Abstract
In Europe the Borrelia burgdorferi sensu lato complex is represented by five distinct genospecies: Borrelia burgdorferi sensu stricto, Borrelia afzelii, Borrelia garinii, Borrelia valaisiana, and Borrelia lusitaniae. These taxonomic entities are known to differ in their specific associations with vertebrate hosts and to provoke distinct clinical manifestations in human patients. However, exceptions to these rules have often been observed, indicating that strains belonging to a single genospecies may be more heterogeneous than expected. It is, therefore, important to develop alternative identification tools which are able to distinguish Borrelia strains not only at the specific level but also at the intraspecific level. DNA from a sample of 370 Ixodes ricinus ticks collected in the Czech Republic was analyzed by PCR for the presence of a ∼230-bp fragment of the rrfA-rrlB intergenic spacer of Borrelia spp. A total of 20.5% of the ticks were found to be positive. The infecting genospecies were identified by analyzing the amplified products by the restriction fragment length polymorphism (RFLP) method with restriction enzyme MseI and by single-strand conformation polymorphism (SSCP) analysis. The two methods were compared, and PCR-SSCP analysis appeared to be a valuable tool for rapid identification of spirochetes at the intraspecific level, particularly when large samples are examined. Furthermore, by using PCR-SSCP analysis we identified a previously unknown Borrelia genotype, genotype I-77, which would have gone unnoticed if RFLP analysis alone had been used.
Lyme borreliosis, the most common vector-borne zoonotic disease in the northern hemisphere, is caused by spirochetes of the Borrelia burgdorferi sensu lato complex. B. burgdorferi is efficiently maintained in nature through enzootic cycles, which involve ticks of the Ixodes ricinus complex and a variety of vertebrate reservoir hosts. In Europe, the recognized vector of Lyme disease is Ixodes ricinus (3, 9, 25).
Data from a number of molecular and phenotypic studies resulted in subdivision of the B. burgdorferi sensu lato complex into different taxonomic entities, named genospecies or genotypes (1, 4, 8, 10, 13, 15, 31, 32, 33, 34, 35, 39, 41, 54, 55, 58). The molecular typing methods, as reviewed by Wang et al. (57), included DNA-DNA hybridization analysis, pulsed-field gel electrophoresis, plasmid fingerprinting, randomly amplified polymorphic DNA analysis, species-specific PCR, PCR followed by restriction fragment length polymorphism (RFLP) analysis, PCR followed by single-strand conformation polymorphism (SSCP) analysis, and nucleic acid sequence analysis.
As a result of the increase in the number of such studies throughout Europe and the gradual implementation of improved molecular tools, the geographic boundaries of each genospecies' distribution are continually revised. According to the latest data, Borrelia afzelii, Borrelia garinii, Borrelia valaisiana, and B. burgdorferi sensu stricto are distributed nearly throughout Europe (1, 11, 12, 14, 17, 24, 27, 28, 30, 40, 43, 47, 48, 49). Unlike the other genospecies, Borrelia lusitaniae seems to occur preferentially in the Iberian Peninsula and is only sporadically detected in other European countries (12). Furthermore, Borrelia bisettii-like spirochetes and strain A14S were isolated from erythema migrans lesions in Slovenia (46) and in The Netherlands (56), respectively.
The three main pathogenic species, B. burgdorferi sensu stricto, B. afzelii, and B. garinii, are known to cause different clinical manifestations in humans (53), and different levels of pathogenicity were recently described for distinct genetic clones within single genospecies (5).
In addition, the European genospecies are considered to be preferentially associated with different reservoir hosts (20, 21, 22, 23, 28, 29). The hypothesized origin of such specific associations is attributed to the fact that different groups of vertebrates (rodents and birds) exhibit distinct types of innate immunity, which may either destroy or tolerate certain Borrelia genospecies (29, 30). However, other observations seem to disagree with these hypotheses (11, 24, 37, 42), implying that within Borrelia genospecies, the interactions with different vertebrate hosts may be more diverse than expected.
Because genetic differences appear to be linked with distinct ecological, epidemiological, and clinical features, it is necessary to develop rapid identification methods in order to identify spirochetes at the intraspecific level. PCR-RFLP analysis of the 5S (rrfA)-23S (rrlB) intergenic spacer sequence is one of the most widely used and rapid methods for classifying Borrelia strains into genospecies (39). This method has the advantage of allowing rapid screening of a large number of samples in epidemiological and medical surveys, but it differentiates strains only at the genospecies level. SSCP analysis is a relatively simple technique which can be used for rapid screening of minimal sequence variations (16, 36, 50). The main aim of this study was to develop and optimize SSCP analysis of the highly variable rrfA-rrlB intergenic spacer of B. burgdorferi sensu lato in order to establish the extent of genetic heterogeneity among and within the genospecies of the B. burgdorferi sensu lato complex detected in I. ricinus ticks collected in the southern part of the Czech Republic.
MATERIALS AND METHODS
Tick collection.
In October 2000, 374 I. ricinus ticks (44 males, 68 females, and 262 nymphs) were collected by flagging the vegetation in a temperate forested area around České Budějovice (48°59′5"N, 14°28′5"E) in the Czech Republic. The ticks were immediately stored in 70% ethanol until DNA was extracted.
DNA extraction.
DNA was extracted from ticks by using a DNeasy tissue kit (Qiagen, Valencia, Calif.) and a previously described modified protocol (6). Immediately prior to extraction, the ticks were vacuum dried for 30 min. Each sample was cut with a disposable sterile scalpel, and proteins were degraded overnight at 56°C in 180 μl of ATL buffer (Qiagen) and 20 μl of a proteinase K solution (14 mg/ml; Boehringer Mannheim, Indianapolis, Ind.). The remaining extraction steps were carried out according to the manufacturer's protocol. The DNA was eluted in 50 μl of deionized water and stored at 4°C.
PCR.
A MasterTaq DNA polymerase kit (Eppendorf, Westbury, N.Y.) was used for all PCRs. A total of 2.5 μl of template DNA was added to a PCR master mixture containing 10.4 μl of deionized water, 5 μl of 5× TaqMaster PCR enhancer, 2.5 μl of 10× Taq buffer (with 15 mM Mg2+), 1.5 μl of a 25 mM solution of magnesium acetate, 0.1 μl of a Taq DNA polymerase solution (5 U/μl), 0.5 μl of a deoxynucleoside triphosphate mixture (containing each deoxynucleoside triphosphate at a concentration of 10 mM) (Eppendorf), and 1.25 μl of a solution of each primer (10 pmol/μl) (Invitrogen, Frederick, Md.). When PCR products were needed for RFLP analysis, the total volume of the PCR mixtures was doubled to 50 μl. Primer sets used in this study are listed in Table 1.
TABLE 1.
Primers used in this study
| Gene | Primer (use) | Sequence (5′-3′) | Reference | Length (bp) |
|---|---|---|---|---|
| Cytochrome b (ticks) | cytb1 (PCR) | TTT TAG CAA TAA ACT TTT CAA G | 620 | |
| cytb2 (PCR) | AAT AAA AAA TAT CAT TCT GG | |||
| rrfA-rrlB intergenic spacer | IGSa (PCR and sequencing) | CGA CCT TCT TCG CCT TAA AGC | ≈222-255 | |
| IGSb (PCR and sequencing) | AGC TCT TAT TCG CTG ATG GTA | |||
| 16S rDNA | 16Sf1 (PCR and sequencing) | ATA ACG AAG AGT TTG ATC CTG GC | 34 | 1,350 |
| 16Sr (PCR and sequencing) | CAG CCG CAC TTT CCA GTA CG | 34 | ||
| 16Sfin1 (sequencing) | GGA ACA CCT CAC AGC ACG AGC TGA | |||
| 16Sfin2 (sequencing) | AAA TGA CAA GGT GAT GAC GTT AAA | |||
| Flagellin | Cf (PCR and sequencing) | GCA GTT CAA TCA GGT AAC GG | 13 | ≈550-580 |
| Dr (PCR and sequencing) | AGG TTT TCA ATA GCA TAC TC | 13 |
In order to verify that DNA had been successfully extracted from each specimen, a ∼620-bp fragment of the mitochondrial cytochrome b gene was amplified from each DNA sample. Primers cytb1 and cytb2 for conserved regions of the cytochrome b genes of Ixodes hexagonus and Rhipicephalus sanguineus (7) were used. The reaction mixtures were subjected to 35 thermal cycles consisting of 94°C for 15 s, 45°C for 50 s, and 65°C for 90 s. The PCR products were electrophoresed on a 1% agarose gel, stained with ethidium bromide, and visualized with a UV transilluminator.
Only positive samples were examined further for the presence of B. burgorferi sensu lato by amplifying a portion of the rrfA-rrlB intergenic spacer. Primers IGSa and IGSb were designed by comparing all homologous nucleotide sequences found in the GenBank database and then selecting two appropriately conserved regions. Each sample was subjected to a touchdown PCR program consisting of five cycles of 94°C for 15 s, 61°C (with the temperature decreasing 0.2°C/cycle) for 25 s, and 72°C for 30 s, followed by five cycles of 94°C for 15 s, 60°C (with the temperature decreasing 0.4°C/cycle) for 25 s, and 72°C for 30 s and then 24 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s.
One of the positive samples, sample I-77, produced unique RFLP and SSCP profiles (see below). To further characterize this spirochete, we also amplified fragments of its 16S rRNA and flagellin genes for sequencing. A fragment of the flagellin gene of I-77 was amplified by using primers Cf and Dr (13). The PCR samples were subjected to 35 cycles, each consisting of 15 s at 94°C, 30 s at 62°C, and 30 s at 72°C. The 16S rRNA gene was amplified with primers 16Sf1 and 16Sr (34). The reactions consisted of 35 thermal cycles of 94°C for 15 s, 64°C for 20 s, and 72°C for 80 s.
PCR samples amplified for sequencing, SSCP, and RFLP analyses were purified by using a QIAquick PCR purification kit (Qiagen), eluted in 25 μl of deionized water, and stored at 4°C.
RFLP analysis.
For each positive sample, 13 μl of amplified DNA (intergenic spacer) was digested at 37°C for 9 h in a solution containing 0.5 μl of MseI (10,000 U/ml; New England Biolabs, Beverly, Mass.) and 1.5 μl of 10× NE buffer 2 (New England Biolabs). The enzyme was heat inactivated at 65°C for 20 min. Electrophoresis was carried out by using 4 to 20% Novex TB gradient gels (Invitrogen, Carlsbad, Calif.) at 150 V for 3 h. The gels were stained with SYBR Gold nucleic acid gel stain (Molecular Probes, Eugene, Oreg.) for 20 min, and bands were visualized with a UV transilluminator. At least two representative samples of each distinct profile were sequenced (see below).
SSCP analysis.
Different running temperatures (5, 15, and 20°C), running times (2, 3, and 4 h), and gel concentrations (4 to 20% polyacrylamide gradient gel and 20% polyacrylamide gel) were tested in order to optimize SSCP conditions. Five microliters of each purified PCR product (intergenic spacer) was mixed with 12.6 μl of 1.25× Tris-borate-EDTA buffer (Eppendorf), 0.4 μl of methylmercury hydroxide (Johnson Matthey, Ward Hill, Mass.), and 2.0 μl of a solution containing 15% (wt/vol) Ficoll, 0.10% (wt/vol) bromophenol blue, and 0.10% (wt/vol) xylene cyanol in distilled water. The solutions were heated at 95°C for 4 min and immediately placed on ice. Samples were loaded on the gels and electrophoresed in a refrigerated vertical electrophoresis chamber (Novex Thermoflow; Electrophoresis Temperature Control System, San Diego, Calif.). Optimization was performed by comparing the profiles of four Borrelia genospecies (B. afzelii, B. garinii, B. burgorferi sensu stricto, and B. valaisiana) previously identified by RFLP and sequence analyses of the initial PCR product. After the SSCP conditions were optimized, all positive PCR products were grouped by genospecies according to the previously obtained RFLP profiles. Each group of SSCP samples was electrophoresed simultaneously on a single gel in order to detect minimal intraspecific differences. After electrophoresis, the gels were stained with SYBR Gold nucleic acid gel stain (Molecular Probes) for 20 min, and the profiles were visualized with a UV transilluminator. At least two representative samples of each unique profile were selected for sequencing.
Mixed infections were suspected when a profile was characterized by more than two single-strand fragments. In these cases, a small portion of each band was punched out separately with a 200-μl pipette tip. This small portion of gel was placed directly into 22.5 μl of PCR master mixture containing primers IGSa and IGSb, and the nucleic acid fragment was reamplified (see above). The PCR products obtained after reamplification of each band were sequenced separately.
Sequencing and sequence analysis.
Sequencing was performed at the W. M. Keck Foundation Biotechnology Resource Laboratory (Yale University). For the intergenic spacer and the flagellin amplicons, both DNA strands were sequenced with the same primers that were used for the initial PCR. After the two extremities of the 16S rRNA gene of I-77 were sequenced with primers 16Sf1 and 16Sr, the central part of the sequence was completed by using primers 16Sfin1 and 16Sfin2 (Table 1).
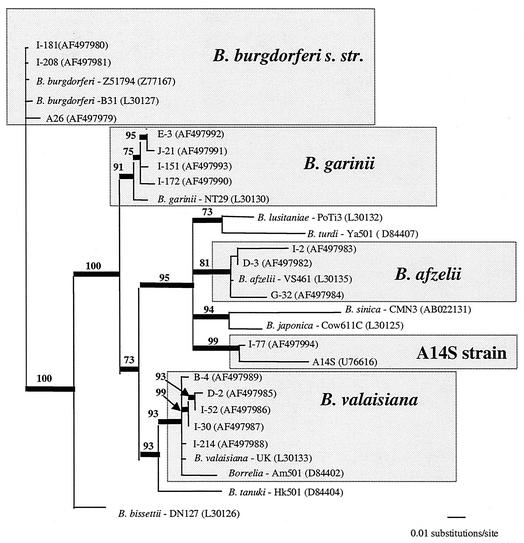
The complementary strands of each sequenced product were assembled by using SeqMan (Lasergene DNA*, Madison, Wis.). When ambiguities could not be solved by assembling the two strands, the PCR was repeated and the strands were sequenced again. For I-77, the flagellin fragment, the intergenic spacer region, and the 16S ribosomal DNA (rDNA) fragment were compared to GenBank entries by performing BLAST searches (Wisconsin Package; Genetics Computer Group, Madison, Wis.) (2). Representatives of each B. burgdorferi sensu lato genospecies were selected from the GenBank database, and their intergenic spacer sequences were used for inferring phylogenies. The GenBank accession numbers of the homologous sequences used and of our genotype sequences are indicated in Fig. 3. Homologous sequences were aligned by using the CLUSTAL X multiple-sequence alignment program (version 1.81) (52). We used a gap creation penalty of 6 and a gap extension penalty of 5 because these intergenic spacer sequences are characterized by frequent indels of variable lengths. Uncorrected “p” pairwise distances were evaluated by using PAUP* 4.0 (51). Phylogenetic reconstructions with the intergenic spacer sequences were inferred by using MrBayes 2.01 (19). Each Markov chain (four simultaneous chains, three cold and one heated) was started from a random tree and run for 200,000 cycles, and trees were sampled every 100th cycle. Among-site variation rates were set to gamma distribution. We used the general time-reversible model of nucleotide substitution. The initial samples, obtained before the likelihood values stabilized, were discarded. The tree in Fig. 3 represents the 50% majority rule consensus (PAUP*) for the remaining sampled trees. The reliability values were the percentages of times that given lineages occurred among the sampled trees. For comparison, a maximum-likelihood tree was inferred from the data set by using PAUP*. A maximum-parsimony tree was used as the starting point for the maximum-likelihood heuristic search and for preliminary estimates of the substitution model (general time reversible), base frequencies, proportions of invariable sites, and among-site variation rates. B. burgdorferi sensu stricto genotypes were used as outgroups for the intergenic spacer tree.
FIG. 3.
Fifty percent majority rule consensus trees resulting from Bayesian analysis of an alignment of the intergenic spacer (269 bp) of representatives of the major Borrelia genospecies and our genotypes. GenBank accession numbers are indicated in parentheses. B. burgdorferi sensu stricto (B. burgdorferi s. str.) genotypes were used as outgroups. The thick lines indicate branches supported in ≥70% of the trees sampled. The reliability values shown at the nodes are the percentages of times that lineages occurred in the trees sampled.
Statistical analysis.
Differences in the prevalence of B. burgdorferi sensu lato among females, males, and nymphs were compared by using the chi-square test. A P value of <0.05 was considered statistically significant.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the nucleotide sequences of the intergenic spacers are as follows: AF497979 for B. burgdorferi sensu stricto genotype A-26, AF497980 for B. burgdorferi sensu stricto genotype I-181, AF497981 for B. burgdorferi sensu stricto genotype I-208, AF497982 for B. afzelii genotype D-3, AF497983 for B. afzelii genotype I-2, AF497984 for B. afzelii genotype G-32, AF497985 for B. valaisiana genotype D-2, AF497986 for B. valaisiana genotype I-52, AF497987 for B. valaisiana genotype I-30, AF497988 for B. valaisiana genotype I-214, AF497989 for B. valaisiana genotype B-4, AF497990 for B. garinii genotype I-172, AF497991 for B. garinii genotype J-21, AF497992 for B. garinii genotype E-3, AF497993 for B. garinii genotype I-151, and AF497994 for genotype I-77. The GenBank accession numbers for the nucleotide sequences of the flagellin gene and the 16S rDNA gene of genotype I-77 are AF497995 and AF497996, respectively.
RESULTS
DNA extraction and PCR.
The cytochrome b primers detected DNA in 370 of 374 ticks examined. Therefore, only these 370 ticks (68 females, 41 males, and 261 nymphs) were analyzed further for the presence of Borrelia by using the IGSa and IGSb primers. The intergenic spacer was amplified in 76 (20.5%) of them. The highest prevalence was found for female ticks (30%), and the values were lower for males (19.5%) and nymphs (18%) (Table 2). The differences in prevalence were found to be significant only between nymphs and females (P ≤ 0.025; df = 1), whereas the differences were not significant between nymphs and males (P ≤ 1; df = 1), between males and females (P ≤ 0.2; df = 1), or between nymphs and total adults (P ≤ 0.1; df = 1).
TABLE 2.
Variability within B. burgdorferi sensu lato genospecies detected by PCR-SSCP and PCR-RFLP analyses
| Stage or gender | Method | Total no. of ticks examined | No. of positive ticks (%) | No. of ticks positive for genospecies (% of positive ticks)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B. afzelii | B. garinii | B. valai- siana | B. burg- dorferi sensu stricto | B. afzelii and B. burgderferi sensu stricto | B. valaisiana and B. garinii | B. garinii and B. garinii | I-77 | ||||
| Nymphs | SSCP | 261 | 47 (18.0) | 27 (57.4) | 9 (19.1)a | 5 (10.6) | 2 (4.2) | 0 (0) | 2 (4.2) | 1 (2.1) | 1 (2.1) |
| Nymphs | RFLP | 261 | 47 (18.0) | 27 (57.4) | 10 (21.3) | 5 (10.6) | 2 (4.2) | 0 (0) | 2 (4.2) | 0 (0) | 1 (2.1) |
| Females | SSCP | 68 | 21 (30.0) | 10 (47.6) | 3 (14.3) | 2 (9.5) | 2 (9.5) | 3 (14.3) | 0 (0) | 0 (0) | 0 (0) |
| Females | RFLP | 68 | 21 (30.0) | 10 (47.6) | 3 (14.3) | 2 (9.5) | 3 (14.3) | 2 (9.5) | 0 (0) | 0 (0) | 0 (0) |
| Males | SSCP | 41 | 8 (19.5) | 6 (75.0) | 1 (12.5) | 0 (0) | 1 (12.5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Males | RFLP | 41 | 8 (19.5) | 6 (75.0) | 1 (12.5) | 0 (0) | 1 (12.5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Total | SSCP | 370 | 76 (20.5) | 43 (56.5) | 13 (17.1) | 7 (9.2) | 5 (6.5) | 3 (3.9) | 2 (2.6) | 1 (1.3) | 1 (1.3) |
| Total | RFLP | 370 | 76 (20.5) | 43 (56.5) | 14 (18.4) | 7 (9.2) | 6 (7.9) | 2 (2.6) | 2 (2.6) | 0 (0) | 1 (1.3) |
Boldface type indicates that the results obtained by PCR-SSCP analysis and the results obtained by PCR-RFLP analysis are different.
PCR-RFLP analysis.
RFLP analysis of the amplified products resulted in characterization of five distinct profiles (Table 2). Combined profiles, corresponding to double infections, were observed in four tick samples. After two representatives of each profile were sequenced, the sequences were compared to homologous DNA fragments in the GenBank database, and each profile was classified accordingly into a genospecies. Of the 76 positive ticks, 43 (56.5%) were infected with B. afzelii, 14 (18.4%) were infected with B. garinii, 7 (9.2%) were infected with B. valaisiana, 6 (7.9%) were infected with B. burgdorferi sensu stricto, 2 (2.6%) were infected simultaneously with B. garinii and B. valaisana, and 2 (2.6%) were infected simultaneously with B. afzelii and B. burgdorferi sensu stricto (Table 2). A unique profile (tick I-77), different from all other profiles, went unnoticed because it was very similar to a B. afzelii-like profile. Minimal differences were detected only after the I-77 sample was electrophoresed again and for a longer time on an RFLP gel. The distributions of the different genospecies between genders did not differ significantly.
PCR-SSCP analysis.
The optimal conditions for PCR-SSCP electrophoresis were 15°C, a 20% Tris-borate-EDTA gel, and 4 h. The PCR-SSCP results largely confirmed the RFLP results in terms of genospecies identification (Table 2). There were, however, a few exceptions. One tick (tick D-5), which was found to be infected with B. burgdorferi sensu stricto by RFLP analysis, proved to be infected with two different strains (B. burgdorferi sensu stricto and B. afzelii) when it was reanalyzed by the PCR-SSCP method. Similarly, a nymphal tick that was infected with B. garinii as determined by RFLP analysis proved to be infected with two different B. garinii strains when it was tested by the SSCP method (Tables 2 and 3).
TABLE 3.
Intraspecific variability of B. burgdorferi sensu lato as detected by PCR-SSCP analysis
| Taxon(a) | Genotype(s) | No. of infected ticks
|
|||
|---|---|---|---|---|---|
| Nymphs | Females | Males | Total | ||
| B. burgdorferi sensu stricto | I-26 | 0 | 1 | 0 | 1 |
| I-181 | 1 | 0 | 0 | 1 | |
| I-208 | 1 | 2 | 1 | 4 | |
| B. afzelii | D-3 | 22 | 10 | 5 | 37 |
| I-2 | 5 | 0 | 0 | 5 | |
| G-32 | 0 | 0 | 1 | 1 | |
| B. afzelii and B. burg- dorferi sensu stricto | I-2 and I-208 | 0 | 1 | 0 | 1 |
| D-3 and I-208 | 0 | 1 | 0 | 1 | |
| I-2 and A-26 | 0 | 1 | 0 | 1 | |
| B. valaisiana | D-2 | 0 | 1 | 0 | 1 |
| I-52 | 3 | 0 | 0 | 3 | |
| I-30 | 1 | 0 | 0 | 1 | |
| I-214 | 1 | 0 | 0 | 1 | |
| B-4 | 0 | 1 | 0 | 1 | |
| B. garinii | I-172 | 1 | 0 | 0 | 1 |
| J-21 | 2 | 0 | 0 | 2 | |
| E-3 | 1 | 3 | 1 | 5 | |
| I-151 | 5 | 0 | 0 | 5 | |
| B. garinii and B. valai- siana | I-214 and E-3 | 1 | 0 | 0 | 1 |
| I-214 and I-151 | 1 | 0 | 0 | 1 | |
| B. garinii and B. garinii | J-21 and E-3 | 1 | 0 | 0 | 1 |
| I-77 | I-77 | 1 | 0 | 0 | 1 |
As determined by SSCP analysis, the Borrelia strains detected were subdivided into 16 different profiles (Table 3), which we call genotypes in this paper. Positive samples that were identified by RFLP analysis as B. burgdorferi sensu stricto were subdivided into three distinct SSCP profiles. The 43 samples characterized by a B. afzelii RFLP profile corresponded to three different SSCP profiles, and one of them (sample D-3) was the most prevalent strain in our overall sample. The seven ticks infected with B. valaisiana were subdivided into five different SSCP profiles. B. garinii, found in 14 specimens, was represented by four different SSCP profiles. One nymph was simultaneously infected with two B. garinii strains (genotypes J-21 and E-3). Other mixed infections were found in three females that were simultaneously infected with B. burgdorferi sensu stricto and B. afzelii and in two nymphs that were simultaneously infected with B. garinii and B. valaisiana. One positive nymph (tick I-77) had a unique SSCP profile. Its RFLP profile was considered to be identical to that of B. afzelii at first. After the SSCP data were analyzed and the amplicon was sequenced, it became clear that this strain was different from all other strains. Examples of PCR-SSCP profiles are shown in Fig. 1 and 2.
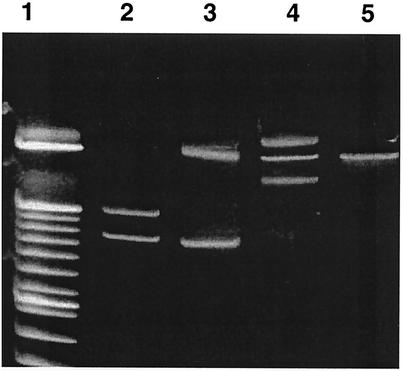
FIG. 1.
PCR-SSCP profiles of the intergenic spacers of B. valaisiana genotype I-52 (lane 2), B. garinii genotype J-21 (lane 3), a mixed infection with B. burgdorferi sensu stricto genotype I-208 and B. afzelii genotype D-3 (lane 4), and B. afzelii genotype D-3 (lane 5). Lane 1 contained a 10-bp ladder marker.
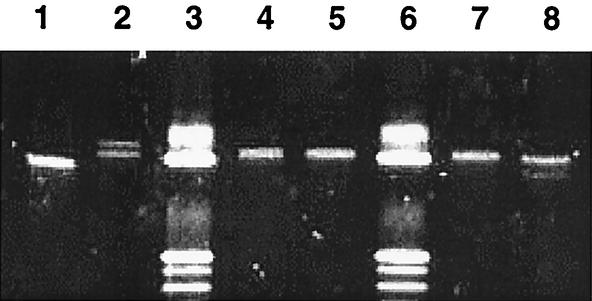
FIG. 2.
PCR-SSCP profiles of genotype I-77 (lane 1), B. afzelii genotype I-2 (lane 2), B. afzelii genotype D-3 (lanes 4, 5, and 7), and B. afzelii genotype G-32 (lane 8). Lanes 3 and 6 contained a 10-bp ladder marker.
In our overall sample, when we included specimens infected simultaneously with two spirochetes, B. afzelii, B. garinii, B. valaisiana, B. burgdorferi sensu stricto, and strain I-77 were found in 12.4, 2.4, 4.6, 2.2, and 0.3% of the questing nymphal and adult ticks, respectively.
Sequence and phylogenetic analyses.
All intergenic spacer sequences corresponding to identical SSCP profiles were found to be identical, and none of the identical sequences corresponded to a different profile. The lengths of our amplicons varied from 222 to 256 bp. The representative nucleotide sequences of the B. afzelii SSCP profiles differed by 7 to 14 bp, the representative nucleotide sequences of the B. burgdorferi sensu stricto SSCP profiles differed by 1 to 4 bp, the representative nucleotide sequences of the B. garinii SSCP profiles differed by 2 to 5 bp, and the representative nucleotide sequences of the B. valaisiana SSCP profiles differed by 1 to 13 bp (indels included). When the sequences were aligned with other homologous sequences from the GenBank database, the total alignment was 269 bp long. The uncorrected p pairwise distance differences varied from 0.01 to 11.4%. The B. burgdorferi sensu stricto genotypes differed by 0.01 to 1.2%, the B. afzelii genotypes differed by 0.4 to 5.3%, the B. garinii genotypes differed by 0.4 to 1.9%, and the B. valaisiana genotypes differed by 0.4 to 1.2%. Genotype I-77 differed from its closest relative, A14S, by 4%.
The pairwise uncorrected p distances between the 16S rDNA of I-77 and the 16S rDNA of other B. burgdorferi sensu lato species varied from 0.7% (B. afzelii) to 2.0% (B. burgdorferi sensu stricto). A14S was found to be the most closely related strain (0.3%). The difference between the I-77 and A14S sequences indicates that these two strains belong to the same genospecies. Indeed, the distance values for members of single genospecies varied from 0.4% (B. afzelii strains) to 0.5% (B. garinii strains). We did not use the 16S rDNA data set for phylogenetic analysis, because the low variability of Borrelia sequences does not provide sufficient resolution within B. burgdorferi sensu lato taxa, as has been repeatedly demonstrated (34, 41, 44).
When the I-77 flagellin sequence was compared with the sequences of homologous representatives of all recognized Borrelia sensu lato genospecies, the uncorrected p distance values varied from 5.5% (Borrelia tanuki or B. afzelii) to 7.1% (Borrelia andersoni). These distance values are higher than those observed between recognized species (2.9% for Borrelia turdi and B. tanuki, 4.7% for B. afzelii and B. valaisiana, or 4.9% for B. garinii and B. afzelii). The p values were higher between B. burgdorferi sensu lato and relapsing fever group spirochetes (14.4 to 18.8%). The flagellin sequence of A14S is unfortunately not available in the GenBank database, and therefore, we could not determine whether this organism is also the closest relative of I-77.
The phylogenetic tree inferred from the intergenic spacer data shows how our genotypes clustered with previously recognized genotypes (Fig. 3).
DISCUSSION
There is increasing evidence that spirochetes belonging to a single genospecies are more heterogeneous than previously expected in terms of their ecology and the clinical manifestations which they cause. Thus, the main aim of this study was to develop simple detection and identification methods to differentiate genotypes within Borrelia genospecies.
As determined by PCR, the overall prevalence of infection in our sample (20.5%) is consistent with that reported for neighboring Germany (18.1%) (30) but lower than that reported for a geographically close Slovak area (40 to 49%) (14, 30). Our results further confirm the fact that prevalence values are very variable even over relatively small geographical areas. The overall prevalence in adults (26.7%) and the overall prevalence in females (30%) were found to be higher than the prevalence in nymphs (18%), which is in agreement with the general pattern of increasing Borrelia prevalence through the life stages of ticks (18). However, the infection prevalence in males (19.5%) did not appear to follow the same pattern as the infection prevalence in females, but the infection rate was comparable to the infection rate in nymphs. This was most likely due to the small sample size for males. However, similar results were recently obtained in the same area by N. Rudenko and L. Grubhoffer (personal communication). In the overall sample of these workers (199 adult ticks), the infection prevalence in males was 14.4% and the infection prevalence in females was 39.8%. The differences in this case were highly significant (df = 1; P ≤ 0.001). We could, therefore, hypothesize that males may differ from females in feeding behavior. Through their life stages, males may switch more easily than females from one type of host to another (for instance, a male may feed on a bird as a larva and on a rodent as a nymph). A spirochete acquired during the first blood meal may be cleared from the tick during the second blood meal by activation of the alternative pathway of rodent complement (23, 29). The sources of several blood meals can be determined in adult ticks by amplifying fragments of cytochrome b of the vertebrate hosts (26). It would certainly be worthwhile to establish whether female and male I. ricinus ticks have the same feeding behavior.
Our PCR-RFLP results show that B. afzelii is the predominant genospecies in the region studied, which is in agreement with some previous reports from Slovakia and Germany (14, 30). This is also the first time that the occurrence of B. valaisiana has been reported in this geographical area. In a single study the workers (48) reported that B. burgdorferi sensu stricto was the prevalent genospecies in the same area of the Czech Republic. However, these results were obtained by isolating spirochetes in BSK medium before they were identified by Western blot analysis. It is likely that isolation techniques preferentially select Borrelia strains (38) which are better adapted than other strains to in vitro growth conditions. Therefore, we believe that these results are not comparable to ours.
The human pathogens, B. burgdorferi sensu stricto, B. afzelii, and B. garinii, are known to be associated with different clinical forms of Lyme disease. B. burgdorferi sensu stricto predominantly causes musculoskeletal disorders, B. afzelii causes cutaneous manifestations, and B. garinii causes neurological disorders (5, 53, 57). In the Czech Republic, a recent clinical survey showed that 65% of the Lyme disease patients had skin lesions, 12% suffered from neuroborreliosis, and 9% had musculoskeletal disorders (D. Janovská, B. Macková, M. Vondrová, and D. Hulínska, Abstr. Tick-Borne Infect. Dis. Other Zoonoses, p. 19, 2001). It is interesting that this pattern is similar to the pattern of our prevalence data for ticks (59.2, 21.1, and 10.5%, respectively).
The frequency of multiple infections in I. ricinus appears to be very variable, depending on the geographical area (11, 24, 30, 37, 40). In our case, the overall prevalence of double infections in positive ticks was relatively low (6.6%) and comparable to previously reported data (8.2%) (30). We believe, however, that when one of the two infecting strains is sufficiently predominant in a tick specimen, the PCR technique fails to detect a mixed infection. The relatively low prevalence of double infections in our sample could also be explained by the habit of frequent host type switching between stages (23, 30).
The combination of B. garinii and B. valaisiana infections in questing nymphs is not surprising. These nymphs must have acquired both infections from their larval hosts, which, if theories about innate immunity are correct, were more likely to be birds, as birds are competent reservoirs for both genospecies (23, 30). B. burgdorferi sensu stricto is known to show variable levels of resistance to different complement types (30). Genotype I-208 was found in two female ticks also infected with B. afzelii, a genospecies more frequently associated with rodents. This may indicate that genotype I-208 is indeed one of the B. burgdorferi sensu stricto genotypes tolerated by rodent complement.
The PCR-SSCP method, which has been applied to analysis of a variety of genes (ospA, P66, and flagellin), has been used for detection of differences within Borrelia genotypes previously (15, 38, 45). We decided to use the rrfA-rrlB intergenic spacer, which is known to be very variable among B. burgdorferi sensu lato strains and has the advantage of having already been widely used by European workers for differentiation of genospecies by RFLP analysis (12, 14, 27, 39). Furthermore, the relatively small size of the amplified fragment (∼230 to 250 bp) is particularly suitable for SSCP analysis. By using SSCP analysis we detected and differentiated 16 different genotypes (Table 3). Advantages of SSCP analysis over RFLP analysis have been emphasized previously (50). PCR-RFLP analysis is certainly a very practical approach for rapidly screening a large data sample and therefore should not be dismissed. However, the SSCP technique requires fewer time-consuming steps and a smaller amount of PCR products (5 instead of 13 μl). In order to obtain a level of discrimination similar to that obtained with RFLP analysis, amplicons should be digested with several enzymes, and each sample should be repeatedly electrophoresed on several different gels. SSCP analysis allowed us to obtain the same amount of information by using a single 25-μl PCR mixture and a single SSCP gel, which in the long run turned out to be a much more economic method. Our results also show that single nucleotide differences can be detected by SSCP analysis; examples of this are the differences between genotypes I-181 and I-208, between genotypes I-30 and I-214, and between genotypes B-4 and I-214. Another obvious advantage of SSCP analysis is the fact that multiple infections can be easily identified by reamplifying and directly sequencing each distinctive band on the gel without having to resort to long cloning procedures. Furthermore, unlike RFLP analysis, double infections with different genotypes of a single genospecies can also be detected, as the rrfA-rrlB intergenic spacer is not a repeated fragment within the rrn ribosomal cluster (57).
PCR-SSCP analysis also helped us identify genotype I-77 as a new genotype. Indeed, the profile was initially identified as B. afzelii by RFLP analysis and could have been missed if we had not compared RFLP and SSCP results. The occurrence of genotype I-77 in a single nymphal tick in this area may have been an accidental unique finding. However, the close relationship of this strain with a recognized human pathogen, A14S, should prompt further studies of this newly described genotype (56). Intergenic spacer and 16S rDNA similarity values seem to indicate that I-77 should belong to the recently recognized A14S genospecies (56). The interest in the intergenic spacer tree results essentially from its ability to cluster genotypes into genospecies. Although most of its lineages appear to be surprisingly fairly well resolved, the global topology of the intergenic spacer tree has to be considered with some caution, because the aligned sequences are very short (269 bp) and the frequencies and lengths of indels are proportionally high.
Our results confirm recent findings concerning the clonal diversity of B. burgdorferi sensu lato (5) strains. It is evident that differentiation at the genospecies level is not sufficient to explain heterogeneous ecological and clinical findings. For instance, among the 16 genotypes, B. afzelii genotype D-3 is the most prevalent genotype in all tick life stages, which clearly suggests that this genotype has more successfully established itself in this region. Further studies should aim at identifying the ecological elements which cause the prevalence of genotype D-3 to be much higher than the prevalence of other B. afzelii genotypes. Moreover, the distribution of different genotypes over time should also be assessed. Temporal changes in genotype distribution may point to ecological changes in spirochete transmission cycles.
The PCR-SSCP technique which we developed provides us with a rapid detection and identification tool for B. burgdorferi sensu lato genotypes. This method could conveniently complement or replace other methods of analysis for large-scale surveys. It should also allow further studies to characterize more precisely relationships between single genotypes and ecological environmental conditions.
Acknowledgments
This research was sponsored by a Fulbright Fellowship to M.D and by grants to D.F. from the G. Harold and Leila Y. Mathers Charitable Foundation and from USDA-ARS (cooperative agreement 58-1265-7-002) and to B.P. from Vega (2/7214/20).
We thank Jean Tsao for reviewing the English of the manuscript and for her constructive comments.
REFERENCES
- 1.Alekseev, A. N., H. V. Dubinina, I. Van De Pol, and L. M. Schouls. 2001. Identification of Ehrlichia spp. and Borrelia burgdorferi in Ixodes ticks in the Baltic regions of Russia. J. Clin. Microbiol. 39:2237-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. F. 1989. Epizootiology of Borrelia in Ixodes tick vectors and reservoir hosts. Rev. Infect. Dis. 11:1451-1459. [DOI] [PubMed] [Google Scholar]
- 4.Baranton, G., D. Postic, I. Saint Girons, P. Boerlin, J. C. Piffaretti, M. Assous, and P. A. Grimont. 1992. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int. J. Syst. Bacteriol. 42:378-383. [DOI] [PubMed] [Google Scholar]
- 5.Baranton, G., G. Seinost, G. Theodore, D. Postic, and D. Dykhuizen. 2001. Distinct levels of genetic diversity of Borrelia burgdorferi are associated with different aspects of pathogenicity. Res. Microbiol. 152:149-156. [DOI] [PubMed] [Google Scholar]
- 6.Beati, L., and J. E. Keirans. 2001. Analysis of the systematic relationships among ticks of the genera Rhipicephalus and Boophilus (Acari: Ixodidae) based on mitochondrial 12S ribosomal DNA gene sequences and morphological characters. J. Parasitol. 87:32-48. [DOI] [PubMed] [Google Scholar]
- 7.Black, W. C., and R. L. Roehrdanz. 1998. Mitochondrial gene order is not conserved in arthropods: prostriate and metastriate tick mitochondrial genomes. Mol. Biol. Evol. 15:1772-1785. [DOI] [PubMed] [Google Scholar]
- 8.Boerlin, P., O. Péter, A. G. Bretz, D. Postic, G. Baranton, and J. C. Piffaretti. 1992. Population genetic analysis of Borrelia burgdorferi isolates by multilocus enzyme electrophoresis. Infect. Immun. 60:1677-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317-1319. [DOI] [PubMed] [Google Scholar]
- 10.Canica, M. M., F. Nato, L. du Merle, J. C. Mazie, G. Baranton, and D. Postic. 1993. Monoclonal antibodies for identification of Borrelia afzelii sp. nov. associated with late cutaneous manifestations of lyme borreliosis. Scand. J. Infect. Dis. 25:441-448. [DOI] [PubMed] [Google Scholar]
- 11.Christova, I., L. Schouls, I. van de Pol, J. Park, S. Panayotov, V. Lefterova, T. Kantardjiev, and J. S. Dumler. 2001. High prevalence of granulocytic ehrlichiae and Borrelia burgdorferi sensu lato in Ixodes ricinus ticks from Bulgaria. J. Clin. Microbiol. 39:4172-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Michelis, S., H.-S. Sewell, M. Collares-Pereira, M. Santos-Reis, L. M. Schouls, V. Benes, E. C. Holmes, and K. Kurtenbach. 2000. Genetic diversity of Borrelia burgdorferi sensu lato in ticks from mainland Portugal. J. Clin. Microbiol. 38:2128-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukunaga, M., K. Okada, M. Nakao, T. Konishi, and Y. Sato. 1996. Phylogenetic analysis of Borrelia species based on flagellin gene sequences and its application for molecular typing of Lyme disease borreliae. Int. J. Syst. Bacteriol. 46:898-905. [DOI] [PubMed] [Google Scholar]
- 14.Gern, L., C. M. Hu, E. Kociánová, V. Výrosteková, and J. Reháèek. 1999. Genetic diversity of Borrelia burgdorferi sensu lato isolates obtained from Ixodes ricinus ticks collected in Slovakia. Eur. J. Epidemiol. 15:665-669. [DOI] [PubMed] [Google Scholar]
- 15.Guttman, D. S., P. W. Wang, I. N. Wang, E. M. Bosler, B. J. Luft, and D. E. Dykhuizen. 1996. Multiple infections of Ixodes scapularis ticks by Borrelia burgdorferi as revealed by single-strand conformation polymorphism analysis. J. Clin. Microbiol. 34:652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hongyo, T., G. S. Buzard, R. J. Calvert, and C. M. Weghorst. 1993. ‘Cold SSCP’: a simple, rapid and nonradioactive method for optimized single strand conformation polymorphism analyses. Nucleic Acids Res. 21:3637-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubálek, Z., and J. Halouzka. 1997. Distribution of Borrelia burgdorferi sensu lato genomic groups in Europe, a review. Eur. J. Epidemiol. 13:951-957. [DOI] [PubMed] [Google Scholar]
- 18.Hubálek, Z., and J. Halouzka. 1998. Prevalence rates of Borrelia burgdorferi sensu lato in host-seeking Ixodes ricinus ticks in Europe. Parasitol. Res. 84:167-172. [DOI] [PubMed] [Google Scholar]
- 19.Huelsenbeck, J. P., and F. R. Ronquist. 2001. MrBayes: Bayesian inference of phylogeny. Biometrics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 20.Humair, P.-F., and L. Gern. 1998. Relationship between Borrelia burgdorferi sensu lato species, red squirrels (Sciurus vulgaris) and Ixodes ricinus in enzootic areas in Switzerland. Acta Trop. 69:213-227. [DOI] [PubMed] [Google Scholar]
- 21.Humair, P.-F., D. Postic, R. Wallich, and L. Gern. 1998. An avian reservoir (Turdus merula) of the Lyme disease spirochetes. Zentralbl. Bakteriol. 287:521-538. [PubMed] [Google Scholar]
- 22.Humair, P.-F., O. Rais, and L. Gern. 1999. Transmission of Borrelia afzelii from Apodemus mice and Clethrionomys voles to Ixodes ricinus ticks: differential transmission pattern and overwintering maintenance. Parasitology 118:33-42. [DOI] [PubMed] [Google Scholar]
- 23.Humair, P.-F., and L. Gern. 2000. The wild hidden face of Lyme borreliosis in Europe. Microbes Infect. 2:915-922. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins, A., B. E. Kristiansen, A. G. Allum, R. K. Aakre, L. Strand, E. J. Kleveland, I. van de Pol, and L. Schouls. 2001. Borrelia burgdorferi sensu lato and Ehrlichia spp. in Ixodes ticks from southern Norway. J. Clin. Microbiol. 39:3666-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, R. C., G. P. Schmid, F. W. Hyde, A. G. Steigerwalt, and D. J. Brenner. 1984. Borrelia burgdorferi sp. nov.: etiologic agent of Lyme disease. Int. J. Syst. Bacteriol. 34:496-497. [Google Scholar]
- 26.Kirstein, F., and J. S. Gray. 1996. A molecular marker for the identification of the zoonotic reservoirs of Lyme borreliosis by analysis of the blood meal in its European vector, Ixodes ricinus. Appl. Environ. Microbiol. 62:4060-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirstein, F., S. Rijpkema, M. Molkenboer, and J. S. Gray. 1997. The distribution and prevalence of B. burgdorferi genomospecies in Ixodes ricinus ticks in Ireland. Eur. J. Epidemiol. 13:67-72. [DOI] [PubMed] [Google Scholar]
- 28.Kurtenbach, K., M. Peacey, S. G. T. Rijpkema, A. N. Hoodless, P. A. Nuttall, and S. E. Randolph. 1998. Differential transmission of the genospecies of Borrelia burgdorferi sensu lato by game birds and small rodents in England. Appl. Environ. Microbiol. 64:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurtenbach, K., H. S. Sewell, N. H. Ogden, S. E. Randolph, and P. A. Nuttall. 1998. Serum complement sensitivity as a key factor in Lyme disease ecology. Infect. Immun. 66:1248-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurtenbach, K., S. De Michelis, H. S. Sewell, S. Etti, S. M. Schafer, R. Hails, M. Collares-Pereira, M. Santos-Reis, K. Hanincová, M. Labuda, A. Bormane, and M. Donaghy. 2001. Distinct combinations of Borrelia burgdorferi sensu lato genospecies found in individual questing ticks from Europe. Appl. Environ. Microbiol. 67:4926-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Fleche, A., D. Postic, K. Girardet, O. Péter, and G. Baranton. 1997. Characterization of Borrelia lusitaniae sp. nov. by 16S ribosomal sequence analysis. Int. J. Syst. Bacteriol. 47:921-925. [DOI] [PubMed] [Google Scholar]
- 32.Liveris, D., G. P. Wormser, J. Nowakowski, R. Nadelman, S. Bittker, D. Cooper, S. Varde, F. H. Moy, G. Forseter, C. S. Pavia, and I. Schwartz. 1996. Molecular typing of Borrelia burgdorferi from Lyme disease patients by PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 34:1306-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marconi, R. T., L. Lubke, W. Hauglum, and C. F. Garon. 1992. Species-specific identification of and distinction between Borrelia burgdorferi genomic groups by using 16S rRNA-directed oligonucleotide probes. J. Clin. Microbiol. 30:628-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuzawa, T., T. Fukui, M. Miyake, H. B. Oh, M. K. Cho, W. H. Chang, Y. Imai, and Y. Yanagihara. 1999. Determination of members of a Borrelia afzelii-related group isolated from Ixodes nipponensis in Korea as Borrelia valaisiana. Int. J. Syst. Bacteriol. 49:1409-1415. [DOI] [PubMed] [Google Scholar]
- 35.Masuzawa, T., N. Takada, M. Kudeken, T. Fukui, Y. Yano, F. Ishiguro, Y. Kawamura, Y. Imai, and T. Ezaki. 2001. Borrelia sinica sp. nov., a lyme disease-related Borrelia species isolated in China. Int. J. Syst. E vol. Microbiol. 51:1817-1824. [DOI] [PubMed] [Google Scholar]
- 36.Michaud, J., L. C. Brody, G. Steel, G. Fontaine, L. S. Martin, D. Valle, and G. Mitchell. 1992. Strand separating conformational polymorphism analysis: efficacy of detection of point mutations in the human ornithine-aminotransferase gene. Genomics 13:389-394. [DOI] [PubMed] [Google Scholar]
- 37.Misonne, M. C., G. van Impe, and P. P. Hoet. 1998. Genetic heterogeneity of Borrelia burgdorferi sensu lato in Ixodes ricinus ticks collected in Belgium. J. Clin. Microbiol. 36:3352-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norris, D. E., B. J. Johnson, J. Piesman, G. O. Maupin, J. L. Clark, and W. C. Black. 1997. Culturing selects for specific genotypes of Borrelia burgdorferi in an enzootic cycle in Colorado. J. Clin. Microbiol. 35:2359-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Postic, D., M. V. Assous, P. A. D. Grimont, and G. Baranton. 1994. Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf (5S)-rrl (23S) intergenic spacer amplicons. Int. J. Syst. Bacteriol. 44:743-752. [DOI] [PubMed] [Google Scholar]
- 40.Postic, D., E. Korenberg, N. Gorelova, Y. V. Kovalevski, E. Bellenger, and G. Baranton. 1997. Borrelia burgdorferi sensu lato in Russia and neighboring countries: high incidence of mixed isolates. Res. Microbiol. 148:691-702. [DOI] [PubMed] [Google Scholar]
- 41.Postic, D., N. M. Ras, R. S. Lane, M. Hendson, and G. Baranton. 1998. Expanded diversity among California Borrelia isolates and description of Borrelia bissetii sp. nov. (formerly Borrelia group DN127). J. Clin. Microbiol. 36:3497-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richter, D., S. Endepols, A. Ohlenbusch, H. Eiffert, A. Spielman, and F. R. Matuschka. 1999. Genospecies diversity of Lyme disease spirochetes in rodent reservoir. Emerg. Infect. Dis. 5:291-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saint Girons, I., L. Gern, J. S. Gray, E. C. Guy, E. Korenberg, P. A. Nuttall, S. G. Rijpkema, A. Schonberg, G. Stanek, and D. Postic. 1998. Identification of Borrelia burgdorferi sensu lato species in Europe. Zentralbl. Bakteriol. 287:190-195. [DOI] [PubMed] [Google Scholar]
- 44.Scoles, G., M. Papero, L. Beati, and D. Fish. 2001. A relapsing group spirochete transmitted by Ixodes scapularis ticks. Vector Borne Zoonot. Dis. 1:21-34. [DOI] [PubMed] [Google Scholar]
- 45.Šitum, M., B. Grahovac, S. Markovic, S. Lipozencic, G. Poje, I. Dobric, B. Marinovic, S. Bolanca-Bumber, and L. Misic-Majerus. 2000. Detection and genotyping of Borrelia burgdorferi sensu lato by polymerase chain reaction. Croat. Med. J. 41:47-53. [PubMed]
- 46.Srle, F., R. N. Picken, Y. Cheng, Y. Cimperman, V. Maraspin, S. Lotric-Furlan, E. Ruzic-Sabljic, and M. M. Picken. 1997. Clinical findings for patients with Lyme borreliosis caused by Borrelia burgdorferi sensu lato with genotypic and phenotypic similarities to strain 25015. Clin. Infect. Dis. 25:273-289. [DOI] [PubMed] [Google Scholar]
- 47.Stanczak, J., B. Kubica-Biernat, M. Racewicz, W. Kruminis-Lozowska, and J. Kur. 2000. Detection of three genospecies of Borrelia burgdorferi sensu lato in Ixodes ricinus ticks collected in different regions of Poland. Int. J. Microbiol. 290:559-566. [DOI] [PubMed] [Google Scholar]
- 48.Štepánová-Tresová, G., J. Kopecký, and M. Kuthejlová. 1999. Identification of Borrelia burgdorferi sensu str., Borrelia garinii and Borrelia afzelii in Ixodes ricinus ticks from southern Bohemia using monoclonal antibodies. Zentralbl. Bakteriol. 289:797-806. [DOI] [PubMed] [Google Scholar]
- 49.Štepánová-Tresová, G., B. Pet'ko, A. Štefančíiková, and D. Nadzamová. 2000. Occurrence of Borrelia burgdorferi sensu str., Borrelia garinii and Borrelia afzelii in the Ixodes ricinus ticks from eastern Slovakia. Eur. J. Epidemiol. 16:105-109. [DOI] [PubMed] [Google Scholar]
- 50.Sunnucks, P., A. C. C. Wilson, L. B. Beheregaray, K. Zenger, J. French, and A. C. Taylor. 2000. SSCP is not so difficult: the application and utility of single-stranded conformation polymorphism in evolutionary biology and molecular ecology. Mol. Ecol. 9:1699-1710. [DOI] [PubMed] [Google Scholar]
- 51.Swofford, D. L. 2000. PAUP*: phylogenetic analysis using parsimony and other methods. Sinauer Associates, Sunderland, Mass.
- 52.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX-Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Dam, A. P., H. Kuiper, K. Vos, A. Widjojokusumo, B. M. de Jongh, L. Spanjaard, A. C. Ramselaar, M. D. Kramer, and J. Dankert. 1993. Different genospecies of Borrelia burgdorferi are associated with distinct clinical manifestations of Lyme borreliosis. Clin. Infect. Dis. 17:708-717. [DOI] [PubMed] [Google Scholar]
- 54.Wang, G., A. P. van Dam, A. Le Fleche, D. Postic, O. Péter, G. Baranton, R. de Boer, L. Spanjaard, and D. Dankert. 1997. Genetic and phenotypic analysis of Borrelia valaisiana sp. nov. (Borrelia genomic groups VS116 and M19). Int. J. Syst. Bacteriol. 47:926-932. [DOI] [PubMed] [Google Scholar]
- 55.Wang, G., A. P. van Dam, L. Spanjaard, and J. Dankert. 1998. Molecular typing of Borrelia burgdorferi sensu lato by randomly amplified polymorphic DNA fingerprinting analysis. J. Clin. Microbiol. 36:768-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, Q., A. P. van Dam, and J. Dankert. 1999. Phenotypic and genetic characterization of a novel Borrelia burgdorferi sensu lato isolate from a patient with Lyme borreliosis. J. Clin. Microbiol. 37:3025-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, Q., A. P. van Dam, I. Schwartz, and J. Dankert. 1999. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin. Microbiol. Rev. 12:633-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilske, B., V. Preaç-Mursic, U. B. Göbel, B. Graf, S. Jauris, E. Soutschek, E. Schwab, and G. Zumstein. 1993. An OspA serotyping system for Borrelia burgdorferi based on reactivity with monoclonal antibodies and OspA sequence analysis. J. Clin. Microbiol. 31:340-350. [DOI] [PMC free article] [PubMed] [Google Scholar]