Abstract
We characterized the effect of deletion of the Trichoderma reesei (Hypocrea jecorina) ace1 gene encoding the novel cellulase regulator ACEI that was isolated based on its ability to bind to and activate in vivo in Saccharomyces cerevisiae the promoter of the main cellulase gene, cbh1. Deletion of ace1 resulted in an increase in the expression of all the main cellulase genes and two xylanase genes in sophorose- and cellulose-induced cultures, indicating that ACEI acts as a repressor of cellulase and xylanase expression. Growth of the strain with a deletion of the ace1 gene on different carbon sources was analyzed. On cellulose-based medium, on which cellulases are needed for growth, the Δace1 strain grew better than the host strain due to the increased cellulase production. On culture media containing sorbitol as the sole carbon source, the growth of the strain with a deletion of the ace1 gene was severely impaired, suggesting that ACEI regulates expression of other genes in addition to cellulase and xylanase genes. A strain with a deletion of the ace1 gene and with a deletion of the ace2 gene coding for the cellulase and xylanase activator ACEII expressed cellulases and xylanases similar to the Δace1 strain, indicating that yet another activator regulating cellulase and xylanase promoters was present.
In filamentous fungi production of the cellulose- and hemicellulose-degrading enzymes, the cellulases and hemicellulases, is controlled at the transcriptional level by the available carbon source. One of the best-studied cellulolytic systems is that of the saprophytic fungus Trichoderma reesei (Hypocrea jecorina). Cellulase genes of T. reesei are repressed in the presence of glucose by the wide-domain carbon catabolite repressor CREI (12, 19, 28, 29), which together with CREA of the aspergilli is the only known repressor of cellulase and hemicellulase genes (for a recent review see reference 4). Cellulase genes are induced in the presence of cellulose or its derivatives or by addition of the disaccharide sophorose to a medium containing a neutral carbon source, such as glycerol (for a review see reference 15). In addition, the monosaccharide l-sorbose was recently reported to induce cellulase genes of T. reesei (22). Most of the hemicellulase genes are also controlled by CREI and induced in the presence of the cellulase inducers cellulose and sophorose (20). Xylobiose, arabitol, and different xylans also induce the expression of many of the hemicellulase genes (20).
Even though the carbon source-dependent expression of cellulase genes has been studied in detail, our knowledge about the molecular mechanisms regulating the cellulase genes in filamentous fungi is rather fragmentary. In addition to CREI/CREA, ACEII in T. reesei and XlnR in Aspergillus niger are known to regulate fungal cellulase promoters. Several genes encoding cellulases and hemicellulases are positively regulated by XlnR, a factor that binds in vitro to a 5′GGCTAA site (8, 35, 36). In T. reesei ACEII binds in vitro to a site containing the 5′GGCTAATAA sequence in the cbh1 promoter and regulates positively all of the main cellulase genes (cbh1, cbh2, egl1, and egl2) and the xylanase gene (xyn2) in cellulose-induced cultures (1). However, deletion of ace2 resulted only in decreased cellulase and xylanase expression, not in complete loss of expression, suggesting that additional activators for these genes exist. In addition, the activity of the cbh2 promoter has been shown to be dependent on the Hap2/3/5 protein complex binding to the CCAAT box, one of the most common elements in eukaryotic promoters (40, 41). Cell-free lysates prepared from T. reesei also bind to CCAAT sequences in the promoters of the xyn1 and xyn2 xylanase genes, suggesting that the Hap2/3/5 complex regulates xylanase promoters as well (42). Furthermore, the Aspergillus nidulans xylanase genes xlnA and xlnB are differentially controlled with respect to ambient pH by the transcription factor PacC, which activates the expression of alkaline-expressed genes and represses the expression of acid-expressed genes (17).
T. reesei ACEI is a factor that was isolated in a yeast-based screening analysis that selected for factors binding to and activating the T. reesei cbh1 promoter in yeast (26), a screening analysis that also resulted in cloning of the ace2 gene (1). ACEI contains three Cys2His2-type zinc fingers and was shown to bind in vitro to eight sites containing the core sequence 5′AGGCA scattered along the 1.15-kb cbh1 promoter. Deletion of the ace1 gene in T. reesei led to reduced colony growth on Solka floc cellulose-containing solid medium, on which cellulases are normally expressed. Growth on glucose plates was not notably affected. ACEI was suggested to be a factor specific for filamentous fungi since no significant sequence similarity to ACEI was found in databases or the Saccharomyces cerevisiae genome (26).
In order to expand our understanding of the regulation of cellulase production, we further investigated the role of ACEI in the regulation of T. reesei cellulase and xylanase expression. We analyzed the effect of deletion of ace1 on the expression of all the main cellulase and xylanase genes under different inducing conditions. Our data indicate that ACEI acts in a repressor-like manner rather than as an activator. We also analyzed the effect of simultaneous deletion of both the ace1 and ace2 genes on cellulase expression.
MATERIALS AND METHODS
Strains.
T. reesei strains ALKO2221 (A. Mäntylä, unpublished data), VTT-D-01850 (strain ALKO2221 with a deletion of the ace1 gene) (26), VTT-D-01849 (another transformant with a deletion of the ace1 gene), and VTT-D-99729 (strain ALKO2221 with a deletion of the ace2 gene) (1) and two transformants with deletions of both the ace1 and ace2 genes (this study) were used.
A T. reesei strain with deletions of both the ace1 and ace2 genes was made by transforming the VTT-D-01850 strain with the ace1 deletion (26) and using the ace2 deletion cassette released from pAS40 (1). In pAS40 1.6 kb of 5′ flanking and 2.2 kb of 3′ flanking sequences of the ace2 gene are cloned at the 3′ and 5′ ends of the hygromycin resistance cassette derived from the selection plasmid pRLMEX30 (18). Transformation was performed as described by Penttilä et al. (23). Southern analysis was used to verify that the transformants contained only one copy of the hygromycin gene and that this copy replaced the coding sequence of the ace2 gene. Two transformants, VTT-D-01851 and VTT-D-01852 (Δace1 Δace2 strain), were studied.
Media and culture conditions.
For RNA isolation and biomass determination all strains were grown in the minimal medium without peptone described by Ilmén et al. (11) supplemented with 2% glycerol, glucose, sorbitol, fructose, or Solka floc cellulose (James River Corp.) as the carbon source; 1 mM α-sophorose (Serva) was added to the glycerol medium to induce cellulase expression. Four hundred milliliters of each growth medium in a 2-liter shake flask was inoculated with 8 × 107 spores of ALKO2221 or a Δace1 or Δace1 Δace2 strain and incubated in a rotary shaker at 250 rpm at 28°C. To analyze the effect of Triton X-100 on the growth of the different strains on Solka floc cellulose, 0.1% Triton X-100 was added to the cellulose media.
To study the growth of the Δace1 and Δace1 Δace2 strains, three parallel shake flasks containing the strains with deletions (VTT-D-01850, VTT-D-01849, VTT-D-01851, VTT-D-01852) and four parallel flasks containing host strain ALKO2221 were inoculated, and the dry weights of the mycelia and pH values of the media were measured for 5 (glucose) or 6 days. The dry weights of the mycelia harvested on filter paper were measured after the preparations were dried at 100°C for 24 h. In Solka floc cellulose medium the growth of a strain was monitored by measuring the amount of the NaOH-extractable protein in the insoluble portion of culture fluid that was washed once with 0.7 NaCl and twice with tap water. NaOH extraction was performed as described by Jayaraman (13), except that the samples were incubated for 24 h at room temperature instead of heated at 90°C. Total protein was measured as described by Lowry et al. (16).
To study the induction of the cellulase genes by cellulose, the Δace1 (VTT-D-01850 and VTT-D-01849), Δace1 Δace2 (VTT-D-01851 and VTT-D-01852), and ALKO2221 strains were grown in shake flasks in triplicate on glycerol medium for 72 h (in the experiment carried out with the Δace1 strain) or for 58 h (in the experiment carried out with the Δace1 Δace2 strain), after which the mycelia from triplicate flasks of each strain were pooled and collected by filtration through Miracloth and transferred into two shake flasks containing cellulose as the sole carbon source and into one shake flask containing glycerol medium as a control. The biomass of the glycerol-grown mycelia was determined to verify that the same amount of mycelia (±6%) from each strain in the experiment was transferred into the cellulose- and glycerol-containing shake flasks. For sophorose induction studies strains were grown on glycerol in two parallel flasks for 72 h, after which 1 mM sophorose was added, and mycelial samples were collected 1, 2, 3, and 6 h after sophorose addition for mRNA analysis.
Northern analysis of cellulase expression.
Total RNA was isolated with the Trizol reagent (Life Technologies, Inc.). The probes used for Northern analysis were the entire cDNAs of cbh1, cbh2 (24), egl1 (25), egl2 (previously called egl3) (27), and ace1 (26) released from vector sequences. The probes for the β-xylanase genes xyn1 and xyn2 were 350-bp fragments prepared by a PCR (20). As an internal loading control the membranes were hybridized with either an actin-encoding (act1) or glyceraldehyde-3-phosphate dehydrogenase-encoding (gpd1) cDNA fragment. The probes were labeled by using a random primed DNA labeling kit (Roche Molecular Biochemicals) and [α-32P]dCTP (Amersham Pharmacia Biotech). Hybridization signals were detected on phosphor screen autoradiographs by using Phosphorimager SI, were quantified by using the ImageQuant software (Molecular Dynamics), and were normalized for the total amount of mRNA loaded by using gpd1 or the actin mRNA as a loading control. The signal intensities of the different blots cannot be compared to each other due to the different specific activities of the probes and the different exposure times used for the various blots. The results of Northern analyses shown in Fig. 2A, 3A, and 4A are the results of exposure to film.
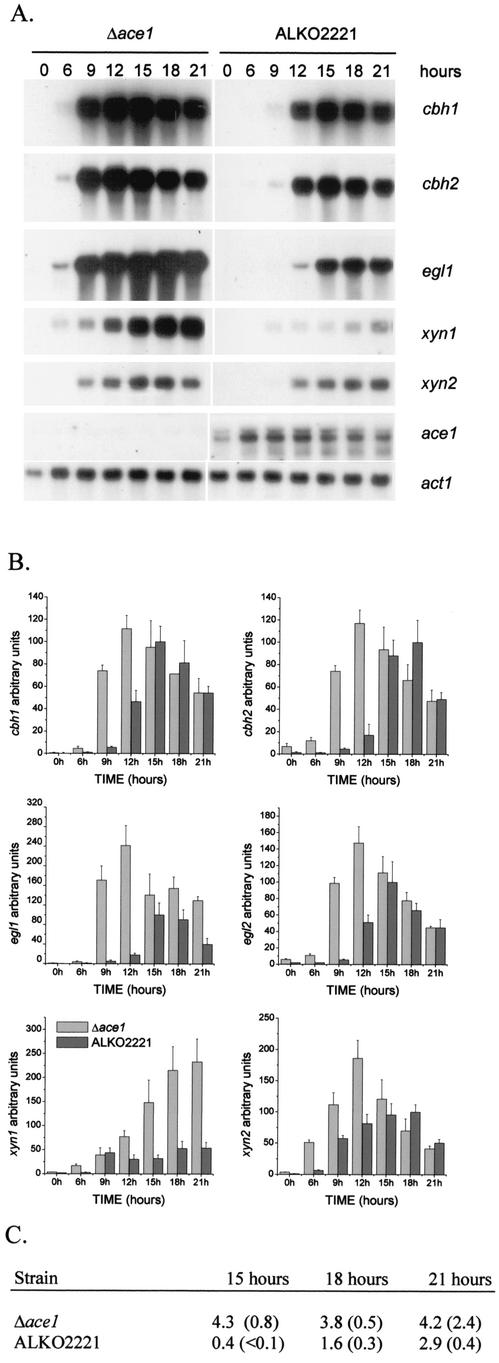
FIG.2.
Effect of disruption of ace1 on expression of the main cellulases, xylanases, and ace1 in cellulose-based cultures. (A) Northern blot analysis of cbh1, cbh2, egl1, xyn1, xyn2, ace1, and act1 (control) mRNAs at different times in Δace1 (VTT-D-01850) and ALKO2221 strains. The probes are indicated on the right. (B) Quantification of the cellulase and xylanase signals normalized with the actin mRNA from two parallel cultures of Δace1 strains VTT-D-01850 and VTT-D-01849 and host strain ALKO2221. The x axis indicates the time after the mycelia were transferred to the cellulose medium. The bars indicate the means of the mRNA signals for two parallel shake flasks. The results are expressed relative to the highest value of the ALKO2221 signal, which was defined as 100. The error bars indicate standard deviations. (C) Cellulase activities (in nanokatals per milliliter; 1 nkat = 1 mmol of methylumbelliferyl released from MUL) in cellulose culture supernatants of the Δace1 strains VTT-D-01849 and VTT-D-01850 and host strain ALKO2221 15, 18, and 21 h after transfer into cellulose-containing media. The values in parentheses are standard deviations.
Enzyme activity assays.
CBHI and EGI activities in the culture supernatants were measured by using 4-methylumbelliferyl-β-d-lactoside (MUL) (Sigma) as the substrate as described by van Tilbeurgh et al. (37); 0.17 mM MUL and 10 min of incubation at pH 5.0 and 50°C were used.
RESULTS
Growth of the Δace1 strain on different carbon sources.
Deletion of the ace1 gene has been shown to result in retardation of the radial growth of colonies on Solka floc and Avicel cellulose-containing plates (26). Repeated plating of the Δace1 strain and host strain ALKO2221 as single-spore colonies on Avicel and Solka floc cellulose plates showed that the diameter of the Δace1 colonies was on average 40% of the diameter of the host strain ALKO2221 colonies (data not shown). A logical explanation for this is that there was reduced cellulase production in the strain with the deletion of the ace1 gene.
In order to study further the role of ACEI in carbon utilization and cellulase production, the Δace1 strain and the Δace1 Δace2 strain (see below) were cultivated together with host strain ALKO2221 in liquid minimal media containing different carbohydrates, glycerol, sorbitol, glucose, and Solka floc cellulose, as the sole carbon sources. The first two of these compounds are neutral carbon sources with respect to cellulase gene expression. Glucose is a repressing carbon source that causes repression of cellulase promoters by CREI (12, 30). Cellulose is an inducing carbon source on which high levels of cellulases are produced. The growth of the strains was monitored by determining biomass and measuring the pH of the culture medium, a common measure of growth in filamentous fungi. The total-protein amounts extracted from cellulose cultures by 1 M NaOH were measured to determine the rates of biomass accumulation of the different strains. Figure 1 shows that reductions in the pH values of the culture media correlated well with the rates of accumulation of fungal biomass.
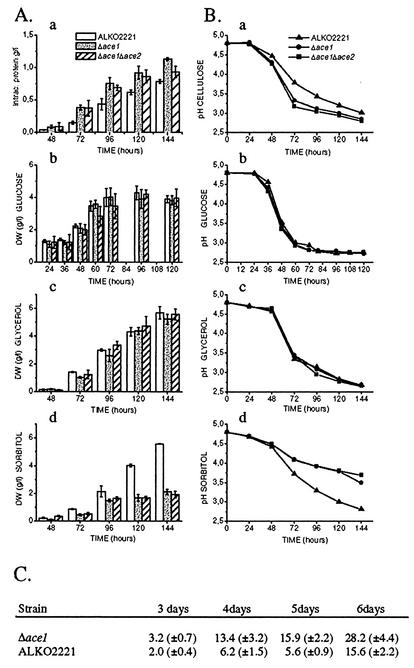
FIG. 1.
(A and B) Growth of the ALKO2221, Δace1 (VTT-D-01850 and VTT-D-01849), and Δace1 Δace2 (VTT-D-01851 and VTT-D-01852) strains on cellulose (panels a), glucose (panels b), glycerol (panels c), and sorbitol (panels d). (A) Mycelial biomasses or total amounts of intracellular protein of the ALKO2221, Δace1, and Δace1 Δace2 strains at different times. The error bars indicate the standard deviations of the mean dry weights (DW) and amounts of total protein. (B) pH values of the culture media of the ALKO2221, Δace1, and Δace1 Δace2 strains. (C) Cellulase activities (in nanokatals per milliliter; 1 nkat =1 mmol of methylumbelliferyl released from MUL) in cellulose culture supernatants of Δace1 strains VTT-D-01849 and VTT-D-01850 and host strain ALKO2221 after 3, 4, 5, and 6 days of growth. The values are means based on samples taken from four parallel shake flasks per strain. The values in parentheses are standard deviations.
In liquid culture media containing Solka floc cellulose as the sole carbon source the Δace1 strain had a higher growth rate than host strain ALKO2221 (Fig. 1). This was somewhat unexpected since growth of the colonies of the Δace1 strain was reduced on cellulose plates. The media on the cellulose plates and the liquid media used differed only with respect to agar, peptone (which is needed for growth on cellulose plates), and Triton X-100 (a detergent used to restrict the colony size on plates). We repeated the liquid cellulose cultivation experiment with addition of 0.1% Triton X-100, and the growth of all strains was similar to the growth of cultures without Triton X-100 (data not shown), suggesting that peptone might affect the growth of the Δace1 strain on cellulose plates. On glycerol and glucose media the growth of the Δace1 strain was similar to that of the host (Fig. 1, panels b and c). However, in sorbitol medium the growth of the Δace1 strain was clearly retarded. During 6 days of growth on sorbitol, the strain with the deletion accumulated only one-half the biomass accumulated by the host strain (Fig. 1d). Because sorbitol may be converted into fructose by the l-iditol 2-dehydrogenase or by sorbitol dehydrogenase (2), we also cultivated the strains on 2% fructose. Growth of the Δace1 strains on fructose was normal (data not shown).
The amounts of cellulase activity produced by the Δace1 and host strains were measured from culture supernatants obtained after 3, 4, 5, and 6 days of growth on Solka floc cellulose-containing medium; MUL was used as the substrate (Fig. 1C). The values resulting from MUL activity measurements reflect mainly the amounts of CBHI and EGI produced by the fungus. The strains with a deletion of the ace1 gene produced more cellobiohydrolase and endoglucanase activities than the host produced. Throughout cultivation the difference in the amounts of MUL activity produced was around twofold, and it was greatest at 5 days. Taken together, the results showed that it is likely that the better growth of the Δace1 strain on cellulose medium resulted primarily from the increased amounts of cellulases produced.
Effect of disruption of the ace1 gene on cellulase gene expression.
To study whether cellulase expression was indeed elevated in the Δace1 strains and what the role of ACEI in cellulase expression is, the levels of the mRNAs of the individual cellulase genes in the Δace1 strains and the host strain were measured under different inducing conditions. First, the Δace1 strains (transformants VTT-D-01849 and VTT-D-01850) and the control strain were grown on glycerol to obtain biomass. Then cellulase expression was induced by transferring equal amounts of the glycerol-grown mycelia of the Δace1 strains and the control strain into a culture medium containing cellulose as the sole carbon source or into glycerol medium, used as a control. The induction of the cellulase genes was monitored by performing Northern analyses of samples collected 6, 9, 12, 15, 18, and 21 h after the transfer. Expression of the main cellulase genes (cbh1, cbh2, egl1, and egl2) was induced earlier and to a higher level in the Δace1 strains than in the host (Fig. 2). Transfer of the mycelia into the glycerol medium did not induce cellulase expression (data not shown). In the Δace1 strains all the cellulases were expressed at a high level as early as 9 h after the transfer into the inducing culture medium, whereas in the host strain high expression was seen only after 15 h. The difference in cellulase transcript levels was generally more profound at the early time points, whereas at the later times the cbh1, cbh2, and egl2 mRNA levels were almost the same in the Δace1 and ALKO2221 strains. Nine hours after transfer of the mycelia to the cellulose media, the Δace1 strains expressed on average 13 times more cbh1 mRNA, 16 times more cbh2 mRNA, 26 times more egl1 mRNA, and 16 times more egll2 mRNA than host strain ALKO2221. Twelve hours after transfer the signals of the main cellulase mRNAs in the Δace1 strains were on average 2.4, 6.8, 11, and 2.8 times higher for the cbh1, cbh2, egl1, and egl2 mRNAs, respectively (Fig. 2B). Enzyme activities were determined for the culture supernatants taken 15, 18, and 21 h after transfer of the mycelia to the cellulose media. In accordance with the increased expression of the cellulase genes, clearly more cellulase activities were detected in the culture media of the Δace1 strains than in the culture media of the host strain (Fig. 2C).
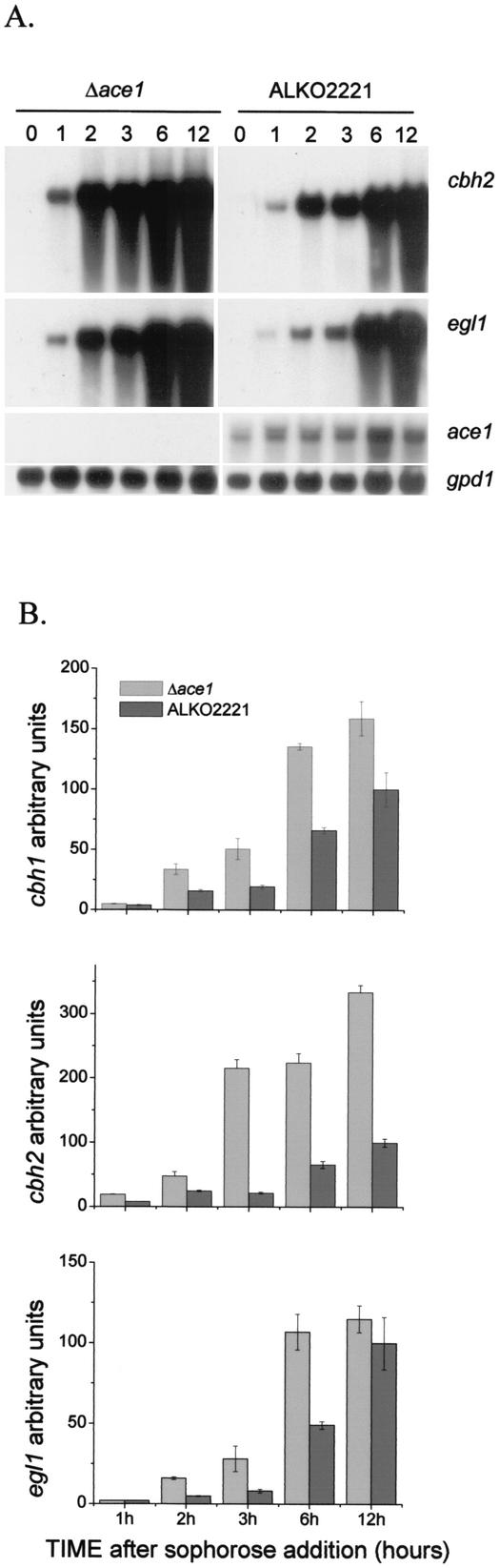
Since deletion of the cellulase regulator gene ace2 has previously been found to affect only cellulose-mediated induction of cellulase expression (1), it is possible that different mechanisms function in cellulase gene induction depending on the inducer. Thus, we were interested in determining whether ace1 also has a role in the induction caused by the known strong and rapid cellulase inducer sophorose. The Δace1 strains (transformants VTT-D-01849 and VTT-D-01850) and host strain ALKO2221 were grown in glycerol medium for 3 days, after which cellulase expression was induced by addition of a small amount of sophorose. The levels of the mRNAs of the three main cellulase genes, cbh1, cbh2, and egl1, were measured 1, 2, 3, 6, and 12 h after sophorose addition. The genes were induced earlier and to higher levels in the strain with a deletion of the ace1 gene than in host strain ALKO2221 (Fig. 3A; data not shown for cbh1). Quantification of the cbh1, cbh2, and egl1 mRNAs showed that a Δace1 strain expressed on average two times more cbh1, three times more cbh2, and two times more egl1 at the different times analyzed than the ALKO2221 strain expressed (Fig. 3B).
FIG. 3.
Effect of ace1 gene deletion on sophorose induction of cellulase genes. (A) Northern analysis of cbh2, egl1, cre1, ace1, and gpd1 (control) mRNAs at 0, 1, 2, 3, 6, and 12 h after sophorose addition for a Δace1 strain (VTT-D-01850) and strain ALKO2221. The probes are indicated on the right. (B) Quantification of cbh1, cbh2, and egl1 signals normalized with the gpd1 mRNA. The x axis indicates thetime after sophorose addition. The bars indicate the means for two parallel cultures. The results are expressed relative to the highest value of the ALKO2221 mRNA signal, which was defined as 100. The error bars indicate standard deviations.
Construction of a strain with deletions of both the ace1 and ace2 genes.
Transcriptional activity often results from the synergistic action of several factors, and thus the removal of one factor does not necessarily lead to dramatic changes in the transcriptional activity, as previously seen after deletion of the ace2 gene (1). Therefore, we wanted to study the effect of simultaneous deletion of the ace1 and ace2 genes. A strain with deletions of the two factors was constructed by deleting the ace2 gene from Δace1 strain VTT-D-01850 (26). In the ace2 deletion cassette the whole protein-encoding region of the ace2 gene was removed and replaced with an expression cassette conferring hygromycin resistance. The transformants were screened by Southern hybridization for single copies of the transformed DNA and correct replacement of the ace2 gene (data not shown). Two transformants, VTT-D-01851 and VTT-D-01852, were used for further studies (referred to below as the Δace1 Δace2 strain).
Growth of the Δace1 Δace2 strain on different carbon sources.
A strain with a deletion of only the ace2 gene has a slightly lower growth rate in liquid cellulose medium than host strain ALKO2221 due to delayed induction and reduced expression of cellulase genes. However, the Δace2 strain grows normally on glucose and glycerol (1). The Δace1 Δace2 strains (transformants VTT-D-01851 and VTT-D-01852) were cultivated together with the Δace1 and ALKO2221 strains on different carbon sources. As Fig. 1 shows, the Δace1 Δace2 strain grew like the strain with a deletion of only the ace1 gene on all of the carbon sources. Similar growth also occurred in a culture containing fructose as the sole carbon source (data not shown). On sorbitol the poor growth of the Δace1 Δace2 strain resulted from deletion of the ace1 gene since the strain with a deletion of only the ace2 gene grew as well as the control strain on sorbitol (data not shown).
Expression of the cellulase genes in the Δace1 Δace2 strain.
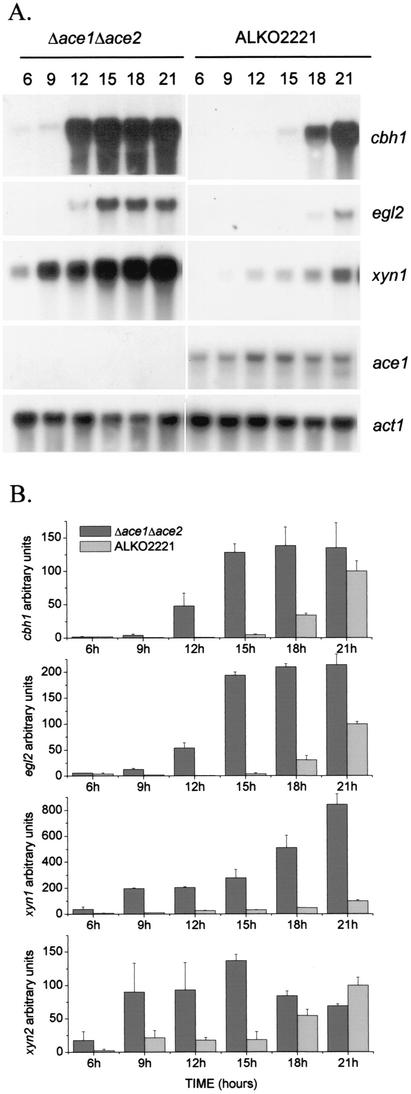
In order to study the combined effect of the ace1 and ace2 deletions on cellulase gene expression, similar cellulose- and sophorose-induced cultures of the Δace1 Δace2 strain (transformants VTT-D-01851 and VTT-D-01852), as were carried out for the Δace1 strain, were grown. The Δace1 Δace2 strains and host strain ALKO2221 were cultivated in glycerol medium, after which the mycelia were collected and transferred into Solka floc cellulose media. The strain with both deletions behaved like the Δace1 strain. cbh1 and egl2 were induced earlier and to higher levels in the Δace1 Δace2 strains than in the host strain (Fig. 4). Figure 4B shows the results of quantification of the cbh1 and egl2 signals at different times after transfer into the cellulose media. These results show that the ace2 deletion in the strain with a deletion of the ace1 gene did not cause a reduction in cellulase expression like that seen in the strain with a deletion of only the ace2 gene (1); instead, an increase in cellulase expression was seen.
FIG. 4.
Induction of cellulase and xylanase genes and expression of ace1 in the Δace1 Δace2 strain on cellulose. (A) Northern blot analysis of cbh1, egl2, xyn1, ace1, and actin (control) mRNAs at 6, 9, 12, 15, 18, and 21 h in the Δace1 Δace2 strain (VTT-D-01851) and host strain ALKO2221. The probes are indicated on the right. (B) Quantification of the cbh1, egl2, xyn1, and xyn2 signals normalized with the actin mRNA from two parallel cultures of each Δace1 Δace2 strain (VTT-D-01851 and VTT-D-01852) and three parallel cultures of host strain ALKO2221. The x axis indicates the time after the mycelia were transferred to the cellulose medium. The bars indicate the means of the mRNA signals. The results are expressed relative to the highest value of the ALKO2221 signal, which was defined as 100. The error bars indicate standard deviations.
In the sophorose induction experiment, the cbh1, cbh2, and egl1 genes were induced to higher levels in the Δace1 Δace2 strain than in host strain ALKO2221 (data not shown). The difference in the amounts of cellulase mRNA in the Δace1 Δace2 strain and the host was similar to the difference observed for the Δace1 strain and the host, as shown in Fig. 3.
Expression of xylanase-encoding genes xyn1 and xyn2 in the Δace1 and Δace1 Δace2 strains.
It has been shown that glucose repression, mediated by CREI, and the regulatory mechanisms involving T. reesei ACEII and A. niger XlnR are at least partially shared by cellulase and hemicellulase genes (1, 8, 20, 35). Therefore, we studied whether the ace1 deletion affects the expression of the xyn1 and xyn2 genes encoding the two main endo-β-xylanases. The same filters that were analyzed for cellulase expression in the cellulose transfer assay described above were hybridized with the xyn1- and xyn2-specific probes. In both the Δace1 and Δace1 Δace2 strains the xyn1 gene was expressed at a higher level than it was expressed in the ALKO2221 strain at all times analyzed after transfer to cellulose medium (Fig. 2A and 4A). The amount of the xyn1 mRNA was up to six times higher in the Δace1 strains than in host strain ALKO2221 at all other times except 9 h after the transfer (Fig. 2). The same was true for the strains with both deletions, and the differences were 4- to 19-fold (Fig. 4). Also, the xyn2 gene was induced to higher levels in the Δace1 and Δace1 Δace2 strains than in host strain ALKO2221. In the Δace1 strains the level of the xyn2 mRNA was 1.3 to 7.8 times higher than the level in host strain ALKO2221 at the early time points (from 6 to 15 h). At the late time points (18 and 21 h), xyn2 was expressed similarly in the Δace1 strain and the host strain (Fig. 2B). In the Δace1 Δace2 strain, expression of the xyn2 gene was 4.2 to 7.3 times greater at the early time points compared to that of the host strain (Fig. 4B).
Expression of the ace1 gene.
To examine if the transcription of ace1 is subject to carbon source-dependent control, total RNA was isolated from mycelia grown on glucose, glycerol, and cellulose media and analyzed for ace1 expression by Northern analysis. Host strain ALKO2221 produced two major ace1 transcripts (3.2 and 3.0 kb) that were detected under all growth conditions studied at approximately the same relative amounts (data not shown for glucose and sorbitol). The expression of ace1 differed only slightly at the different times after sophorose addition (Fig. 3A) and increased slightly after transfer from glycerol medium to cellulose medium (Fig. 2A and 4A). We also analyzed the expression of ace1 in a strain with a deletion of the ace2 gene (1) in similar experiments, as shown in Fig. 2 and 4, and expression of ace1 was similar to expression in the host strain, suggesting that ACEII is not involved in the regulation of ace1 expression. Furthermore, a 1,000-bp ace1 promoter contains 11 sites fitting the CREI/CreA binding consensus sequence (5′SYGGRG). We studied whether ace1 is under the control of CREI by performing a Northern analysis of the ace1 expression in the Rut-C30 strain expressing a truncated mutant form of the cre1 gene and the Rut-C30 strain transformed with the full-length cre1 gene (12). ace1 mRNA analysis of cultures grown on glucose (1 and 2 days), sorbitol, glycerol, and cellulose (3 and 6 days) showed that ace1 expression was similar in the two strains under the growth conditions studied, suggesting that expression of ace1 is not subject to glucose repression and is not affected by CREI (data not shown).
Sequence comparisons of the ACEI protein.
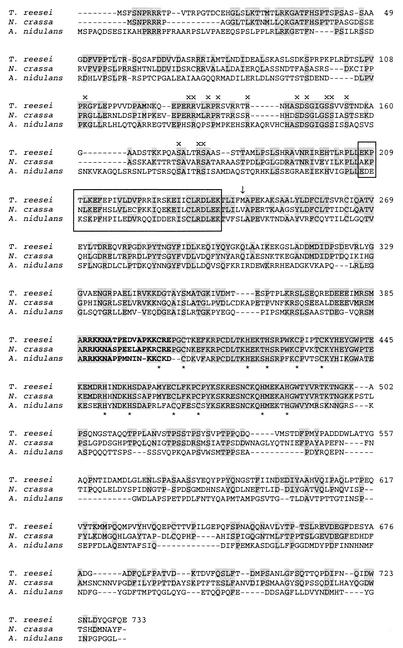
We have previously reported the presence of sequences resembling the amino acid sequence of ACEI in the A. nidulans and Neurospora crassa expressed sequence tag databases (26). More recent sequence similarity searches performed with the BLAST program resulted in identification of a novel A. nidulans gene, stzA (accession number AF202995), whose product has 58% overall amino acid similarity to the ACEI protein. The database information indicated that stzA is a gene that encodes a C2H2 zinc finger protein that alleviates sensitivity to salt and DNA-damaging agents. Furthermore, the sequences in the recently released N. crassa genome database (2nd release; Neurospora Sequencing Project, Whitehead Institute/MIT Center for Genome Research [www-genome.wi.mit.edu]) contain a sequence coding for a polypeptide that has 48% identity and 75% overall similarity with the ACEI protein. The zinc finger regions of the Neurospora and Aspergillus proteins exhibit 93 and 69% identity with the zinc finger region of ACEI, respectively, suggesting that at least the predicted Neurospora protein binds to similar sequences. Comparison of these three proteins allowed us to identify possible meaningful amino acid sequences in addition to the zinc fingers within the ACEI protein. The region encompassing the putative nuclear localization signal, amino acids 387 to 403 in ACEI (Fig. 5), is nearly identical in the three proteins. In addition, the N-terminal parts contain two very similar stretches of amino acids, a region rich in arginine and serine residues and a region where basic and acidic amino acids alternate (Fig. 5). Meaningful sequence similarities of these regions to regions in other proteins are difficult to identify from databases due to the short lengths of the amino acid stretches.
FIG. 5.
Alignment of ACEI (T. reesei), the protein encoded by the stzA gene (StzA) (A. nidulans), and the protein deduced from the N. crassa genome database (N. crassa). Amino acids identical to T. reesei ACEI amino acids are indicated by a grey background. The predicted bipartite nuclear targeting signal is indicated by boldface type, and the zinc-coordinating Cys and His residues are indicated by asterisks below the sequence. The first methionine in the original ace1 clone recovered in the yeast screening analysis and sufficient for activation in yeast (26) is indicated by an arrow. The conserved regions in the three proteins are indicated by x and by boxes. The alignment was constructed by using the ClustalW (1.74) software. Two putative intron sequences that were located at the same positions as the known introns in ace1 were removed from the Neurospora sequence prior to translation on the basis of the general consensus splicing signals suggested for filamentous fungi (32).
DISCUSSION
In fungi, the production of cellulolytic enzymes is subject to transcriptional regulation by the available carbon source. So far, the known cellulase regulators include the CREI/CreA carbon catabolite repressors from T. reesei and aspergilli and the activators ACEII from T. reesei and XlnR from A. niger. We have previously reported cloning of the transcription factor ACEI that binds to and activates the promoter of the main cellulase gene (cbh1) of T. reesei in S. cerevisiae. ACEI bound in vitro to eight sites in the cbh1 promoter, and deletion of the ace1 gene led to reduced colony growth on cellulose-containing plates (26), suggesting that ACEI regulates cellulase gene expression in T. reesei.
Deletion of the ace1 gene clearly affected the growth of T. reesei in cultures containing cellulose as the sole carbon source. The growth of the Δace1 strain was enhanced compared to the growth of the host strain, suggesting that the Δace1 strain was able to degrade cellulose more efficiently than the host strain. This observation was in accordance with the Northern analysis of the cellulase gene expression in cellulose-induced cultures, which showed that transcription of all the major cellulase genes, cbh1, cbh2, egl1, and egl2, was induced earlier and to a higher level in the Δace1 strain than in the host strain (Fig. 2). The increases in the amounts of the individual cellulase mRNAs varied from 2- to 30-fold at the early time points (6 to 12 h) after the transfer to cellulose media. Furthermore, the enzyme activity measurements obtained from the culture supernatants of the cellulose cultures showed that more cellulases accumulated in the culture supernatant of the Δace1 strain, both during direct cultivation on cellulose (Fig. 1C) and during the transfer experiments in which cellulase expression was induced by transferring mycelia pregrown on glycerol to cellulose media (Fig. 2C).
Cellulose induction and sophorose induction of cellulase genes could be mediated by the same mechanism since cellobiose, a disaccharide released from cellulose, can be converted to sophorose by a β-glucosidase (33). However, our recent results indicated that deletion of the ace2 gene encoding the activator ACEII affected the induction mediated by cellulose but not the induction mediated by sophorose, thus suggesting that at least partially different mechanisms operate in cellulose-mediated induction and sophorose-mediated induction (1). ACEI, however, seems to function in both. The transcription of the cellulase genes was enhanced in the Δace1 strain both in the cellulose-based cultures and when the genes were induced by sophorose.
Our current knowledge concerning xylanase gene expression suggests that it involves mechanisms that are partially similar to mechanisms involved in cellulase gene induction, while there is also evidence of a separate mechanism (20, 42). Expression of the two xylanase genes (xyn1 and xyn2) increased in the strain with a deletion of the ace1 gene, indicating that ACEI also regulates the xylanase genes (Fig. 2). In this respect ACEI behaves like all the other (hemi)cellulase regulatory factors studied so far, including CREI, ACEII, and XlnR. The data presented in this paper clearly show that although originally isolated based on its ability to activate transcription, ACEI in fact negatively regulates, either directly or indirectly, cellulase and xylanase expression in Trichoderma.
Knowledge concerning the DNA binding repressors of filamentous fungi is limited to a few isolated factors, including the glucose repressor CREI/CreA from various different fungi (5, 6, 12, 28, 30, 31, 38) and NREB involved in nitrogen control in Penicillium chrysogenum (9). Repressors for fungal extracellular enzyme-encoding genes other than CREI/CreA have not been reported previously. Promoter regions reported to mediate repression under inducing conditions have been identified for the promoter of the cgxA gene encoding a xylanase of Chaetomium gracile and for a cutinase gene of Fusarium solani (14, 21). In A. nidulans, creA mutation increased arabinase production during growth on arabitol, suggesting that CreA down regulates arabinase genes also under inducing conditions (34).
The amino acid sequence of ACEI shows similarity to an amino acid sequence (a putative ACEI equivalent) deduced from the N. crassa genome database and to the product of the A. nidulans stzA gene (accession number AF202995) that has been deposited in a database as a C2H2 zinc finger protein that alleviates sensitivity to salt and DNA-damaging agents. The sequence comparison in Fig. 5 shows that in addition to the putative nuclear targeting signal, the N-terminal halves of the proteins contain at least two other highly conserved regions. Based on the similarity of these regions, it can be assumed that they are important for function. ACEI was originally cloned based on its activator function in yeast (26). The cDNA clone recovered, however, was truncated and lacked most of these N-terminal regions (Fig. 5), suggesting that the transcription-regulating function of the truncated form of ACEI might differ from that of the full-length ACEI. It is also possible that ACEI functions in a context-dependent manner and that it may act either as a repressor or as an activator, perhaps depending on the interactions with another factor(s) that influences its function.
Cellulase promoters, especially that of cbh1, are among the strongest eukaryotic promoters known. Certain strains have been reported to produce up to 35 g of extracellular protein per liter, and the major part of this consists of CBHI encoded by a single gene (7). Therefore, it is possible that that there are mechanisms that down regulate the cellulase and xylanase promoters also under inducing conditions in order to maintain the balance between the amount of mRNA transcribed and the protein translation and/or secretion rate. This type of regulation could be mediated by ACEI. Furthermore, it is possible that the strong induction of the cellulase genes is somehow controlled by the amount of glucose (or disaccharides) released from cellulose, although such carbon catabolite repression is likely to be mediated by CREI. No significant amounts of glucose, however, accumulated in the supernatants of the cellulose cultures of the Δace1 and ALKO2221 strains (data not shown).
It is still not known why the ace1 deletion reduced growth on cellulose plates but on the other hand improved growth in liquid cultures containing cellulose as the sole carbon source. Furthermore, our observation that on sorbitol the growth of the Δace1 strain was impaired was unexpected. This suggests that ace1 has a role in the regulation of expression of some other genes in addition to those encoding cellulases and hemicellulases. The activator of the cellulase and xylanase genes, XlnR of A. niger, also regulates the xyrA gene encoding d-xylose reductase, an intracellular enzyme involved in the utilization of xylose as a carbon source (10). So far, the genes involved in sorbitol utilization in filamentous fungi have not been characterized. Sorbitol utilization may be mediated by nonspecific dehydrogenases/reductases, one of which is the xylitol dehydrogenase which in A. niger also uses sorbitol as a substrate (39). It is also possible that deletion of ace1 leads to the accumulation of a harmful metabolite during growth on sorbitol or somehow sensitizes the mycelia to sorbitol, which at high concentrations has been shown to cause an osmotic shock to the cells (3). It remains to be determined at what level disruption of the ace1 gene affects the growth on sorbitol.
In accordance with a more general regulator role, the ace1 gene was found to be transcribed under all the conditions studied in this work, as is the case with the glucose repressor CREI (12). Similarly, expression of ace1 appeared to be similar in a strain expressing a mutant glucose repressor CREI and in a strain with a deletion of the ace2 gene encoding the cellulase activator ACEII (data not shown), suggesting that expression of ace1 is not regulated by these factors. Expression of cre1 encoding the glucose repressor CREI appeared to be similar in the strain with the deletion and the host strain in experiments whose results are shown in Fig. 2 and 3 (data not shown), suggesting that ACEI does not regulate cre1 expression under the conditions studied.
In eukaryotic organisms, appropriate transcriptional regulation often requires the combinatorial and synergistic action of different repressors and activators bound at multiple sites in the promoter. The transcriptional response to these factors can be either graded or binary, only modulating the level of expression or completely turning expression off or on. We analyzed the effect of simultaneous deletion of ace1 and ace2 on the induction of the cellulase and xylanase genes in order to see whether the activator ACEII mediates the increased cellulase and xylanase expression seen in the strain with a deletion of the ace1 gene. Deletion of the ace2 gene in the Δace1 strain allowed expression of cellulases at high levels that were comparable to those seen in the Δace1 strain, indicating that the repressive function of ACEI is dominant and/or that the increase in cellulase and xylanase expression seen in the Δace1 strain is mediated not by ACEII but by another factor or factors. According to the data now available on the regulation of cellulase and xylanase expression in T. reesei, it can be concluded that the level of cellulase and xylanase gene expression is determined by the balance of both positively and negatively acting regulators, including ACEII, ACEI, and CREI. The effect of these factors is graded, so that they are individually or together only partially responsible for complete regulation of the cellulase and hemicellulase genes.
Acknowledgments
We warmly thank Seija Nordberg for her skilled technical assistance.
This work was supported by the Helsinki Graduate School in Molecular Biology and Biotechnology, the Finnish Cultural Foundation, Roal Oy, and the Jenny and Antti Wihuri Foundation. This work is part of the research program “VTT Industrial Biotechnology” (Academy of Finland, Finnish Centre of Excellence programme, 2000-2005, project no. 64330).
REFERENCES
- 1.Aro, N., A. Saloheimo, M. Ilmén, and M. Penttilä. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276:24309-24314. [DOI] [PubMed] [Google Scholar]
- 2.Bergmeyer, H. U. 1974. Methods of enzymatic analysis, 2nd ed., vol. 2. Academic Press, New York, N.Y.
- 3.Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760-1763. [DOI] [PubMed] [Google Scholar]
- 4.de Vries, R. P., and J. Visser. 2001. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 65:497-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dowzer, C. E., and J. M. Kelly. 1991. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol. Cell. Biol. 11:5701-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drysdale, M. R., S. E. Kolze, and J. M. Kelly. 1993. The Aspergillus niger carbon catabolite repressor encoding gene, creA. Gene 130:241-245. [DOI] [PubMed] [Google Scholar]
- 7.Durand, H., H. Clanet, and G. Tiraby. 1988. Genetic improvement of Trichoderma reesei for large scale cellulase production. Enzyme Microbiol. Technol. 10:341-346. [Google Scholar]
- 8.Gielkens, M. M., E. Dekkers, J. Visser, and L. H. de Graaff. 1999. Two cellobiohydrolase-encoding genes from Aspergillus niger require d-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl. Environ. Microbiol. 65:4340-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas, H., K. Angermayr, I. Zadra, and G. Stoffler. 1997. Overexpression of nreB, a new GATA factor-encoding gene of Penicillium chrysogenum, leads to repression of the nitrate assimilatory gene cluster. J. Biol. Chem. 272:22576-22582. [DOI] [PubMed] [Google Scholar]
- 10.Hasper, A. A., J. Visser, and L. H. de Graaff. 2000. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates d-xylose reductase gene expression. Mol. Microbiol. 36:193-200. [DOI] [PubMed] [Google Scholar]
- 11.Ilmén, M., A. Saloheimo, M. L. Onnela, and M. E. Penttilä. 1997. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ilmén, M., C. Thrane, and M. Penttilä. 1996. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol. Gen. Genet. 251:451-460. [DOI] [PubMed] [Google Scholar]
- 13.Jayaraman, J., C. Cotman, H. R. Mahler, and C. W. Sharp. 1966. Biochemical correlates of respiratory deficiency. VII. Glucose repression. Arch. Biochem. Biophys. 116:224-251. [DOI] [PubMed] [Google Scholar]
- 14.Kamper, J. T., U. Kamper, L. M. Rogers, and P. E. Kolattukudy. 1994. Identification of regulatory elements in the cutinase promoter from Fusarium solani f. sp. pisi (Nectria haematococca). J. Biol. Chem. 269:9195-9204. [PubMed] [Google Scholar]
- 15.Kubicek, C. P., and M. Penttilä. 1998. Regulation of production of plant polysaccharide degrading enzymes by Trichoderma, vol. 2. Taylor & Francis Ltd., London, United Kingdom.
- 16.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 17.MacCabe, A. P., M. Orejas, J. A. Perez-Gonzalez, and D. Ramon. 1998. Opposite patterns of expression of two Aspergillus nidulans xylanase genes with respect to ambient pH. J. Bacteriol. 180:1331-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mach, R. L., M. Schindler, and C. P. Kubicek. 1994. Transformation of Trichoderma reesei based on hygromycin B resistance using homologous expression signals. Curr. Genet. 25:567-570. [DOI] [PubMed] [Google Scholar]
- 19.Mach, R. L., J. Strauss, S. Zeilinger, M. Schindler, and C. P. Kubicek. 1996. Carbon catabolite repression of xylanase I (xyn1) gene expression in Trichoderma reesei. Mol. Microbiol. 21:1273-1281. [DOI] [PubMed] [Google Scholar]
- 20.Margolles-Clark, M., M. Ilmén, and M. Penttilä. 1997. Expression patterns of ten hemicellulase genes from filamentous fungus Trichoderma reesei on various carbon sources. J. Biotechnol. 57:167-179. [Google Scholar]
- 21.Mimura, S., U. Rao, S. Yoshino, M. Kato, and N. Tsukagoshi. 1999. Depression of the xylanase-encoding cgxA gene of Chaetomium gracile in Aspergillus nidulans. Microbiol. Res. 153:369-376. [DOI] [PubMed] [Google Scholar]
- 22.Nogawa, M., M. Goto, H. Okada, and Y. Morikawa. 2001. l-Sorbose induces cellulase gene transcription in the cellulolytic fungus Trichoderma reesei. Curr. Genet. 38:329-334. [DOI] [PubMed] [Google Scholar]
- 23.Penttilä, M., H. Nevalainen, M. Rättö, E. Salminen, and J. Knowles. 1987. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155-164. [DOI] [PubMed] [Google Scholar]
- 24.Penttilä, M. E., L. Andre, P. Lehtovaara, M. Bailey, T. T. Teeri, and J. K. Knowles. 1988. Efficient secretion of two fungal cellobiohydrolases by Saccharomyces cerevisiae. Gene 63:103-112. [DOI] [PubMed] [Google Scholar]
- 25.Penttilä, M. E., L. Andre, M. Saloheimo, P. Lehtovaara, and J. K. Knowles. 1987. Expression of two Trichoderma reesei endoglucanases in the yeast Saccharomyces cerevisiae. Yeast 3:175-185. [DOI] [PubMed] [Google Scholar]
- 26.Saloheimo, A., N. Aro, M. Ilmén, and M. Penttilä. 2000. Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J. Biol. Chem. 275:5817-5825. [DOI] [PubMed] [Google Scholar]
- 27.Saloheimo, M., P. Lehtovaara, M. Penttilä, T. T. Teeri, J. Ståhlberg, G. Johansson, G. Pettersson, M. Claeyssens, P. Tomme, and J. K. Knowles. 1988. EGIII, a new endoglucanase from Trichoderma reesei: the characterization of both gene and enzyme. Gene 63:11-22. [DOI] [PubMed] [Google Scholar]
- 28.Strauss, J., R. L. Mach, S. Zeilinger, G. Hartler, G. Stoffler, M. Wolschek, and C. P. Kubicek. 1995. Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS Lett. 376:103-107. [DOI] [PubMed] [Google Scholar]
- 29.Takashima, S., H. Iikura, A. Nakamura, H. Masaki, and T. Uozumi. 1996. Analysis of Cre1 binding sites in the Trichoderma reesei cbh1 upstream region. FEMS Microbiol. Lett. 145:361-366. [DOI] [PubMed] [Google Scholar]
- 30.Takashima, S., A. Nakamura, M. Hidaka, H. Masaki, and T. Uozumi. 1998. Isolation of the creA gene from the cellulolytic fungus Humicola grisea and analysis of CreA binding sites upstream from the cellulase genes. Biosci. Biotechnol. Biochem. 62:2364-2370. [DOI] [PubMed] [Google Scholar]
- 31.Takashima, S., A. Nakamura, H. Iikura, H. Masaki, and T. Uozumi. 1996. Cloning of a gene encoding a putative carbon catabolite repressor from Trichoderma reesei. Biosci. Biotechnol. Biochem. 60:173-176. [DOI] [PubMed] [Google Scholar]
- 32.Unkles, S. E. 1992. Gene organization in industrial filamentous fungi, vol. 1. Blackie Academic & Professional, London, United Kingdom.
- 33.Vaheri, M., M. Leisola, and V. Kauppinen. 1979. Transglycosylation products of cellulase system of Trichoderma reesei. Bio/Technology 1:696-699. [Google Scholar]
- 34.van der Veen, P., H. N. Arst, Jr., M. J. Flipphi, and J. Visser. 1994. Extracellular arabinases in Aspergillus nidulans: the effect of different cre mutations on enzyme levels. Arch. Microbiol. 162:433-440. [DOI] [PubMed] [Google Scholar]
- 35.van Peij, N. N., M. M. Gielkens, R. P. de Vries, J. Visser, and L. H. de Graaff. 1998. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 64:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Peij, N. N., J. Visser, and L. H. de Graaff. 1998. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol. Microbiol. 27:131-142. [DOI] [PubMed] [Google Scholar]
- 37.van Tilbeurgh, H., M. Claeyssens, and C. K. de Bruyne. 1982. The use of 4-methylumbelliferyl and other chromophoric glycosides in the study of cellulolytic enzymes. FEBS Lett. 149:152-156. [Google Scholar]
- 38.Vautard, G., P. Cotton, and M. Fevre. 1999. The glucose repressor CRE1 from Sclerotinia sclerotiorum is functionally related to CREA from Aspergillus nidulans but not to the Mig proteins from Saccharomyces cerevisiae. FEBS Lett. 453:54-58. [DOI] [PubMed] [Google Scholar]
- 39.Witteveen, C. F. B., F. Weber, R. Busink, and J. Visser. 1994. Isolation and characterization of two xylitol dehydrogenases from Aspergillus niger. Microbiology 140:1679-1685. [Google Scholar]
- 40.Zeilinger, S., A. Ebner, T. Marosits, R. Mach, and C. P. Kubicek. 2001. The Hypocrea jecorina HAP 2/3/5 protein complex binds to the inverted CCAAT-box (ATTGG) within the cbh2 (cellobiohydrolase II-gene) activating element. Mol. Genet. Genomics 266:56-63. [DOI] [PubMed] [Google Scholar]
- 41.Zeilinger, S., R. L. Mach, and C. P. Kubicek. 1998. Two adjacent protein binding motifs in the cbh2 (cellobiohydrolase II-encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J. Biol. Chem. 273:34463-34471. [DOI] [PubMed] [Google Scholar]
- 42.Zeilinger, S., R. L. Mach, M. Schindler, P. Herzog, and C. P. Kubicek. 1996. Different inducibility of expression of the two xylanase genes xyn1 and xyn2 in Trichoderma reesei. J. Biol. Chem. 271:25624-25629. [DOI] [PubMed] [Google Scholar]