°C with shaking until the A600 was approximately 0.5 (3 to 4 days for M. smegmatis and 3 weeks for M. bovis BCG). One hundred microliters of each culture was plated onto a 190-mm nonselective agar plate (medium without antibiotics) and incubated at 37°C to obtain a lawn of growth. Serial dilutions of the culture were plated onto selective and nonselective plates to obtain single cells in order to determine the proportion of antibiotic-resistant (plasmid-carrying) cells. Cells were scraped from the lawn on a nonselective plate into 1 ml of medium. Dilutions were plated onto selective and nonselective plates. Each plating was referred to as a round. This procedure was repeated through rounds of nonselective plating until no colonies could be counted on the antibiotic plates (i.e., when undiluted cells did not give rise to any colonies); no more than three rounds were necessary. Cells were always scraped from a nonselective plate having a lawn of growth. The relative stabilities of the two plasmids were monitored by calculating the reduction in plasmid numbers at each round from the colony counts and plotting the reductions in plasmid number against the rounds of plating.
The loss factor (LF) (12) was calculated by using the equation LF = 1 − (V1/V2)1/g, where V2 is the total number of cells (cells growing on nonselective medium) after g generations and V1 is the number of cells that retained the plasmid (cells growing on selective medium) after g generations. We estimated that each round was about 23 generations (the time necessary for one cell to grow into a colony of 107 cells). The average loss factor was calculated from three separate experiments and was expressed as the mean ± standard deviation.
Isolation of mutants.
The method used to isolate mutants was essentially the same as the method used to assess plasmid stability. However, following growth of a cotransformant in antibiotic-containing medium, 109 cells were initially plated onto an agar plate supplemented with gentamicin to obtain sufficient numbers of plasmid-carrying colonies for mutant isolation. Cells were then scraped and subjected to nonselective rounds of plating by plating dilutions onto plates containing gentamicin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) in order to isolate white gentamicin-resistant colonies (i.e., colonies containing the mutant pyrF allele but lacking the plasmid-borne lacZ gene).
Analysis of M. smegmatis mutants.
White colonies were initially tested for uracil auxotrophy and resistance to 1 mg of 5′-fluoroorotic acid (5′-FOA) (Sigma) per ml on minimal medium (M9 minimal salts [Difco], 20% [wt/vol] glucose, 2 ml of glycerol per liter, 0.2 mM uracil). DNA was prepared from 5-ml liquid cultures (2, 20) and used to confirm mutant generation by Southern blotting and PCR analysis.
To assess the frequency of mutant generation, serial dilutions were plated four times, and three independent experiments were performed. Phenotypically, pyrF mutants should have produced white colonies, been uracil auxotrophs, been resistant to 5′-FOA, and grown on plates containing 10% (wt/vol) sucrose (sacB was carried on pINC52).
RESULTS AND DISCUSSION
Incompatibility between plasmids can provide the basis for a delivery system.
The essence of any delivery system is that a DNA molecule carrying a transposon or altered gene is introduced and then lost. Efficient loss is essential, as the events being sought are rare and it is necessary to select for them. Thus, if a researcher is looking for gene replacement with an allele carrying an antibiotic resistance marker, the vector carrying this marker must be lost. This is most straightforwardly done by using a nonreplicating vector. The use of replicating vectors should theoretically increase the frequency of mutant isolation compared with that obtained with suicide vectors since (i) replicating vectors have an increased time for homologous recombination events to occur and (ii) DNA replication and recombination occur simultaneously in the cell, but replicating vectors are then more difficult to remove. Conditionally replicating vectors have been used for gene replacement with a temperature-sensitive origin of replication (4, 17). Temperature-sensitive vectors are, however, weakly thermosensitive in slowly growing members of the M. tuberculosis complex because of their narrow temperature range (17). These vectors therefore have to be combined with a counterselectable marker, such as sacB, to eliminate clones that still contain vector DNA. Growth at the permissive temperature is very slow, while colonies grown at the nonpermissive temperature also take longer to grow and have been reported to be microscopic (unpublished results).
Having shown previously that incompatibility could in principle be used (23), we carried out experiments using different plasmid pairs in order to optimize this system.
Model A: use of codependent plasmids.
We previously showed that when M. smegmatis is transformed with two pAL5000-based plasmids carrying different markers, selection for one plasmid causes the second plasmid to be lost due to incompatibility pressure (23). This is not an ideal delivery system, in part because it requires a functional plasmid to integrate into the chromosome, which appears to be lethal for the pAL5000 replicon in M. tuberculosis (7).
We therefore used the fact that repA and repB act in trans (23) to develop a system in which we used a pair of codependent plasmids, each containing the pAL5000 minimal origin of replication (SmaI fragment in Fig. 1) but lacking one replication gene. This minimal origin contains regions involved in incompatibility and is referred to below as oriM/inc. As neither plasmid can replicate independently in mycobacteria, selection for one plasmid automatically selects for both, while growth in the absence of selection should result in the loss of both plasmids and thus the plasmid carrying the mutated target gene should be lost.
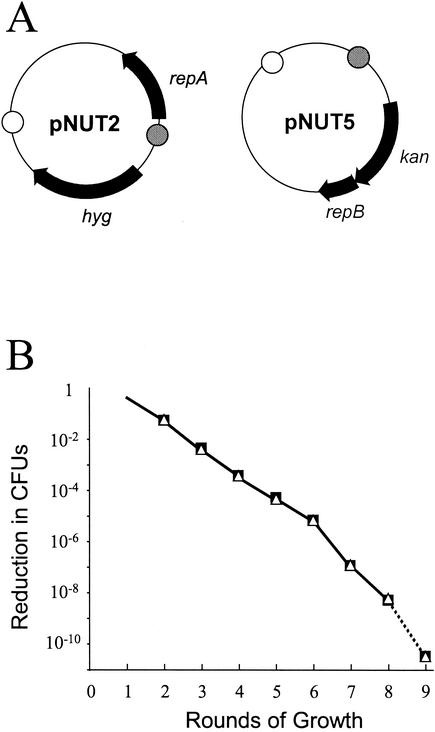
We constructed an Hygr plasmid carrying repA only (pNUT2) and a Kanr plasmid that carried only repB (pNUT5). We called this pairing model A (Fig. 2A). The plasmids were cotransformed into M. smegmatis mc2155 and selected on plates containing both hygromycin and kanamycin. Plasmid loss was determined during serial culture on Lemco agar in the absence of antibiotics. Plasmids were lost at a steady rate of 10−1 per round with a loss factor of 0.095 ± 0.016 (Fig. 2B). When model A cotransformants were obtained, the colony size was also small but recovered after restreaking onto fresh plates, possibly because this allowed a pool of the Rep proteins to be established, although we cannot rule out the possibility that there were compensatory mutations.
FIG. 2.
Loss kinetics with model A plasmids. (A) Plasmid constructs. kan, kanamycin resistance gene; hyg, hygromycin resistance gene; repA and repB, replication genes from pAL5000; open circle, E. coli ColE1 oriE; cross-hatched circle, pAL5000 oriM. (B) Plasmid loss kinetics in M. smegmatis. Symbols: ▪, Hygr colonies (pNUT2); ▵, Kanr colonies (pNUT5). The dotted line indicates that the limit of sensitivity was reached.
Effects of different antibiotic resistance genes.
A combination of antibiotic resistance genes cloned into the two incompatible plasmids was shown to be important in terms of loss rates. We have previously shown that plasmids carrying the kan gene are less stable than plasmids carrying hyg (23). In order to extend these observations, we compared the stability of pAL5000-based plasmids carrying gm with the stability of the same plasmids that had gm replaced with kan or hyg at the same site and in the same orientation; hence, the plasmids were identical except for the resistance gene. No difference in stability between plasmids carrying gm and plasmids carrying hyg was observed. However, plasmids carrying kan were less stable than both of these. Therefore, it may be beneficial to include kan on one plasmid. A reason proposed for this is that the hyg gene from Streptomyces hygroscopicus is GC rich and so may be more compatible with mycobacteria than the transposon-derived kan genes having lower G+C contents (23).
Model B: addition of a second oriM/inc region.
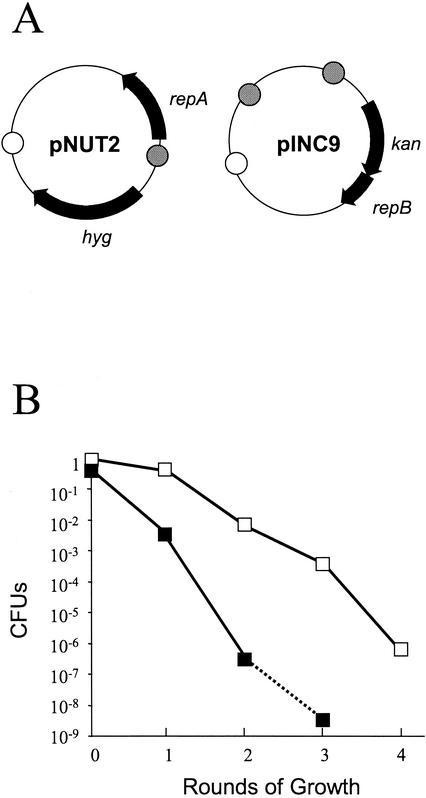
In order to increase the rate of loss, we used the observation made in previous work that a second copy of the replication origin/incompatibility region increased incompatibility if it was cloned close to the other copy in the same orientation (23). We therefore adapted model A plasmids by adding a second oriM/inc region to pNUT5, producing pINC9. We called the pairing of pINC9 (repB kan oriM/inc ×2) with pNUT2 (repA hyg oriM/inc) model B (Fig. 3A). Colonies were obtained at a frequency that was about 1% of the frequency obtained with a single replicating plasmid. The colonies were very small in the initial transformation but recovered after restreaking.
FIG. 3.
Loss kinetics with model B plasmids. (A) Plasmid constructs. kan, kanamycin resistance gene; hyg, hygromycin resistance gene; repA and repB, replication genes from pAL5000; open circle, E. coli ColE1 oriE; cross-hatched circle, pAL5000 oriM. (B) Plasmid loss kinetics in M. smegmatis and in M. bovis BCG. Symbols: ▪, Hygr M. smegmatis(pATB12); □, Hygr M. bovis BCG. The dotted line indicates that the limit of sensitivity was reached.
Additionally, we found using other models a significant degree of plasmid rearrangement in the ColE1 origin regions (data not shown). We therefore used different Escherichia coli origins (ColE1 and P15A) for the remaining experiments, and this removed the problem.
Plasmid loss was measured in the absence of selection. Both plasmids were simultaneously lost at a rate of approximately 10−3 per round of plating, corresponding to a loss factor of 0.213 ± 0.068 (Fig. 3B). This rate was higher than the rate seen with model A. Again, we looked for interplasmid recombination but did not observe any (data not shown).
The same experiment was repeated with M. bovis BCG (Fig. 3B). The plasmids were lost rapidly (approximately 10−2 per round of plating), although the loss factor (0.178 ± 0.066) was slightly lower than that seen in M. smegmatis (Fig. 3B). We concluded that we could introduce pairs of plasmids and maintain them by antibiotic selection but we could cure the plasmids efficiently by removing the selection. This provided the basis for an efficient delivery system. The most promising system was model B, and we carried out mutagenesis experiments using this model.
Targeted mutagenesis of M. smegmatis pyrF by using an incompatibility-based delivery system.
Having shown that model B complementing plasmids were lost efficiently when selection was removed, we tested them as a delivery system for targeted mutagenesis. The M. smegmatis pyrF gene encodes orotidine monophosphate decarboxylase, which allows growth in media deficient in uracil but is lethal to cells grown in the presence of 5′-FOA. Mutants lacking this gene have been generated by other methods, making it a suitable test system.
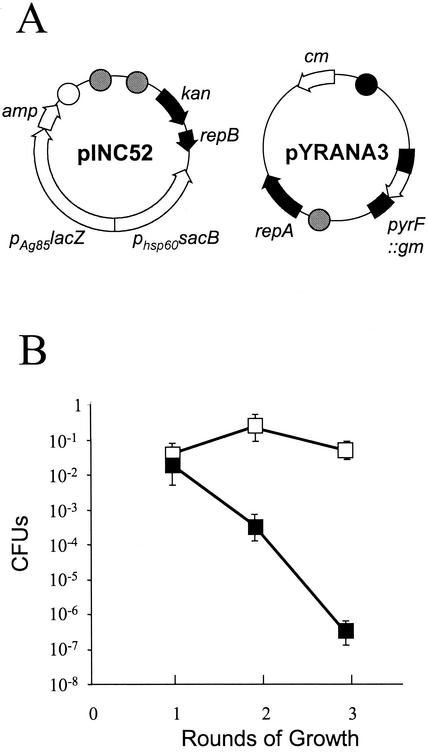
A 3.2-kb SphI fragment containing the pyrF gene interrupted by gm was cloned into pNUT12, producing pYRANA3 (Fig. 4A). pYRANA3 and pINC52 were cotransformed into M. smegmatis mc2155, with selection for gentamicin and kanamycin. The cells were grown in selective liquid media (containing gentamicin and kanamycin) before they were plated on agar containing gentamicin. Cells were then scraped and plated onto antibiotic-free plates and taken through rounds of plating. As we were looking for relatively rare events, such as homologous recombination (11), the number of cells used to seed each plate had to be high, and we used 109 cells. The rationale of the screening procedure was that the selection with gentamicin would select for cells carrying the mutant pyrF gene, either on the plasmid or integrated into the chromosome. To eliminate the former possibility, we plated the cells in the presence of X-Gal, which distinguished between colonies carrying the pINC52 (lacZ+) plasmid (blue) and plasmid-free colonies (white). Thus, cells were plated onto plates containing gentamicin and X-Gal at each round to identify mutants, and white colonies were picked as potential mutants for further analysis. Note that as pINC52 carries the sacB gene (which provides an additional screen), plasmid-carrying cells are sucrose sensitive, while mutants lacking the plasmid are also sucrose resistant.
FIG. 4.
Frequency of pyrF mutant generation in M. smegmatis. (A) Plasmid constructs. kan, kanamycin resistance gene; cm, chloramphenicol resistance gene; gm, gentamicin resistance gene; amp, ampicillin resistance gene; repA and repB, replication genes from pAL5000; open circle, E. coli ColE1 oriE; cross-hatched circle, E. coli P15A oriE; solid circle, pAL5000 oriM. (B) Plasmid loss and mutant isolation. Symbols: ▪, total Gmr colonies (plasmid loss); □, white Gmr colonies (pyrF mutants). The error bars indicate standard deviations from three separate experiments.
After the first round of plating, white colonies were observed at an average frequency of 2.6% ± 0.9%. During the second round, the frequency of mutants increased to 22% ± 29% (one experiment yielded 64% mutants), but the frequency of mutants fell during the third round to 5.5% ± 0.50% (Fig. 4B). All of the white colonies (n = 95) were tested phenotypically; 90 of these had the expected phenotype of a pyrF mutant (uracil auxotrophs and those resistant to 5′-FOA and sucrose). Five colonies were not auxotrophic and still retained plasmids; restriction analysis showed that they had undergone rearrangements in the lacZ gene (data not shown). Thirty-one randomly selected auxotrophic colonies were also tested genotypically by PCR and Southern blot analysis and were confirmed to be double-crossover mutants.
Fitness of pyrF mutants.
One surprise was that there was not a gradual accumulation of mutants when organisms were plated on gentamicin. Gmr bacteria were seen either because they had both plasmids or because one plasmid had integrated into the chromosome as a single or double crossover. As the former was constantly decreasing and the latter should have been stable and increasing, the number of white colonies should have increased with every round. In fact, an increase was seen initially, but then there was a decrease in the subsequent round; this pattern was observed in nine separate experiments. The fact that there was not an accumulation was likely because of the greater fitness of the wild-type cells during competitive growth (21, 25). We confirmed this by growing mutant and wild-type cells in a mixed culture (data not shown). This procedure could be manipulated by making plasmid carriage more stressful. For example, we could use kan as the selectable marker in the target gene as it increased loss; if it was placed on the other plasmid, the competitive advantage of the wild-type cells might have been decreased. However, it is likely that most mutations result in some disadvantage, and this suggests that taking the bacteria through more than two rounds of nonselective growth is unlikely to provide great benefit.
Improved flexibility of plasmid pairs.
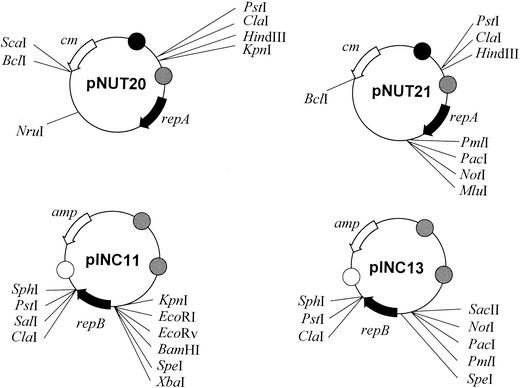
Having shown that the plasmids could be used to efficiently isolate mutants, we addressed the lack of cloning sites in the vectors, which severely restricted their utility. Derivatives of pNUT12 and pINC52 were constructed which had an increased number of convenient restriction sites (Fig. 5). The pINC series of plasmids all contained repB, two oriM/inc regions that were adjacent and in the same orientation, and an amp gene, and oriE was derived from ColE1. The complementing pNUT series contained repA, a single oriM/inc region, and a cm gene, and oriE was derived from P15A. The two-plasmid series contained different unique restriction enzymes for cloning (Fig. 5). As two plasmids are needed for this delivery system, the mutated gene can be cloned into either plasmid depending on which plasmid carries the most convenient sites for the target gene.
FIG. 5.
Flexible vectors for cloning genes for mutagenesis. The target gene is cloned into one of these plasmids (either the pNUT or pINC series depending on which series has the most convenient restriction sites for cloning of the target gene) and mutated (or a mutated gene can be cloned in directly). If the gene is cloned in pNUT20 or pNUT21, the preparation should be cotransformed with a pINC-based plasmid (i.e., pINC12, which contains lacZ for screening and repB); if it is cloned into pINC11 or pINC13, the preparation should be cotransformed with a pNUT-based plasmid (i.e., pNUT22, which contains lacZ for screening and repA). A cotransformant can be grown in selective media and then plated onto nonselective media (to allow plasmid loss by incompatibility). Mutants can be identified by plating preparations onto plates containing X-Gal plus the relevant antibiotic (mutants produce white colonies). cm, chloramphenicol resistance gene; amp, ampicillin resistance gene; repA and repB, replication genes from pAL5000; open circle, E. coli ColE1 oriE; cross-hatched circle, E. coli P15A oriE; solid circle, pAL5000 oriM.
In addition, a PacI site was incorporated into both plasmids so that PacI marker cassettes in the pGOAL series of plasmids (16) could be easily cloned onto either vector. For example, the pGOAL19 cassette contains hyg, PAg85lacZ, and Phsp60sacB. We also constructed plasmids that contained the markers kan and PAg85lacZ/kan for both plasmids of the incompatible pair since these cassettes are not available as PacI fragments in the pGOAL series.
In order to confirm that these plasmids were suitable delivery plasmids, combinations were tested by using pyrF as the target gene as described above. pyrF::gm was cloned into pINC11, and this plasmid was cotransformed with pNUT22 (Table 1). pyrF mutants were again isolated after one round of plating on nonselective agar, and the frequency was 4 to 16% of the colonies on a plate.
Conclusions.
In this work a two-plasmid incompatibility system was developed as a delivery system for mycobacteria. Initially, the loss kinetics of different pairs of incompatible plasmids that had distinct properties, including deletions in the rep genes, different selectable markers, and additional copies of the oriM/inc region, were monitored. Both models examined showed that incompatibility could be used to efficiently eliminate plasmids from cells under nonselective conditions and therefore had the potential for use in a delivery system.
Model B was tested by using the pyrF gene of M. smegmatis, for which a high mutation frequency (10−1 to 10−2) was obtained. This system is not only efficient and simple but rapid, since mutants were isolated during the first round of plating. Thus, M. smegmatis mutants could be obtained in as little as 9 days by using model B once plasmids had been constructed. Other groups have used pyrF in gene replacement experiments with nonreplicating plasmids and have obtained uracil auxotrophs at efficiencies of 40% (18), 1 to 10% (19), and 5% (8). Our results are comparable to these results; our average frequency was 10%, and the frequency was 64% on one occasion.
Although only used here for allelic replacement, this delivery system should also be suitable for transposon mutagenesis. Plasmid incompatibility has been used to deliver a transposon on a replicating vector in Erwinia herbicola. The incompatibility pressure converted a stable transposon-carrying vector into a suicide vector after introduction of an incompatible plasmid (26). In Pasteurella haemolytica, gene replacement was forced through extended growth of cells when a pair of incompatible plasmids was made to coexist (3). A negative selection strategy had to be used in the last stage to exclude plasmids, as transformants mainly contained two plasmids. Strong incompatibility between plasmids was also used to deliver a stable replicative plasmid into the chromosome of Corynebacterium glutamicum via homologous recombination, although the plasmid remained in the chromosome (9).
The approach taken in this work not only should contribute to the tools used for mycobacterial genetics since it provides a simple and efficient delivery system but also should be useful for other organisms for which plasmids have been characterized and transformation techniques have been described. As more information concerning other mycobacterial plasmids becomes available, this principle could be used in other mycobacterial species in which pAL5000 is unable to replicate, such as the members of the Mycobacterium avium-Mycobacterium intracellulare-Mycobacterium scrofulaceum complex.
Acknowledgments
C.A.P. and T.P. were supported by the GlaxoSmithKline Action TB initiative.
We thank Pelle Stolt for donation of plasmid vectors.
REFERENCES
- 1.Bardarov, S., J. Kriakov, C. Carriere, S. Yu, C. Vaamonde, R. A. McAdam, B. R. Bloom, G. F. Hatfull, and W. R. Jacobs, Jr. 1997. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10961-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belisle, J. T., and M. G. Sonnenberg. 1998. Isolation of genomic DNA from mycobacteria. Methods Mol. Biol. 101:31-44. [DOI] [PubMed] [Google Scholar]
- 3.Fedorova, N. D., and S. K. Highlander. 1997. Generation of targeted nonpolar gene insertions and operon fusions in Pasteurella haemolytica and creation of a strain that produces and secretes inactive leukotoxin. Infect. Immun. 65:2593-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gavigan, J. A., C. Guilhot, B. Gicquel, and C. Martin. 1995. Use of conjugative and thermosensitive cloning vectors for transposon delivery to Mycobacterium smegmatis. FEMS Microbiol. Lett. 127:35-39. [DOI] [PubMed] [Google Scholar]
- 5.Geissler, S., and M. Drummond. 1993. A counterselectable pACYC184-based lacZ alpha-complementing plasmid vector with novel multiple cloning sites; construction of chromosomal deletions in Klebsiella pneumoniae. Gene 136:253-255. [DOI] [PubMed] [Google Scholar]
- 6.Guilhot, C., B. Gicquel, and C. Martin. 1992. Temperature-sensitive mutants of the Mycobacterium plasmid pAL5000. FEMS Microbiol. Lett. 77:181-186. [DOI] [PubMed] [Google Scholar]
- 7.Hatfull, G. F. 1996. The molecular genetics of Mycobacterium tuberculosis. Curr. Top. Microbiol. Immunol. 215:29-47. [DOI] [PubMed] [Google Scholar]
- 8.Husson, R. N., B. E. James, and R. A. Young. 1990. Gene replacement and expression of foreign DNA in mycobacteria. J. Bacteriol. 172:519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda, M., and R. Katsumata. 1998. A novel system with positive selection for the chromosomal integration of replicative plasmid DNA in Corynebacterium glutamicum. Microbiology 144:1863-1868. [DOI] [PubMed] [Google Scholar]
- 10.Martin, C., J. Timm, J. Rauzier, R. Gomez Lus, J. Davies, and B. Gicquel. 1990. Transposition of an antibiotic resistance element in mycobacteria. Nature 345:739-743. [DOI] [PubMed] [Google Scholar]
- 11.McFadden, J. 1996. Recombination in mycobacteria. Mol. Microbiol. 21:205-211. [DOI] [PubMed] [Google Scholar]
- 12.Nordstrom, K., and S. J. Austin. 1989. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 23:37-69. [DOI] [PubMed] [Google Scholar]
- 13.Novick, R. P. 1987. Plasmid incompatibility. Microbiol. Rev. 51:381-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parish, T., B. G. Gordhan, R. A. McAdam, K. Duncan, V. Mizrahi, and N. G. Stoker. 1999. Production of mutants in amino acid biosynthesis genes of Mycobacterium tuberculosis by homologous recombination. Microbiology 145:3497-3503. [DOI] [PubMed] [Google Scholar]
- 15.Parish, T., and N. G. Stoker. 1998. Electroporation of mycobacteria. Methods Mol. Biol. 101:129-144. [DOI] [PubMed]
- 16.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969-1975. [DOI] [PubMed] [Google Scholar]
- 17.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. Jacobs, Jr., B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol. Microbiol. 20:919-925. [DOI] [PubMed] [Google Scholar]
- 19.Sander, P., A. Meier, and E. C. Bottger. 1995. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol. Microbiol. 16:991-1000. [DOI] [PubMed] [Google Scholar]
- 20.Santos, A. R., A. B. De Miranda, L. M. Lima, P. N. Suffys, and W. M. Degrave. 1992. Method for high yield preparation in large and small scale of nucleic acids from mycobacteria. J. Microbiol. Methods 15:83-94.
- 21.Smeulders, M. J., J. Keer, R. A. Speight, and H. D. Williams. 1999. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181:270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 23.Stolt, P., and N. G. Stoker. 1996. Functional definition of regions necessary for replication and incompatibility in the Mycobacterium fortuitum plasmid pAL5000. Microbiology 142:2795-2802. [DOI] [PubMed] [Google Scholar]
- 24.Stolt, P., and N. G. Stoker. 1996. Protein-DNA interactions in the ori region of the Mycobacterium fortuitum plasmid pAL5000. J. Bacteriol. 178:6693-6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thatcher, J. W., J. M. Shaw, and W. J. Dickinson. 1998. Marginal fitness contributions of nonessential genes in yeast. Proc. Natl. Acad. Sci. USA 95:253-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vakhlu, J., S. Johri, V. Verma, and G. N. Quazi. 1998. Strategy based on plasmid incompatibility for TN5 mutagenesis in Erwinia herbicola ATCC 21998. Curr. Sci. 74:627-630.