Abstract
The interaction between Escherichia coli O157:H7 and its specific bacteriophage PP01 was investigated in chemostat continuous culture. Following the addition of bacteriophage PP01, E. coli O157:H7 cell lysis was observed by over 4 orders of magnitude at a dilution rate of 0.876 h−1 and by 3 orders of magnitude at a lower dilution rate (0.327 h−1). However, the appearance of a series of phage-resistant E. coli isolates, which showed a low efficiency of plating against bacteriophage PP01, led to an increase in the cell concentration in the culture. The colony shape, outer membrane protein expression, and lipopolysaccharide production of each escape mutant were compared. Cessation of major outer membrane protein OmpC production and alteration of lipopolysaccharide composition enabled E. coli O157:H7 to escape PP01 infection. One of the escape mutants of E. coli O157:H7 which formed a mucoid colony (Mu) on Luria-Bertani agar appeared 56 h postincubation at a dilution rate of 0.867 h−1 and persisted until the end of the experiment (∼200 h). Mu mutant cells could coexist with bacteriophage PP01 in batch culture. Concentrations of the Mu cells and bacteriophage PP01 increased together. The appearance of mutant phage, which showed a different host range among the O157:H7 escape mutants than wild-type PP01, was also detected in the chemostat culture. Thus, coevolution of phage and E. coli O157:H7 proceeded as a mutual arms race in chemostat continuous culture.
It has been suggested that most Escherichia coli O157:H7 infections in humans are food-borne illnesses and that dairy and beef cattle are reservoirs of E. coli O157:H7 (10). In addition, E. coli O157:H7 seems to persist in food processing because of its acid and heat tolerance (18). Contamination with E. coli O157:H7 may frequently occur at various stages of food processing and drive up the potential for human infection (7). Thus, the elimination of E. coli O157:H7 from the animal intestine might be effective for the prevention of the infection. In previous reports, dietary manipulations, such as a fasting followed by a reseeding or administration of Lactobacillus casei, induced the clearance of E. coli O157:H7 from animal gastrointestinal tracts (13, 17, 22).
Previous reports also discussed the role of bacteriophage in reducing enteropathogenic bacteria in live animals and gastrointestinal models (2, 4, 12, 19, 23, 25). It is known that the elevated levels of virulent phage in human feces correlate with diseased conditions (8). Thus, phage may play an important role in affecting pathogenic bacteria in intestinal environments.
The emergence of infectious disease caused by drug-resistant bacteria requires alternatives to conventional antibiotics (1, 3, 6, 26). Phage therapy is one possible option, and it can provide an economical tool for controlling pathogens in the intestinal tract without affecting the viability of other normal flora (14, 15). Three virulent E. coli O157 antigen-specific phages, designated KH1, KH4, and KH5, have been analyzed in an attempt to control E. coli O157:H7 in batch culture (14). However, the population dynamics of the phage and its host organism were not analyzed. The relationships between predators and prey may bring about coevolution as the result of an endless arms race between host cell defenses and phage counterdefenses. In fact, some of the failures of phage therapy were due to bacterial mutations leading to resistance to phage infection (1, 3, 6, 26).
However, chemostat cultures have been used as in vitro models of the gut microbial ecosystem and can reproduce many important properties of the intestinal flora (5, 15, 16, 27). In this study, we used a continuous culture of E. coli O157:H7 with its specific virulent phage, named PP01 (21), to investigate the coevolutionary change in E. coli O157:H7 and a virulent bacteriophage specific to it.
MATERIALS AND METHODS
Bacteria, strains, and bacteriophages.
E. coli O157:H7 ATCC 43888 bacteria were used as the host cells for phage. This strain does not produce Shiga toxin 1 or 2 because it lacks the genes for these toxins, but it possesses an envelope structure similar to that of enterohemorrhagic E. coli O157:H7. The bacteriophage PP01, isolated from swine stool and infectious to E. coli O157:H7 strains with high specificity and lytic activity (21), was employed as the predator for E. coli O157:H7.
Phage infection in continuous and batch culture.
In continuous culture, bacterial and phage cultures were grown in Luria-Bertani (LB) broth. A peristaltic pump was used to supply fresh medium and remove spent medium from the culture vessel at the same flow rate. The culture volume was approximately 30 ml, and the dilution rate was altered by changing the pump running speed. After inoculation with E. coli O157:H7 ATCC 43888, the culture was kept at 37°C with stirring and maintained overnight to establish a steady-state condition. Bacteriophage PP01 was then added to the culture with a multiplicity of infection (MOI) of approximately 0.01.
In batch culture, E. coli O157:H7 ATCC 43888 was cultured overnight in 2 ml of LB broth at 37°C with shaking (120 rpm). Three hundred microliters of the initial culture broth was inoculated into 30 ml of fresh LB broth. The optical density of the medium at 600 nm (OD600) was measured to estimate cell lysis. Bacteriophage PP01 infection with an MOI of 2 was performed at an OD600 of 0.1. Additionally, this method was applied to the evaluation of PP01-induced lysis of the mutant cells mentioned below.
Determination of bacterium and phage concentration.
The continuous culture was periodically sampled to determine the concentrations of bacteria and phage. The samples were centrifuged at 12,000 × g for 5 min at 4°C to separate the supernatant and cell pellet. The titer of the phage in the supernatant was determined by serial dilution with sterile SM buffer (10 mM MgSO4, 100 mM NaCl, 0.01% gelatin, 50 mM Tris-HCl [pH 7.5]) followed by plaque assay on lawns of E. coli O157:H7 ATCC 43888. The cell pellets were washed, resuspended, and diluted with phosphate-buffered saline, and the viability of cells was determined by spreading on LB agar plates. All assays were done in triplicate.
Isolation and characterization of mutant cells in the continuous culture.
Several morphologically distinct types of colonies were apparent on the plates used for determining the bacterial cell count. Representative samples of each were transferred with toothpicks into liquid LB broth. After confirming that these cultures were phage free, these isolates were analyzed for resistance to bacteriophage PP01 by the spot test assay. Plaques formed on susceptible isolates and did not form on resistant strains. In susceptible isolates, the efficiency of plating (EOP) of bacteriophage PP01 was estimated by comparing the number of plaques with those of wild-type (WT) E. coli O157:H7 ATCC 43888.
Outer membrane proteins of the isolated mutants were purified as previously described (21), separated on a 12% polyacrylamide gel containing sodium dodecyl sulfate (SDS) and 4 M urea, and stained with Coomassie brilliant blue R-250. Lipopolysaccharide (LPS) was prepared by using the protocol described by Slauch et al. (24). LPS was separated on a 15% polyacrylamide gel containing SDS and detected with two-dimensional Silver Stain II (Daiichi Pure Chemicals). For O-antigen detection, the same LPS samples were run on a 15% polyacrylamide gel containing SDS and electroblotted on an Immobilon-P transfer membrane (Millipore). Peroxidase-labeled affinity-purified antibody to the E. coli O157:H7 antigen (Kirkegaard & Perry Laboratories, Inc.) and the ECL Western blotting detection reagent (Amersham Pharmacia Biotech) were used for light emission detection.
Analysis of phage host-range mutation and isolation of host-range mutants.
The host range of phages in the continuous culture was examined by a series of plaque assays with the mutant isolates. Supernatants from samples of the continuous culture were used as phage lysate, and the number of plaques formed on lawns of the mutant bacterial strains was compared with that of the WT E. coli O157:H7 ATCC 43888. The relative phage titer was defined as the ratio of plaques on each mutant lawn to that on the WT E. coli strain. At time points of 56, 70, and 215 h, analysis revealed phage with novel properties compared to phage from previous time points. Plaques from these time points were isolated by picking the plaque with a micropipette tip, culturing the phage-containing agar overnight in liquid LB medium, and replating the liquid medium on LB agar plates with lawns of susceptible bacteria. In this way, the phages M01a, M01b, and M01f were isolated from the continuous culture at 56, 70, and 215 h, respectively. The host range of each of these phage mutants against the various bacterial mutants derived from E. coli O157:H7 ATCC 43888 was determined by the spot test assay.
RESULTS
Fate of E. coli O157:H7 and PP01 in continuous culture.
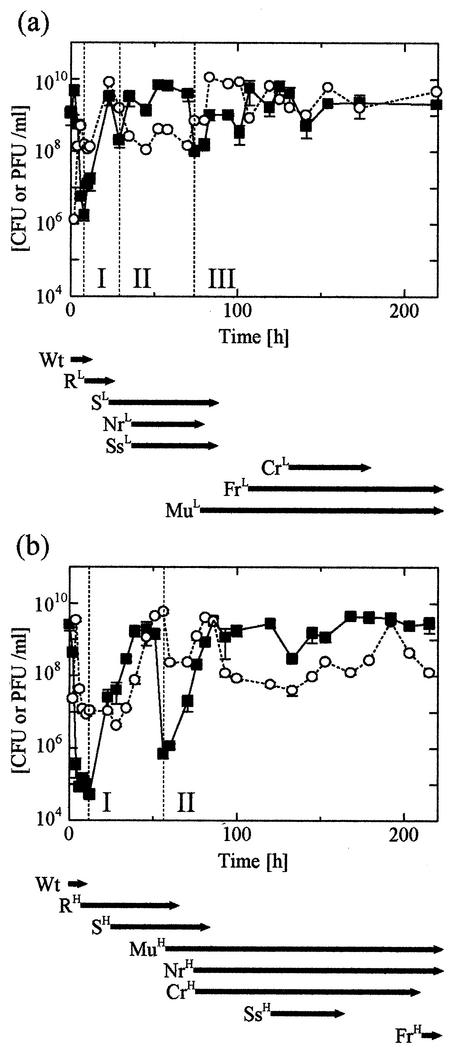
The continuous culture was operated at two different dilution rates (D of 0.327 and 0.876 h−1) to study the effect of bacterial growth on lytic phage activity (Fig. 1). Immediately following phage infection, deep oscillations of the bacterium and phage concentrations were observed. The oscillation amplitude was larger at a D of 0.876 h−1 than at a D of 0.327 h−1. In the first oscillation, the bacterium concentration fell by a factor of approximately 104-fold at the higher dilution rate while an approximately 103-fold decrease was observed at the lower dilution rate. The higher dilution rate and resultant higher growth rate induced more efficient phage-mediated bacteriolysis than the lower one did (9). Eventually, the oscillations in the phage concentration synchronized with the oscillations in the bacterium concentration because phage propagation resulted from host cell lysis. However, the oscillations decayed gradually in both cases, suggesting that bacterium and phage concentrations were moving towards equilibrium. The reproducibility of the results was confirmed by three independent continuous cultures.
FIG. 1.
Bacterium and phage concentration in continuous culture. (a) D = 0.327 h−1; (b) D = 0.876 h−1. Closed squares indicate E. coli O157:H7, and open circles indicate PP01 and derivative phage. The appearance of alternate colony shapes on LB agar was used to track mutant bacterial strains. The arrows below each graph represent the spans of time for which a given bacterial mutant was detectable in the culture. Abbreviations used for the mutant strains reflect their shapes: strains ending in the letter S represent entire, round colonies, and strains ending in the letter R had irregular colonies. The Mu strains were highly mucoid colonies. Wt indicates WT E. coli O157:H7. The dilution rate from which a given strain was isolated was indicated by a superscript L (D = 0.327 h−1) or H (D = 0.876 h−1), which represents the dilution rate from which the strains were isolated. Strains present as less than 1% of the total bacterial CFU were not detectable. Bacterium and phage concentrations are measured on the y axes.
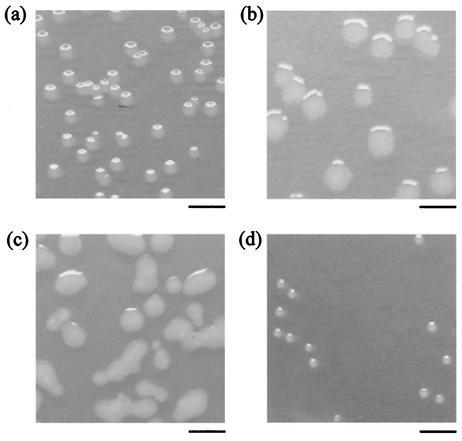
The colonies on LB plates of cells harvested from the culture had various morphologies, which indicated the appearance of phage-resistant mutants. In total, eight distinct strains including the WT (E. coli O157:H7 ATCC 43888) were observed at various times in the continuous culture at both dilution rates. The colony shape and the dilution rate from which it was isolated were used to classify the strains. WT and S mutant cells formed entire, round colonies, but irregular colonies were observed in 4 mutants, designated R, Nr, Cr, and Fr. The Ss mutant formed small, round colonies, and the Mu mutant formed mucoid colonies (Fig. 2 shows photographs of representative colonies). Superscript H's (high, D = 0.876 h−1) and L's (low, D = 0.327 h−1) were used to denote the dilution rate from which the strains were isolated (Fig. 1 and 2). Several distinct phases of phage-bacterium population behavior were apparent in the experiments for which data are shown in Fig. 1. In Fig. 1a, phase I is marked by the emergence and subsequent disappearance of strain RL, which coincided with the initial recovery in the bacterial concentration. In phase II, the SL, NrL, and SsL mutants appeared only to be replaced by the MuL strain, whose emergence marked the onset of the final phase III.
FIG. 2.
Digital photographs of representative mutant cell colonies taken after 18 h of incubation. (a) WT; (b) RH mutant; (c) MuH mutant; (d) SsH mutant. Bars, 5 mm.
In Fig. 1b, two clearly distinct phases were apparent. Phase I was marked by the emergence of the RH and SH strains. A rapid decrease in bacterial concentration coincided with the disappearance of these strains and the emergence of the MuH, NrH, and CrH strains. This marked the onset of phase II. In the final phase of both the low- and high-dilution-rate experiments, oscillations of both the bacterium and phage concentrations were greatly reduced in magnitude. In both cases, this damping occurred with the emergence of the Mu strains, which suggests that these strains can stably coexist with PP01 and PP01-derived mutant bacteriophage.
PP01 sensitivity and bacterial envelope profile of dominant cells.
Mutants from the high-dilution-rate experiment were selected for further study via phage sensitivity analysis and envelope profiling. The dominant cells showed various EOP values toward bacteriophage PP01 (Table 1). The EOP values for the SH, NrH, SsH, and FrH mutants were too low (<10−7) to be evaluated. In both the CrH and MuH mutants, the value was ca. 0.5, and in the RH mutant, the value was 0.05. In addition, the plaques on the RH, CrH, and MuH mutants were turbid and distinctly different from the clear plaque on the WT strain.
TABLE 1.
Susceptibility of mutant cells to WT PP01
| Strain | Appearing time (h of incubation) | Colony shape | EOPa |
|---|---|---|---|
| WT | 0 | Round | 1.00 |
| RH | 4-70 | Irregular | 0.05 |
| SH | 28-76 | Round | <10−7 |
| MuH | 60-215 | Mucoid | 0.52 |
| NrH | 76-215 | Irregular | <10−7 |
| CrH | 76-192 | Irregular | 0.47 |
| SsH | 120-153 | Round, small | <10−7 |
| FrH | 251 | Irregular | <10−7 |
Plaque titers were expressed relative to that obtained on ATCC 43888, which was 108 PFU/ml.
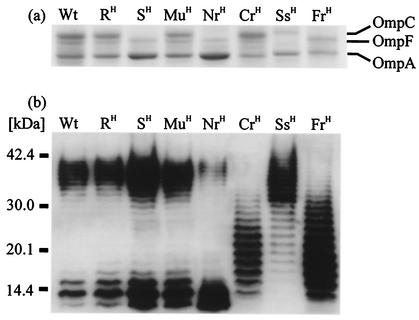
Outer membrane protein analysis by SDS-polyacrylamide gel electrophoresis showed three distinct bands in the WT strain near 40 kDa, which corresponded to OmpC, OmpF, and OmpA (Fig. 3a). However, the production of OmpC was not detected in the SH, NrH, and FrH mutants, all of which showed complete resistance to PP01. The ability of PP01 to bind to these strains was a factor of 20 lower than to the WT strain in a 10-min binding experiment (data not shown). Thus, it seems that OmpC served as a receptor protein for PP01, and its production affected the affinity of bacteriophage PP01 for the cell surface (21).
FIG. 3.
Bacterial envelope profiling of mutant cells. (a) Outer membrane proteins detected by Coomassie brilliant blue R-250 staining; (b) LPS detected by immunoblotting with antibody to E. coli O157:H7.
O157-antigenic LPS was found in both ranges of high molecular mass (>30 kDa) and low molecular mass (<20 kDa) in the WT and RH strains. LPS patterns for the SH and MuH mutants were not remarkably different (Fig. 3b). The production of high- and low-molecular-mass LPS was reduced in the NrH and SsH mutants, respectively. Furthermore, the CrH and FrH mutants produced abundant middle-molecular-mass (20 to 30 kDa) LPS, which was scarcely detected in other strains. The CrH mutant showed lower sensitivity to bacteriophage PP01 but had normal OmpC production, suggesting that effective infection might require normal LPS, not just OmpC. NrH mutants appeared after the decline of the SH mutant and differed from SH only by lacking OmpC. Thus, the NrH mutant seemed to originate from the SH mutant by alternation of LPS structure.
Silver staining of LPS gels enabled detection of O-antigen-free LPS, i.e., only the R core region and the lipid A complex. This analysis revealed that the R core and lipid A complex in the Ss mutant were smaller than those of normal strains (data not shown). The Ss mutant showed complete resistance to PP01 in spite of OmpC production. According to the results mentioned above, not only is OmpC required for PP01 infection but the R core and lipid A components of LPS are also required for PP01 infection.
Mu mutant lysis by bacteriophage PP01 in batch culture.
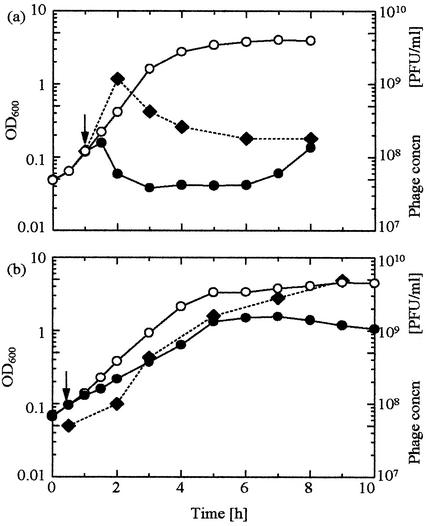
An approach to a seeming equilibrium of bacterium and phage populations was observed after the emergence of the MuH mutant, which suggested that the MuH mutant has an ability to coexist with PP01. MuH cells appeared at 56 h postinfection (at a D of 0.867 h−1) and persisted until the end of the experiment (∼200 h) (Fig. 1b). In batch culture, the culture turbidity of the WT strain decreased after PP01 addition at an MOI of 2 (Fig. 4a), and phage concentration increased during this time. However, with the MuH mutant, the culture turbidity increased after PP01 infection, though more slowly than without PP01 (Fig. 4b). The phage concentration steadily increased together with that of the bacteria, in stark contrast to the WT case. Thus, MuH mutants seemed to grow with only partial cell lysis by the bacteriophage PP01. This observation may explain the population equilibration mentioned above.
FIG. 4.
E. coli O157:H7 lysis by bacteriophage PP01. (a) WT; (b) MuH mutant. The culture was incubated at 37°C and infected with bacteriophage PP01 (closed circles) at an MOI of 2 at the time indicated by the arrow. Open circles are negative controls with no phage. The bacteriophage PP01 concentration (concn) was also measured (closed diamonds).
PP01 host-range mutants in continuous culture.
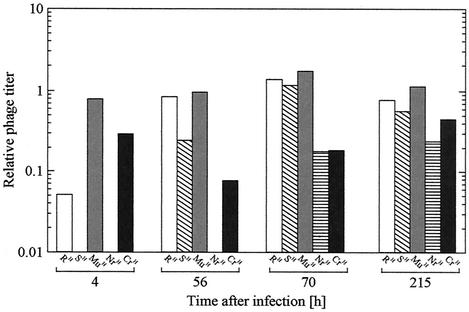
Initially the SH mutant, completely resistant to PP01, became dominant. However, this mutant did not persist in the culture and was detected only through 100 h of incubation. It is suggested that the SH mutant suffered lysis by a mutant phage and was washed out of the culture. The decrease in bacterial concentration after the first 50 h (Fig. 1b) might be explained by the lysis of mutant cells by a PP01 host-range mutant. To confirm this hypothesis, the supernatant of the culture, which was sampled periodically, was subjected to plaque assay with each mutant cell (Fig. 5). The first sample (taken 4 h after infection) did not contain phage which could infect the SH mutant, but a sample taken after 56 h of incubation had phage-infected SH. Concomitantly, the number of RH-infective phage also increased. At this time, RH and SH mutants were dominant in the culture, which suggested that a mutation of PP01 occurred so as to infect the dominant cells. Additionally, phage infectious to the NrH mutant was not observed until the NrH cell became dominant. Thus, PP01 mutated sequentially to become infectious to the new mutant cells, except for the MuH mutant. The relative titer of the phage in the continuous culture supernatants of MuH was near 1 throughout the 200-h experiment.
FIG. 5.
Analysis of phage host-range mutation. Samples of a continuous culture of E. coli O157:H7 and bacteriophage PP01 were taken at the indicated times, and the supernatants were stored and used for plaque assay on the mutant bacterial strains also isolated from the continuous culture. The titer of the WT strain (see text) was defined as 1.0 at all time points.
To analyze this series of mutations of PP01, the isolation of PP01 host-range mutants and the determination of phage infectivity were carried out (Table 2). The host-range mutants, M01a, M01b, and M01f, were isolated from the 56-, 70-, and 215-h culture media, respectively. PP01 could infect the WT, RH, MuH, and CrH strains, and M01a could infect the SH strain in addition to those strains. Moreover, the NrH strain that resisted PP01 and M01a showed susceptibility to the M01b host-range mutant. The M01f phage could infect all strains except for the FrH and CrH strains. From the results mentioned above, it is apparent that the mutational evolution of phage broadened its host range.
TABLE 2.
Turbidity of plaque formation of PP01 and its host-range mutants by spot test assay
| Strain | Resulta for phage:
|
|||
|---|---|---|---|---|
| PP01 | M01a | M01b | M01f | |
| WT | ++ | ++ | ++ | + |
| RH | + | ++ | ++ | + |
| SH | − | ++ | ++ | + |
| MuH | + | + | + | + |
| NrH | − | − | + | + |
| CrH | ++ | + | + | − |
| SsH | − | − | − | + |
| FrH | − | − | − | − |
++, clear plaque; +, turbid plaque; −, no plaque.
DISCUSSION
The ideal phage-based therapeutic should specifically eliminate its target pathogen in a predictable, reliable manner. The emergence of phage-resistant mutants is undesirable. The present study employs a continuous culture to investigate sequential mutations of both phage and its host cells. The bacteriophage PP01, previously shown to efficiently and specifically lyse E. coli O157:H7 (21), was chosen as a model phage.
Bacterial and phage mutants evolve in a coevolutionary arms race.
Following an initial, rapid decrease in viable cell count, resistant bacteria appeared and began to flourish. But susceptibility to phage was not completely eliminated. Phage continued to coexist with the bacteria in the culture. This initial coexistence was not stable, and oscillations in the concentrations of both phage and bacteria were observed. Analysis revealed the recurring appearance of new, distinct bacterial mutants (Fig. 2 and 3 and Table 1) at times spread throughout the 200-h time period of the culture. Phage host-range mutants were also observed (Fig. 5). The bacterial RH and then the MuH mutant strains were dominant at different times in the 200 h of continuous culture at the higher dilution rate, despite both expressing OmpC, having a similar LPS profile to the WT strain, and being partially susceptible to phage lysis. The FrH mutant strain, completely resistant to both PP01 and its derivative bacteriophages, either may have not had enough time to establish dominance or may have grown too slowly to do so, perhaps because its altered LPS and Omp profiles were unfavorable for growth. The results show that OmpC and LPS are not the only factors involved in PP01 adsorption and infection to the bacterial surface. This is consistent with previous results with E. coli with T-even phages, some of which can use both OmpC and LPS as receptors (11).
Both the phage and bacteria seemed to be continuously evolving in a mutual, ever-escalating arms race. Previous chemostat studies have not shown this behavior (5, 15, 16). Prior studies mainly used minimal medium, with glucose as the sole carbon source, at a concentration of less than 1 g liter−1. They were focused on modeling bacterium-phage systems in natural ecosystems, where resources are relatively scarce. In contrast, resources are relatively abundant in the human gastrointestinal tract. Bacterial density is very high (20). To represent this fact, LB broth, a very rich medium that can support very high cell densities, was used in this study. Therefore, in environments of high biodensity, such as the human gastrointestinal tract, a mutual arms race may be a significant factor in phage and bacterial evolution.
Mechanisms of coevolution.
The first bacterial mutants to appear were the R and S strains, which had altered colony shapes, LPS profiles, and OmpC expression compared to the WT strain. Phage host-range mutants appeared which could lyse the R and S strains, causing these strains to disappear from the culture. The fact that these mutants could lyse the non-OmpC-expressing strain S implies that the receptor of these host-range mutants is most likely altered. The Mu, Nr, and Cr bacterial mutants appeared next in the culture. The LPS expression of all of these strains was different from that of the WT, especially that of strain Cr. Strain Nr also had a different Omp expression profile (Fig. 3). Both Mu and Cr expressed OmpC. This is further evidence that LPS is involved in the binding of PP01 to E. coli O157:H7. Altered LPS or other cell envelope structures may act as a barrier, preventing phage access to OmpC.
These findings show that coevolution of phage and the clinically relevant bacterium E. coli O157:H7 can and sometimes does proceed as a mutual arms race. Several phenotypically different O157:H7 bacterial mutants may appear. Development of a successful phage therapeutic against this enteropathogen must address the emergence of all of these mutant strains. Additionally, this study shows that at least in some cases, phage binding and infection of E. coli O157:H7 is not controlled only by a single receptor but that many features of the target cell envelope may be important. Future studies should address the DNA alterations responsible for the altered phenotypes of the mutants. Also, to test the generality of these results, our experiments should be extended to other O157:H7-specific phages. Only through understanding and controlling the emergence of phage-resistant bacteria can bacteriophage become a clinically useful tool.
Acknowledgments
This research was financially supported by a grant (no. 13450340) from the Japanese Ministry of Education, Science, Sports, and Culture.
REFERENCES
- 1.Alisky, J., K. Iczkowski, A. Rapoport, and N. Troitsky. 1998. Bacteriophages show promise as antimicrobial agents. J. Infect. 36:5-15. [DOI] [PubMed] [Google Scholar]
- 2.Barrow, P., M. Lovell, and A. Berchieri, Jr. 1998. Use of lytic bacteriophage for control of experimental Escherichia coli septicemia and meningitis in chickens and calves. Clin. Diagn. Lab. Immunol. 5:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrow, P. A., and J. S. Soothill. 1997. Bacteriophage therapy and prophylaxis: rediscovery and renewed assessment of potential. Trends Microbiol. 5:268-271. [DOI] [PubMed] [Google Scholar]
- 4.Biswas, B., S. Adhya, P. Washart, B. Paul, A. N. Trostel, B. Powell, R. Carlton, and C. R. Merril. 2002. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect. Immun. 70:204-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohannan, B. J. M., and R. E. Lenski. 1997. Effect of resource enrichment on a chemostat community of bacteria and bacteriophage. Ecology 78:2303-2315. [Google Scholar]
- 6.Carlton, R. M. 1999. Phage therapy: past history and future prospects. Arch. Immunol. Ther. Exp. 47:267-274. [PubMed] [Google Scholar]
- 7.Dean-Nystrom, E. A., B. T. Bosworth, W. C. Cray, Jr., and H. W. Moon. 1997. Pathogenicity of Escherichia coli O157:H7 in the intestines of neonatal calves. Infect. Immun. 65:1842-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuse, K., S. Osawa, J. Kawashiro, R. Tanaka, A. Ozawa, S. Sawamura, Y. Yanagawa, T. Nagao, and I. Watanabe. 1983. Bacteriophage distribution in human faeces: continuous survey of healthy subjects and patients with internal and leukaemic diseases. J. Gen. Virol. 64:2039-2043. [DOI] [PubMed] [Google Scholar]
- 9.Hadas, H., M. Einav, I. Fishov, and A. Zaritsky. 1997. Bacteriophage T4 development depends on the physiology of its host Escherichia coli. Microbiology 143:179-185. [DOI] [PubMed] [Google Scholar]
- 10.Hancock, D. D., T. E. Besser, and D. H. Rice. 1998. Ecology of Escherichia coli O157:H7 in cattle and impact of management practices, p. 85-91. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, D.C.
- 11.Heller, K. J. 1992. Molecular interaction between bacteriophage and the gram-negative cell envelope. Arch. Microbiol. 158:235-248. [DOI] [PubMed] [Google Scholar]
- 12.Koo, J., D. L. Marshall, and A. DePaola. 2001. Antacid increases survival of Vibrio vulnificus and Vibrio vulnificus phage in a gastrointestinal model. Appl. Environ. Microbiol. 67:2895-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kudva, I. T., P. G. Hatfield, and C. J. Hovde. 1995. Effect of diet on the shedding of Escherichia coli O157:H7 in a sheep model. Appl. Environ. Microbiol. 61:1363-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kudva, I. T., S. Jelacic, P. I. Tarr, P. Youderian, and C. J. Hovde. 1999. Biocontrol of Escherichia coli O157 with O157-specific bacteriophages. Appl. Environ. Microbiol. 65:3767-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenski, R. E., and B. R. Levin. 1985. Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am. Nat. 125:585-602. [Google Scholar]
- 16.Levin, B. R., F. M. Stewart, and L. Chao. 1977. Resource-limited growth, competition, and predation: a model and experimental studies with bacteria and bacteriophage. Am. Nat. 111:3-24. [Google Scholar]
- 17.Magnuson, B. A., M. Davis, S. Hubele, P. R. Austin, I. T. Kudva, C. J. Williams, C. W. Hunt, and C. J. Hovde. 2000. Ruminant gastrointestinal cell proliferation and clearance of Escherichia coli O157:H7. Infect. Immun. 68:3808-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao, Y., M. P. Doyle, and J. Chen. 2001. Insertion mutagenesis of wca reduces acid and heat tolerance of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:3811-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merril, C. R., B. Biswas, R. Carlton, N. C. Jensen, G. J. Creed, S. Zullo, and S. Adhya. 1996. Long-circulating bacteriophage as antibacterial agents. Proc. Natl. Acad. Sci. USA 93:3188-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minekus, M., M. Smeets-Peeters, A. Bernalier, S. Marol-Bonnin, R. Havenaar, P. Marteau, M. Alric, G. Fonty, and J. H. J. Huis in't Veld. 1999. A computer-controlled system to simulate conditions of the large intestine with peristaltic mixing, water absorption and absorption of fermentation products. Appl. Microbiol. Biotechnol. 53:108-114. [DOI] [PubMed] [Google Scholar]
- 21.Morita, M., Y. Tanji, K. Mizoguchi, T. Akitsu, N. Kijima, and H. Unno. 2002. Characterization of a virulent bacteriophage specific for Escherichia coli O157:H7 and analysis of its cellular receptor and two tail fiber genes. FEMS Microbiol. Lett. 211:77-83. [DOI] [PubMed] [Google Scholar]
- 22.Ogawa, M., K. Shimizu, K. Nomoto, M. Takahashi, M. Watanuki, R. Tanaka, T. Tanaka, T. Hamabata, S. Yamasaki, and Y. Takeda. 2001. Protective effect of Lactobacillus casei strain Shirota on Shiga toxin-producing Escherichia coli O157:H7 infection in infant rabbits. Infect. Immun. 69:1101-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, S. C., I. Shimamura, M. Fukunaga, K. Mori, and T. Nakai. 2000. Isolation of bacteriophages specific to a fish pathogen, Pseudomonas plecoglossicida, as a candidate for disease control. Appl. Environ. Microbiol. 66:1416-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slauch, J. M., M. J. Mahan, P. Michetti, M. R. Neutra, and J. J. Mekalanos. 1995. Acetylation (O-factor 5) affects the structural and immunological properties of Salmonella typhimurium lipopolysaccharide O antigen. Infect. Immun. 63:437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, H. W., M. B. Huggins, and K. M. Shaw. 1987. The control of experimental Escherichia coli diarrhoea in calves by means of bacteriophages. J. Gen. Microbiol. 133:1111-1126. [DOI] [PubMed] [Google Scholar]
- 26.Sulakvelidze, A., Z. Alavidze, and J. G. Morris, Jr. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkinson, M. H. F. 2001. Predation in the presence of decoys: an inhibitory factor on pathogen control by bacteriophages or bdellovibrios in dense and diverse ecosystems. J. Theor. Biol. 208:27-36. [DOI] [PubMed] [Google Scholar]