Abstract
A quantitative competitive PCR (QC-PCR) assay targeting the phlA gene of Pseudomonas fluorescens CHA0 was developed and tested in vitro. Statistically significant, positive correlations were found between QC-PCR and both CFU and total cell number when studying cells in log or stationary phase. The correlations disappeared when considering stressed cells.
The root-colonizing bacterium Pseudomonas fluorescens CHA0, which can protect plants from soil-borne fungal diseases, has become a model for studying the behavior of biocontrol inoculants in the soil ecosystem (2-4, 10, 11, 15). When introduced into the environment, CHA0 may under certain conditions persist as mixed populations of culturable and nonculturable cells (2, 4, 15), which limits the usefulness of colony counts.
Recently, quantitative competitive PCR (QC-PCR) has been successfully used as a rapid and sensitive means to detect and enumerate bacteria in pure culture, food, and environmental samples (5, 9). The method is based on the coamplification, with the same set of primers, of the DNA sequence to be quantified (the target) and a known amount of a similar sequence of slightly different size (the competitor). It allows quantification of the target, which in turn enables an estimation of the number of corresponding cells.
The objectives of this work were to determine if QC-PCR could be applied to enumerate P. fluorescens CHA0 cells in vitro and to assess the effect of stress on the performance of the assay. We chose the Pseudomonas gene phlA, implicated in the synthesis of the biocontrol metabolite 2,4-diacetylphloroglucinol, as the target.
Experimental samples.
Cells were obtained at 27°C with shaking (160 rpm) after growth for 14 to 16 h (log cells; 33 samples) or incubation for 48 h to 7 days (stationary-phase cells; 25 samples) in tryptic soy broth (Difco, Detroit, Mich.), Luria-Bertani broth (LB) (13), King's B broth (KB) (7), or M9 broth medium containing 0.1% glucose (10). Stress conditions (for 81 samples in total) were obtained by resuspending LB- or KB-grown log cells as follows: (i) 4.5 h to 7 days in M9 medium containing 0.1% glucose and 1.5 M NaCl (NaCl stress [10]) (17 samples), (ii) 1 to 21 days under nitrogen atmosphere in M9 medium containing 0.1% glucose and 50 mM potassium hexacyanoferrate (oxygen limitation and low [230 mV] redox potential [10]) (13 samples), (iii) 0.5 h to 14 days in 60 mM citric acid-80 mM Na2HPO4 (pH 4) or 50 mM glycine-11 mM HCl (pH 3; acidity stress) (17 samples), (iv) overnight in 0.9% NaCl containing 0.5 to 50 mM CuSO4 (metal stress) (18 samples), or (v) by prolonged incubation (7 to 30 days) in KB or LB (16 samples).
Culturable cells of CHA0 were enumerated on King’s B agar (for all 137 samples) and total cells were counted by immunofluorescence microscopy (3, 10) (for 102 of 137 samples). Total cell counts and CFU were similar in the absence of stress, whereas nonculturable cells were found in large amounts under stress conditions.
Since modifications in DNA packing and conformation resulting from exposure to stress may affect PCR efficacy (12), we assessed the effects of antibiotics inhibiting de novo protein synthesis, i.e., chloramphenicol (200 μg/ml), or DNA supercoiling, i.e., ciprofloxacin (20 μg/ml), on PCR amplification. This was done after incubating CHA0 cells with or without the antibiotics at pH 7 (control) and 4 (acidity stress) in the buffers described above. Antibiotics were used at concentrations enabling growth of CHA0 in LB.
Competitor oligonucleotide.
The 20-bp primers PhlA-1f and PhlA-1r (Table 1) were designed for amplification of the target oligonucleotide based on the published (14) phlA sequence. Primer specificity was verified by BLAST alignment of the 418-bp amplicon and confirmed by PCR analysis of selected Phl-producing and non-Phl-producing bacteria. The competitor oligonucleotide was constructed by PCR using PhlA-1r and PhlA-1c. Primer PhlA-1c, designed as described by Celi et al. (1), is 40 bp long and has a 20-bp region that binds 41 bp downstream of the PhlA-1f binding site. The remaining overhanging 20-bp sequence is identical to PhlA-1f. Thus, PCR yields a 377-bp competitor which can be amplified by the same primers as the target. The competitor was separated on 2% agarose and purified with a QIAquick gel extraction kit (Qiagen, Basel, Switzerland). Its concentration was determined both spectrophotometrically and visually on agarose by comparison with DNA standards. Correct sequences of both target and competitor were verified by sequencing.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ → 3′)a | Source or reference |
|---|---|---|
| Primers for phlAb | ||
| PhlA-1f | TCA GAT CGA AGC CCT GTA CC | This work |
| PhlA-1r | GAT GCT GTT CTT GTC CGA GC | This work |
| PhlA-1c | TCA GAT CGA AGC CCT GTA CCT CGA TCA TCC TGG AAA TGC T | This work |
| RAPD primersc | ||
| GAC | CCG TTA TTG CGC CCG G | 6 |
| D7 | TTG GCA CGG G | 6 |
| Primers for rrsd | ||
| PSMG | CCT TCC TCC CAA CTT | 5 |
| 9-27 | GAG TTT GAT CCT GGC TCA G | 5 |
The sequence in bold is the overhanging end used for the construction of the competitor.
Primers PhlA-1f and PhlA-1r enabled amplification of phlA sequences from fluorescent pseudomonads from amplified 16S ribosomal DNA restriction analysis (ARDRA) groups 1 (e.g., CHA0) and 3 (e.g., F113) but not from strains from ARDRA group 2 (e.g., Q2-87). No PCR product was obtained when testing Phl-negative pseudomonads.
RAPD analysis was carried out using primer GAC or D7, as described by Keel et al. (6).
PCR with primers PSMG and 9-27 was performed as described by Johnsen et al. (5).
DNA extraction and PCRs.
Various methods were tested to extract and amplify DNA from CHA0. Direct amplification of cell lysate was carried out, as it proved to give reproducible results which were as good as when purified DNA was used (data not shown). Cell suspensions (5 μl) were heated for 10 min at 99°C with 95 μl of lysis buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3], 0.1% Tween 20) in a PTC-100 thermal cycler (MJ Research, Waltham, Mass.). PCR amplification was carried out in 20-μl reaction mixtures containing 4 μl of cell lysate, 4 μl of internal standard (for QC-PCR only), and 1× PCR buffer. The latter contained 100 μM (each) dATP, dCTP, dGTP, and dTTP, 0.07 U of Taq polymerase (Amersham Pharmacia Biotech, Piscataway, N.J.) per μl, and 0.20 μM (each) primers. Bovine serum albumin (0.25 mg/ml; Biofinex, Praroman, Switzerland) and 5% dimethyl sulfoxide were used as PCR enhancers when amplifying phlA. During competitor construction, PCR was performed under the following conditions: 5 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C; and then 10 min at 72°C. QC-PCR was performed as described above but with only 24 cycles. All PCR products were separated by electrophoresis in 2% agarose.
QC-PCR in the absence of stress.
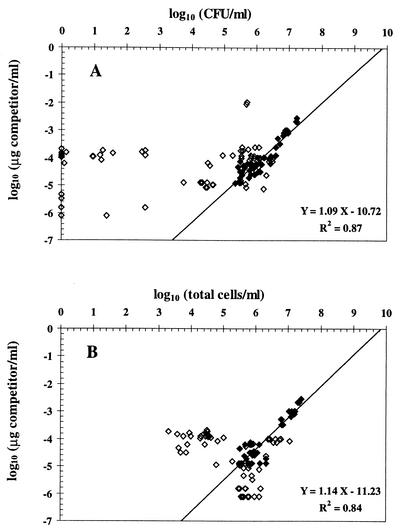
A linear correlation was obtained, as expected, between cell density and the competitor concentration needed to have the same amplification of both fragments when cell suspensions of CHA0 and competitor solutions were diluted twofold repeatedly (illustrated with log cells in Fig. 1). Similar results were obtained when purified cell DNA was used instead of cell lysate (data not shown). Next, QC-PCR was performed on independent samples consisting of log or stationary-phase cells. The logarithm of the concentration of competitor needed to have two bands of equal intensity was plotted against the logarithm of cell concentration. Statistically significant correlations were found between QC-PCR results (derived from eight serial twofold dilutions) and CFU or the total number (most of them were culturable) of CHA0 cells (Fig. 2).
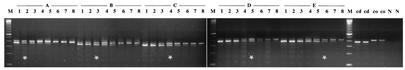
FIG. 1.
QC-PCR carried out on log cells of P. fluorescens CHA0 (A, 4 × 106 cells/ml; B, 2 × 106 cells/ml; C, 1 × 106 cells/ml; D, 5 × 105 cells/ml; E, 2.5 × 105 cells/ml) against twofold dilutions of competitor oligonucleotide. Lanes: 1, 2.5 × 10−4 μg/ml; 2, 1.25 × 10−4 μg/ml; 3, 6.25 × 10−5 μg/ml; 4, 3.13 × 10−5 μg/ml; 5, 1.57 × 10−5 μg/ml; 6, 7.81 × 10−6 μg/ml; 7, 3.92 × 10−6 μg/ml; 8, 1.96 × 10−6 μg/ml; M, 100-bp ladder; cd, cell DNA alone (left, 4 × 106 cells/ml; right, 2.5 × 105 cells/ml); co, competitor oligonucleotide alone (left, 2.5 × 10−4 μg/ml; right, 3.13 × 10−5 μg/ml); N, negative control (blank). Reactions yielding two bands of the same intensity are indicated by an asterisk.
FIG. 2.
Effect of stress on the relationship between QC-PCR results and the number of CFU (A) and total cells (B) of P. fluorescens CHA0. The result of QC-PCR is represented by the concentration of competitor oligonucleotide needed to yield the same amount of amplification product as the corresponding target bacterial DNA in the reaction. ◊, data obtained with stressed cells; ⧫, data derived from log or stationary-phase cells (i.e., control). CFU are arbitrarily plotted as zero when below the detection limit (0.92 log CFU/ml). For CFU and for total cells, statistically significant positive correlations with QC-PCR results were found when log and stationary-phase cells were considered, and regression parameters are thus indicated.
QC-PCR in the presence of stress.
No correlation was obtained when considering samples of CHA0 cells subjected to stress, regardless of whether QC-PCR results were compared with the number of total cells or with CFU (Fig. 2). A proportion of the nonculturable cells in some of these samples responded to Kogure's cell elongation test (8), yielding as many as 6.2 log nutrient-responsive (i.e., presumably viable) but nonculturable cells/ml in certain cases, but no correlation was found between Kogure's counts of nutrient-responsive cells (done as described previously [3, 10]) and QC-PCR results based on 31 stress samples studied (data not shown).
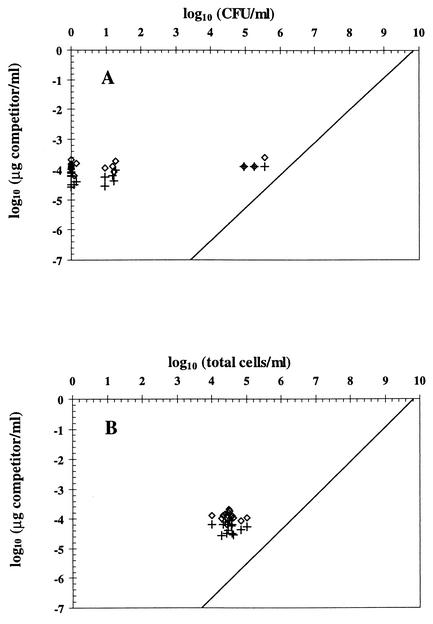
In Fig. 2A, most of the points derived from stress situations are located left of the regression line obtained in the absence of stress, meaning that cell numbers estimated by QC-PCR were higher than CFU. This excess DNA might have belonged to nonculturable cells (including dead cells) and/or corresponded to extracellular DNA (probably released by natural lysis of dead cells). The presence of extracellular DNA was indicated by the fact that cell-free samples obtained by filtration through a 0.2-μm-pore-size Minisart-plus membrane (Sartorius, Göttingen, Germany) still enabled PCR amplification of phlA, whereas no amplification took place when cell-free samples were incubated for 2 h with 5 U (in a 30-μl volume) of restriction enzyme Sau3AI (cuts immediately before the sequence GATC, which predictably occurs five times in the target sequence) prior to PCR (data not shown). Yet, elimination of extracellular DNA by Sau3AI digestion (which had no effect on the cell culturability of CHA0) was not sufficient to bring QC-PCR results down to the level of culturable cells (Fig. 3). This means that extracellular DNA could not explain the discrepancy between both types of results. In Fig. 2B, the points for cells subjected to stress are found on both sides of the regression line obtained in the absence of stress. One implication is that not all of the DNA present in some of the samples was amplified by PCR, although complete cell lysis took place (confirmed by microscopy). Perhaps this was caused by a particular conformation of DNA in stressed cells.
FIG. 3.
Contribution of extracellular DNA to QC-PCR results and comparison of the latter with the numbers of CFU (A) and total cells (B) of P. fluorescens CHA0 exposed to CuSO4 stress (18 samples). In each case, half the sample was digested with Sau3AI to eliminate extracellular DNA (+), whereas the other half was not digested (⋄). CFU are arbitrarily plotted as zero when below the detection limit (0.92 log CFU/ml). The line of best fit for control samples obtained for Fig. 2 is also represented.
Effect of antibiotics affecting DNA conformation and packing.
Oliver and Warner (12) have shown that the loss of random amplified polymorphic DNA (RAPD) signal in starved Vibrio vulnificus, presumably linked to modifications in DNA packing and conformation, could be prevented by inhibiting de novo protein synthesis using chloramphenicol or DNA supercoiling using ciprofloxacin. Incubation of CHA0 cells in the presence of either antibiotic had no effect on phlA PCR efficacy at pH 7, even in the case of ciprofloxacin, which caused loss of colony-forming ability. The phlA PCR signal disappeared shortly after exposure of cells to pH 4, along with a decrease in the number of culturable cells, even when chloramphenicol or ciprofloxacin was used. Similar results were obtained when amplifying rrs instead of phlA or when performing RAPD analysis (Table 2). In summary, the two antibiotics were unable to prevent the disappearance of the PCR signals under acidic stress.
TABLE 2.
Effect of chloramphenicol (200 μg/ml) and ciprofloxacin (20 μg/ml) on survival (CFU) and PCR amplification of P. fluorescens CHA0 (used at 7.7 log CFU/ml) incubated at pH 7 (control) or pH 4 (acidity stress)
| Antibiotic and time of exposure | Survival of cells (log CFU/ml)a at pH:
|
phlA PCRb at pH:
|
rrs PCR at pH:
|
D7 RAPD PCR at pH:
|
GAC RAPD PCR at pH:
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 4 | 7 | 4 | 7 | 4 | 7 | 4 | 7 | 4 | |
| None | ||||||||||
| 0.5 h | 7.47 ± 0.14 | 4.92 ± 0.19 | + | 0 | + | + | + | (+) | (+) | 0 |
| 4 h | 6.99 ± 0.14 | 2.39 ± 0.25 | + | (+) | ++ | + | ++ | (+) | + | 0 |
| 1 day | 7.41 ± 0.52 | BD | + | (+) | ++ | (+) | ++ | 0 | ++ | 0 |
| Chloramphenicol | ||||||||||
| 0.5 h | 6.95 ± 0.06 | 5.92 ± 0.19 | + | 0 | + | (+) | + | (+) | (+) | 0 |
| 4 h | 6.24 ± 0.20 | 0.92c | + | (+) | ++ | + | ++ | (+) | + | 0 |
| 1 day | 7.08 ± 0.03 | BD | + | (+) | ++ | + | ++ | 0 | ++ | 0 |
| Ciprofloxacin | ||||||||||
| 0.5 h | BD | BD | + | 0 | + | (+) | + | 0 | + | 0 |
| 4 h | BD | BD | + | (+) | ++ | (+) | + | 0 | + | 0 |
| 1 day | BD | BD | + | (+) | ++ | (+) | + | 0 | + | 0 |
Results are means ± standard deviations. BD, below detection (detection limit = 0.92 log CFU/ml).
Efficiency of PCR amplification methods was rated as follows: 0, no amplification; (+), weak or nonspecific amplification; +, normal amplification; ++, strong amplification.
From 0 to 2 colonies were recovered per plate.
Search for cytoplasmic PCR inhibitors.
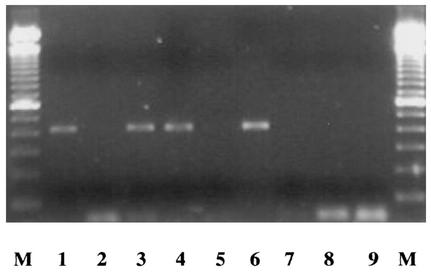
Another explanation for the incomplete PCR amplification might be the presence of cytoplasmic PCR inhibitors released during PCR lysis of stressed cells. To test this hypothesis, CHA0 samples from pH 7 cultures were mixed with pH 4 CHA0 samples, either before or after having implemented cell lysis, and PCR was performed. No significant loss of PCR amplification was found (Fig. 4), ruling out this hypothesis. In fact, the same fading of the PCR signal observed when progressively increasing the relative proportion of pH 4 samples, from 50:50 up to a 1:99 (vol/vol) ratio, was obtained when using water or pH 4 buffer instead (data not shown).
FIG. 4.
PCR amplification of phlA gene in P. fluorescens CHA0 (cells from 1-day-old cultures; used at 7.7 log CFU/ml) following the addition (50:50 [vol/vol]) of cell samples obtained after incubation for 1 day at pH 4 (2.09 ± 0.36 log CFU/ml). Lanes: M, 100-bp ladder; 1, pH 7 cells; 2, pH 4 cells; 3, pH 7 cells and pH 4 cells mixed before lysis; 4, pH 7 cells and pH 4 cells mixed after lysis; 5, pH 4 cells and pH 7 buffer mixed before lysis; 6, pH 7 cells and pH 4 buffer mixed before lysis; 7, pH 7 buffer; 8, pH 4 buffer; 9, water blank. Identical results were obtained when PCR was performed after DNA purification of pH 7 and pH 4 samples (cell lysates) by direct ethanol precipitation (16) or phenol-chloroform-isoamyl alcohol extraction (followed by precipitation with 1 volume of 2 M NaCl and 2 volumes of ice-cold 95% ethanol) instead of with cell lysates directly (data not shown). Each treatment was replicated at least four times.
Conclusion.
Successful detection of P. fluorescens CHA0 by PCR depended strongly on the physiological state of the cells, and QC-PCR gave effective results in vitro only when applied to samples from which most CHA0 cells where culturable. When cells were stressed, the amount of DNA estimated by QC-PCR was higher than the total amount of DNA present in culturable cells, which at least in the case of CuSO4 stress was likely due to the contribution of nonculturable cells rather than that of extracellular DNA. However, the amount of DNA estimated by QC-PCR did not necessarily correspond to that present in all CHA0 cells either, and the loss of amplification efficacy following stress was not due to PCR inhibitors synthesized by stressed cells. Overall, it appears that stress negatively affected the ability of cell DNA to be amplified by PCR. These findings may limit the usefulness of PCR amplification to monitor pseudomonads in stressful environments (e.g., bulk soil). They also have implications when studying the genetic diversity of bacterial communities by culture-independent methods, which often take for granted the fact that DNA can be amplified equally from cells of different taxonomic and/or physiological status.
Acknowledgments
We thank Fabio Mascher and Carsten Hase for helpful discussion.
This work was supported by the Swiss Biotechnology SPP (project 5002-04502311), the Swiss Federal Office for Education and Science (COST Action 830), the Fachverein für Arbeit und Umwelt (FAU, Zürich, Switzerland), and the French Embassy in Switzerland (France-Switzerland research grant).
REFERENCES
- 1.Celi, F. S., M. E. Zenilman, and A. R. Shuldiner. 1993. A rapid and versatile method to synthesize internal standards for competitive PCR. Nucleic Acids Res. 21:1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Défago, G., C. Keel, and Y. Moënne-Loccoz. 1997. Fate of released Pseudomonas bacteria in the soil profile: implications for the use of genetically modified microbial inoculants, p. 403-418. In J. T. Zelikoff, J. M. Lynch, and J. Shepers (ed.), Ecotoxicology: responses, biomarkers and risk assessment. SOS Publications, Fair Heaven, N.J.
- 3.Hase, C., F. Mascher, Y. Moënne-Loccoz, and G. Défago. 1999. Nutrient deprivation and the subsequent survival of biocontrol Pseudomonas fluorescens CHA0 in soil. Soil Biol. Biochem. 31:1181-1188. [Google Scholar]
- 4.Hase, C., M. Hottinger, Y. Moënne-Loccoz, and G. Défago. 2000. Survival and cell culturability of biocontrol Pseudomonas fluorescens CHA0 in the rhizosphere of cucumber grown in two soils of contrasting fertility status. Biol. Fertil. Soils 32:217-221. [Google Scholar]
- 5.Johnsen, K., O. Enger, C. S. Jacobsen, L. Thirup, and V. Torsvik. 1999. Quantitative selective PCR of 16S ribosomal DNA correlates well with selective agar plating in describing population dynamics of indigenous Pseudomonas spp. in soil hot spots. Appl. Environ. Microbiol. 65:1786-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keel, C., D. M. Weller, A. Natsch, G. Défago, R. J. Cook, and L. S. Thomashow. 1996. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl. Environ. Microbiol. 62:552-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 8.Kogure, K., U. Simidu, and U. Taga. 1979. A tentative direct microscopic method for counting living marine bacteria. Can. J. Microbiol. 25:415-420. [DOI] [PubMed] [Google Scholar]
- 9.Li, W., and M. A. Drake. 2001. Development of a quantitative competitive PCR assay for detection and quantification of Escherichia coli O157:H7 cells. Appl. Environ. Microbiol. 67:3291-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mascher, F., C. Hase, Y. Moënne-Loccoz, and G. Défago. 2000. The viable-but-nonculturable state induced by abiotic stress in the biocontrol agent Pseudomonas fluorescens CHA0 does not promote strain persistence in soil. Appl. Environ. Microbiol. 66:1662-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Natsch, A., C. Keel, H. A. Pfirter, D. Haas, and G. Défago. 1994. Contribution of the global regulator gene gacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHA0 introduced into soil microcosms. Appl. Environ. Microbiol. 60:2553-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver, J. D., and J. M. Warner. 1998. Randomly amplified polymorphic DNA analysis of starved and viable but nonculturable Vibrio vulnificus cells. Appl. Environ. Microbiol. 64:3025-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Schnider-Keel, U., A. Seematter, M. Maurhofer, C. Blumer, B. Duffy, C. Gigot-Bonnefoy, C. Reimmann, R. Notz, G. Défago, D. Haas, and C. Keel. 2000. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 182:1215-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Troxler, J., M. Zala, A. Natsch, Y. Moënne-Loccoz, C. Keel, and G. Défago. 1997. Predominance of nonculturable cells of the biocontrol strain Pseudomonas fluorescens CHA0 in the surface horizon of large outdoor lysimeters. Appl. Environ. Microbiol. 63:3776-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, C., E. Knill, B. Glick, and G. Défago. 2000. Effect of transferring 1-aminocyclopropane-1-carboxylic (ACC) deaminase genes into Pseudomonas fluorescens strain CHA0 and its gacA derivative CHA96 on their growth-promoting and disease-suppressive capacities. Can. J. Microbiol. 46:898-907. [DOI] [PubMed] [Google Scholar]