Abstract
Bacterium-bacterium interactions occur at intimate spatial scales on the order of micrometers, but our knowledge of interactions at this level is rudimentary. Antagonism is a potential interaction in such microenvironments. To study the ecological role of antibiosis, we developed a model system involving an antibiotic-producing isolate (SWAT5) derived from a marine particle and its dominant antibiotic product, 2-n-pentyl-4-quinolinol (PQ). This system was used to address questions about the significance of this antibiotic for microbial ecology and carbon cycling on particles. We characterized the chemical and inhibitory properties of PQ in relation to the mechanisms used by particle-associated bacteria in interacting with particles and with other attached bacteria. PQ was produced by SWAT5 only on surfaces. When SWAT5 was grown in polysaccharide matrices, PQ diffused within the matrices but not into the surrounding seawater. SWAT5 might thus be able to generate a localized zone of high antibiotic concentration on particles suspended or sinking through seawater. Target bacterial respiration was most sensitive to PQ (75 nM), while inhibition of DNA synthesis, protein synthesis, and bacterial motility required higher (micromolar) PQ levels. The presence of PQ altered the composition of the bacterial community that colonized and developed in a model particle system. PQ also inhibited Synechococcus and phytoplankton growth. Our results suggest that antibiosis may significantly influence community composition and activities of attached bacterial and thus regulate the biogeochemical fate of particulate organic matter in the ocean.
Since Burkholder et al. (7) and Lovell (29) determined the first novel structure of an antibiotic produced by marine bacteria, there has been growing interest in marine bacteria as a potential source of natural products (10, 20, 21, 39, 41). Numerous novel chemical structures have been discovered in marine bacteria. However, our understanding of antibiosis as an ecological phenomenon in the marine pelagic environment is quite limited, and our current knowledge is based upon antagonistic interactions of cultured isolates (24). Recently, 86 phylogenetically characterized marine bacterial isolates were examined to determine their antagonistic interactions with other isolates within the group (26) (rather than their inhibition of human pathogens). In this study it was found that production of antibiotics by marine bacteria that inhibit other marine bacteria is a much more common phenotype than had previously been recognized. While antibiotic production was distributed throughout all major cultivable heterotrophic phylogenetic groups, the most potent producers belonged to the γ-proteobacteria. Furthermore, the isolates derived from particles, such as marine snow, were more potent inhibitors than free-living bacteria, although antibiotic production was quite common among free-living bacteria as well. In this previous study the hypothesis that there is pervasive antibiosis which creates distinct microspatial patterns of species distribution in the pelagic realm was presented. The study also showed that particle-derived bacteria had more intense and broader-spectrum antagonistic interactions than free-living bacteria (26). This may suggest that the particle realm in the ocean offers microenvironments where antibiosis is most intense.
Recent findings have shown that bacterial interactions with particulate organic matter, both living and detrital, can have important consequences for the biogeochemistry of the ocean and the biosphere. Particle-associated bacteria expressing high ectoproteolytic activity on diatom detritus critically control silica remineralization (5). Differential activities of different ectohydrolytic enzymes of bacteria on marine snow can change the C/N and C/P ratios of sinking particles (47). It has been suggested that bacteria attached to dimethylsulfoniopropionate-producing algae cause rapid transformation of dimethylsulfoniopropionate to dimethylsulfide, a cloud-nucleating volatile compound (M. Scarratt, G. Cantin, M. Levasseur, and S. Michaud, Abstr. 10-09, Am. Soc. Limnol. Oceanogr. 2000, 2000). Bacterial ectohydrolase activity on transparent organic particles (e.g., transparent exopolymer particles [1] and Coomassie blue-stained particles [25]) and on phytoplankton cells may reduce the stickiness of the particles and thereby inhibit aggregation (48). All these examples involve spatially intimate small-scale interactions of bacteria with organic particles. These and other considerations have led to the notion that microspatial interactions of bacteria play important roles in regulating ocean-basin-scale biogeochemical dynamics (2, 27).
Molecular studies of the species composition of attached and free-living bacteria have shown that bacterial species diversity and its dynamics are much greater than had previously been recognized. Numerous investigators have speculated about the concept of particle specialist bacteria (as distinct from free-living bacteria) based on the kinetics of their nutrient uptake systems (3, 37), their metabolic differences (11), and phylogenetic analyses (6, 9, 38, 44). Presumed older particles and marine snow have bacterial assemblages that are distinct from those in the surrounding water mass (9, 44). Unique or different bacterial assemblages can, in turn, lead to both qualitative and quantitative differences in the nature and rate of degradation of organic matter, since different marine bacteria are likely to have different abilities to degrade organic matter (33). Thus, processes causing variations in bacterial species richness and diversity are relevant to the biogeochemical fates of organic particles in the ocean.
To address our hypothesis that antibiosis may regulate bacterial interactions with particulate organic matter, we used a model system involving antibiosis by particle-attached bacteria. SWAT5, an alteromonad isolated from a marine particle, in mixed-culture assays was found to inhibit 26 of 86 isolates examined (26). Upon further investigation, it was determined that this bacterium produced its inhibitory substances only when it was grown on surfaces. In the study described here, we purified and elucidated the structures of two antibiotics from SWAT5 and then developed a model system based on 2-n-pentyl-4-quinolinol (PQ), the dominant compound produced by SWAT5. We characterized the chemical and biological (physiological and metabolic) inhibitory properties of the molecule in relation to the assumed lifestyle of particle-associated bacteria. We tested the hypothesis that the antibiotic could alter bacterial colonization at the species level of a model particle system using a culture-independent method. In addition to its effect on bacterium-particle interactions, we also addressed whether the antibiotic could modify the interactions of bacteria with diatoms.
MATERIALS AND METHODS
Isolation and culture of bacteria.
Pelagic marine bacteria were isolated on ZoBell 2216E plates (each liter of GF/F-filtered seawater contained 5 g of peptone, 1 g of yeast extract, and 15 g of Bacto Agar [Difco]; FeCl3 was excluded from the recipe). Isolates were initially grown at the ambient sea surface temperature at the time of isolation, which ranged from 12.5 to 25°C. The majority of the isolates were obtained from southern California coastal waters (off Scripps Institution of Oceanography, La Jolla, Calif., and University of California, Santa Barbara, Santa Barbara, Calif.). We operationally defined attached or particle-associated bacteria as bacteria that were isolated from visible particles in our samples. The particles were individually picked and rinsed with filtered (pore size, 0.22 μm) seawater several times. A particle was then spread across a ZoBell plate. Free-living bacteria were defined as either bacteria that were gravity filterable through a 1.0-μm-pore-size filter or isolated in bulk or bacteria in unfiltered seawater that contained no visible particles.
Bacterial isolates were grown in ZoBell 2216E broth (each liter of GF/F-filtered seawater contained 5 g of peptone and 1 g of yeast extract) without FeCl3. Isolates were grown at room temperature (20 ± 2°C) on a rotary shaker at 100 rpm. The concentrations of isolate suspensions were adjusted to an optical density at 600 nm (OD600) of 1 with GF/F-filtered autoclaved seawater.
Reagents.
All of the solvents used were freshly distilled from reagent-grade preparations before use.
Instrumentation.
1H nuclear magnetic resonance (1H-NMR) spectra were obtained with a Varian 300-MHz spectrometer. 13C-NMR spectra were obtained with a Varian 400-MHz spectrometer operating at 100 MHz. Samples were dissolved in deuterated methanol or dimethyl sulfoxide. High-performance liquid chromatography (HPLC) was carried out by using a Waters M-6000 pump attached to a Waters R401 differential refractometer and an ISCO model 229 UV/VIS detector. Electrospray ionization mass spectrometry was carried out with a Finnigan MAT LCQ spectrometer.
Extraction and purification of the alteromonad's inhibitory compound.
Lawns (ZoBell top agar, 0.6%) of the alteromonad were poured onto ZoBell agar plates and incubated for 3 days at 20°C. The lawns were then scraped off and discarded, the underlying agar was diced and extracted with ethyl acetate, and the extract was concentrated by solvent evaporation by using a Rotavapor. The crude extract was chromatographed with a C18 Sep-Pak cartridge (Whatman) by using a solvent gradient ranging from 100% water to 100% methanol in 20% methanol steps. Fractions whose compositions were similar, as shown by 1H-NMR and biological activity (see below), were combined and further purified by HPLC with a C18 column (Dynamax-60 Å, 5 μm, two 10- by 250-mm columns in series); 70% methanol with 0.05% trifluoroacetic acid was used as the solvent, and three fractions were found to be biologically active. The structures of these fractions were determined by electrospray ionization mass spectrometry and 1H- and 13C-NMR spectroscopy.
Synthesis of PQ.
The method used for synthesis of PQ was the method described by Wratten et al. (50). However, the ethyl-3-oxooctanoate intermediate was prepared as follows. Diisopropylamine (6.06 g) was dissolved in freshly distilled tetrahydrofuran (50 ml) under N2. The solution was cooled to −78°C, and n-butyl lithium in hexane (5 ml of a 10.0 M solution) was added dropwise with stirring. The solution was warmed to −5°C for 20 min and then cooled back to −78°C, and freshly distilled ethyl acetate (2.45 ml) was added dropwise via a syringe. After the solution was stirred for another 10 min, hexanoyl chloride (5 ml) was added dropwise with stirring. After stirring for a further 10 min, hydrochloric acid (3 N, 25 ml) was added. The residue was partitioned against ether, dried (with MgSO4), and evaporated. The oily residue was distilled under a vacuum (1.7 mm of Hg) to obtain 1.4 g of the desired product, ethyl-3-oxooctanoate (22% yield).
Characterization of PQ.
The absorption spectrum of PQ was measured with a Perkin-Elmer lamba 4a UV/VIS spectrophotometer. Fluorescent spectra were measured with a FluoroMax-2 spectrofluorometer (Instruments S.A., Inc.).
(i) Testing the diffusion properties of PQ in agar.
Aliquots (150 μl) of 0.1 and 0.8% agar were amended with PQ (0.01 to 2 mM) in a 96-well plate. After the agar solidified, fluorescence due to PQ was measured with a Perkin-Elmer HTS700 plate reader with 340- and 635-nm excitation and emission filters, respectively. The plate was then submersed in filtered (Whatman GF/F) autoclaved seawater for 18 h. The water was removed from the plate, and the fluorescence was measured again to determine the change in the PQ concentration in the agar. Controls for fluorescence due to agar leachates and ethanol were also included.
(ii) Assay of antibacterial activity.
Burkholder inhibition assays were performed by overlaying lawns of target cells (30 μl of a suspension having an OD600 of 1) in 2.5 ml of ZoBell soft agar onto ZoBell agar plates. The test solutions were either directly spotted onto the lawns or placed on antibiotic test discs (Becton Dickinson 231039), which were then placed on the lawns. The plates were scored for zones of inhibition after 1 or 2 days depending on the growth rate of the target cells.
Effects of PQ on bacterial metabolic and physiological parameters.
A variety of parameters were measured to ascertain the effect that PQ might have in direct bacterium-bacterium interactions, as well as to determine the MICs necessary to cause an effect on isolates and natural assemblages collected off Scripps Pier. The concentration gradients of PQ for the following experiments ranged from 500 pM to 1 mM, and controls for ethanol and agar were used when appropriate.
(i) Inhibition of isolates grown in liquid.
Target isolates were grown at 22°C in ZoBell broth, and OD600 were measured with a spectrophotometer (Spectronic 20).
(ii) Inhibition of isolates grown on solid media.
Target isolates were grown at 22°C on ZoBell agar plugs by using a 96-well plate format. Eighty microliters of ZoBell hard agar was amended with PQ, and a lawn (20 μl of ZoBell soft agar seeded with ∼100 target cells) was overlaid. The plate was incubated overnight, and inhibition was scored when no colonies or lawns were observed. Variations of this approach were used to examine diffusion of PQ from one matrix to another matrix and to examine the effects of the location of a target strain and its susceptibility to PQ.
(iii) Inhibition of bacterial protein and DNA production in isolates and natural assemblages.
Protein production and DNA production were measured by using [3H]thymidine and [3H]leucine techniques (12, 22), as modified for centrifugation (46). Cells were preincubated with PQ and then diluted 10-fold into fresh media containing either 20 nM [3H]leucine or 20 nM [3H]thymidine and incubated for 15 to 60 min. Incubations were conducted at the ambient sea surface temperature at the location from which the natural assemblage was collected.
(iv) Inhibition of respiration of isolates.
Respiration rates were measured with a microprocessor-controlled oxygen electrode (Corning 317) in 125-ml biological oxygen demand bottles. Test cells were grown overnight in ZoBell broth, pelleted, washed with filtered (pore size, 0.2 μm) seawater, resuspended in filtered seawater at a concentration of 107 cells ml−1, and treated with PQ. Replicate samples were measured at zero time and 20 h.
(v) Effect on growth of autotrophic bacteria.
Stationary and exponentially growing Synechococcus sp. cultures were inoculated into SN media amended with PQ, incubated for 4 days under low light, and examined for effects on abundance, size (side scattering), and autofluorescence (phycouribilin) with a flow cytometer (Fac Sort; Becton Dickinson).
(vi) Inhibition of bacterial motility.
Motility of heterotrophic isolates was examined by using dark-field microscopy. Cells were resuspended in filtered autoclaved seawater, and subsamples were treated with PQ in a microtiter plate, viewed at a magnification of ×40, and qualitatively scored for reduction of motility. For Synechococcus motility assays, exponentially growing cells were treated with PQ. After 10 min of incubation, wet mounts were examined at a magnification of ×400 by phase-contrast microscopy.
(vii) Inhibition of ectoenzyme activities.
Enzymatic activities were measured by using the fluorogenic substrate analogs (18) methylumbelliferyl (MUF) phosphate and leucine aminomethylcoumarin (AMC) (Sigma Chemical Co.). Isolates were preincubated for 30 to 60 min with PQ in filtered autoclaved seawater. Substrate (final concentration, 100 μM) was then added, and hydrolysis was monitored for 5 min. Hydrolysis rates were determined by determining the change in fluorescence over time with a Hoefer TKO-100 fluorometer. The fluorometer was calibrated with MUF and AMC standards, and enzymatic activity was expressed in terms of the amount of MUF or AMC produced over time and was normalized to the activity of the control.
(viii) Change in cell surface charge.
A bacterial adhesion to hydrocarbons (BATH)-xylene cell surface charge assay (45) was performed. Isolates were incubated with subinhibitory and inhibitory concentrations of PQ. After 30 min of incubation, cells were examined for changes in the cell surface charge.
Colonization experiment.
Five-microliter capillary tubes (Drummond Scientific Co.) were filled with ZoBell agar amended with PQ (0 to 100 μM) in quadruplicate. The capillary tubes were plugged at one end, and excess agar was cut away. The tubes were then placed into an incubation chamber containing seawater collected off Scripps Pier that had been gravity filtered through a 3.0-μm-pore-size polycarbonate filter (Poretics). Controls for agar and ethanol (in which PQ was dissolved) were also tested. The capillary tubes were incubated for 5 h at 20°C.
A second colonization experiment was performed by using Telfon-coated microscope slides with 6-mm-diameter wells imprinted on them (Electron Microscopy Science). The wells were coated with 3 μl of 0.5% low-melting-point agarose amended with treatments as described above. Quadruplicate sets were incubated for 22 h at 20°C. Three of the sets were used for bacterial community analysis, and the fourth set was used for microscopic observation of bacterial colonization of the particles.
Bacterial DNA extraction and PCR amplification.
To collect bacterial community DNA and to produce PCR products for denaturing gradient gel electrophoresis (DGGE) analysis, a modification of the protocol developed by Long and Azam (27) was used. Capillary tubes were transferred to 200-μl PCR tubes containing 5 μl of Lyse-N-Go (Pierce). The samples were enclosed in a sterile box and placed on a shaker for 1 h at 20°C. The capillary tubes were then removed, and the PCR tubes were placed in a thermocycler. The thermocycling protocol for lysis was carried out according to the manufacturer's instructions (65°C for 30 s, 8°C for 30 s, 65°C for 90 s, 97°C for 180 s, 8°C for 60 s, 65°C for 180 s, 97°C for 60 s, 65°C for 60 s, and kept at 80°C until the addition of PCR reagents). After completion of the lysis cycle, 20 μl of a PCR master mixture (1 U of Qiagen Taq DNA polymerase, each primer at a concentration of 1 μM, each deoxynucleoside triphosphate at a final concentration of 0.8 mM) was added to each tube, and a touchdown PCR (see below) was performed by using a universal primer complementary to base positions 517 to 534 (5′-ATTACCGCGGCTGCTGG-3′) and a eubacterial primer complementary to base positions 341 to 358 with a 40-bp GC clamp (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGGCACGGGGGGCCTACCGG AGGCAGC-3′) (35). The touchdown PCR protocol consisted of an initial 94°C denaturing step for 5 min, followed by 30 cycles of amplification (3 min of denaturation at 94°C, 1 min of annealing starting at 65°C for the first cycle and reduced 0.5°C per cycle to 50°C, and 3 min of extension at 72°C) and then five additional cycles of amplification (3 min at 94°C, 1 min at 50°C, and 3 min at 72°C) and a final extension of 10 min at 72°C. Negative controls were run with sterile filtered water in place of template. PCR products were separated by electrophoresis on a 1.5% agarose gel and stained with ethidium bromide to confirm amplification of the desired product size.
DGGE analysis.
The DGGE techniques and reagents used were modified slightly from those of Muyzer et al. (35). Ten microliters of PCR product was loaded onto a 8% polyacrylamide gel (ratio of acrylamide to N,N′-methylenebisacrylamide, 37:1) in 0.5× TAE (20 mM Tris, 10 mM acetate, 0.5 mM Na2 EDTA; pH 8.2) with a top-to-bottom 35 to 50% denaturant gradient (100% denaturant was 40% [vol/vol] deionized formamide and 7 M urea.). Electrophoresis was conducted at 10 V/cm for 5.5 h at 60°C with a hot-bath DGGE unit (CBS Scientific, San Diego, Calif.) and a running buffer consisting of 0.5× TAE. Gels were stained with 1× SYBR Green I in 0.5× TAE (Molecular Probes, Eugene, Oreg.) for 30 min in the dark and then destained in 0.5× TAE for 10 min. The gels were then visualized and documented by using UVP Epi-chemi Darkroom with a charge-coupled device camera.
Algicidal activity of PQ.
Cultures of Thalassiosira weissflogii, Chaetoceros simplex, and Cylindrotheca fusiformis were inoculated into f/20 medium (16) amended with PQ or into ethanol controls and grown with a cycle consisting of 12 h of light and 12 h of darkness at 16 to 17°C. Cell density and autofluorescence were monitored for 2 days by flow cytometry. Bacterial interactions with the diatoms were visualized by staining the samples with 4′,6-diamidino-2-phenylindole (DAPI) (final concentration, 1 μg ml−1) for 15 min and then filtering the preparations onto 0.2- or 3.0-μm-pore-size black polycarbonate filters (Poretics), and the samples were examined with UV excitation at a magnification of ×1,250 by using epifluorescence microscopy (42).
RESULTS
Isolation and elucidation of the active compounds.
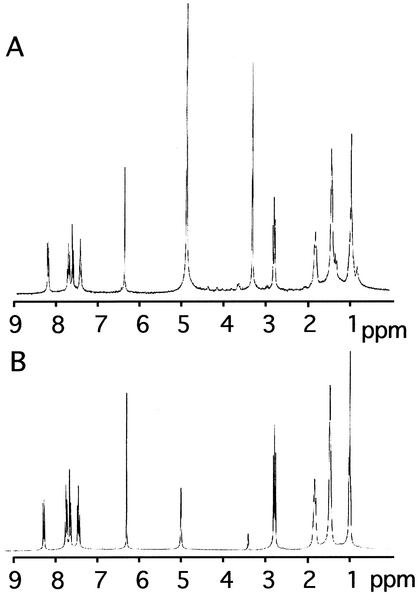
HPLC yielded three biologically active fractions. Two of the compounds isolated were determined to be PQ and 2-n-heptyl-4-quinolinol, both of which have been described previously (50). The third, minor compound was not identified. The dominant compound, PQ, was synthesized, and its structure and purity were confirmed by 1H-NMR (Fig. 1).
FIG. 1.
1H-NMR comparison of natural and synthetic PQ. (A) HPLC fraction with PQ. (B) Synthesized PQ. The peaks at 4.88 and 3.34 ppm are water and methanol, respectively.
Characterization of PQ.
PQ has a fluorescence excitation peak at 341 nm and an emission peak at 671 nm. Using the fluorescent properties of PQ, we examined its diffusion properties in agar (a complex polysaccharide matrix). It readily diffused within the gel matrix. However, it did not diffuse from agar into filtered autoclaved seawater.
Metabolic effects on isolates and a natural assemblage.
The effect of synthetic PQ on the isolates originally inhibited by SWAT5 was examined. PQ inhibited 23 of the 26 isolates inhibited by SWAT5, as assayed by the Burkholder antibiotic disk assay, as well as both Escherichia coli and Dietzia sp. strain SB96A.
The concentrations causing 50% inhibition (ID50) of both protein synthesis and DNA synthesis in the isolates were approximately 10 μM, while 1 mM PQ inhibited synthesis of both protein and DNA almost completely (the values were 8 and 4% of the control values, respectively). The MIC (the concentration at which statistically significant reduction of incorporation was observed) was generally the same as the ID50 (Table 1).
TABLE 1.
Inhibition of bacterial physiology
| Parameter | PQ concna | Activity normalized to controlb |
|---|---|---|
| Respirationc | 10 nM | 111 |
| 25 nM | 106 | |
| 50 nM | 106 | |
| 75 nM | 20 | |
| 100 nM | 11 | |
| Protein synthesis | 10 nM | 91 |
| 100 nM | 106 | |
| 1 μM | 105 | |
| 10 μM | 63 | |
| 100 μM | 30 | |
| 1 mM | 8 | |
| DNA synthesis | 10 nM | 109 |
| 100 nM | 98 | |
| 1 μM | 102 | |
| 10 μM | 56 | |
| 100 μM | 15 | |
| 1 mM | 4 |
Ethanol controls had values similar to the control values.
The control values were defined as 100.
Respiration was also tested by using PQ concentrations up to 25 μM, and the levels of inhibition were similar to the level of inhibition observed with 100 nM.
The assays determining incorporation of [3H]leucine and [3H]thymidine are sensitive assays to examine the effect of PQ on bacterial production in a natural assemblage. The natural assemblage examined was more sensitive to PQ than the isolates were. The MIC of PQ was 10 nM for protein synthesis, while the ID50 was 1 μM. DNA synthesis was not as sensitive; MIC for DNA synthesis was 1 μM.
The effect of PQ on the respiration of isolates was examined by measuring oxygen consumption. PQ had no effect until at a concentration of 75 nM it abruptly reduced oxygen consumption to 20% of the control level (Table 1). Incubation with higher concentrations of PQ (up to 25 μM) did not significantly inhibit respiration further.
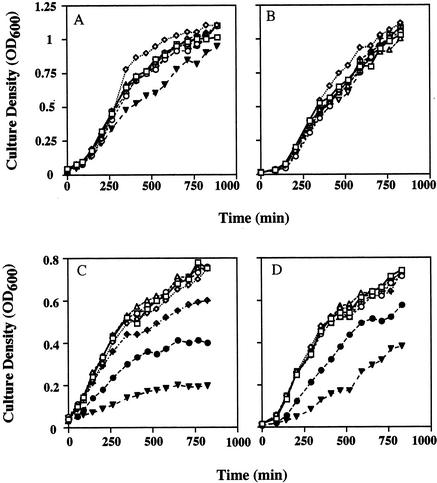
To assess whether PQ was bacteriostatic or bactericidal, a recovery experiment was performed with bacterial isolates. We studied the effect of PQ on the growth rates, as measured by the optical densities of SB1 and SB10 cultures growing in ZoBell broth. After 1 h of exposure to PQ, subcultures were diluted 10-fold, and the abilities of the isolates to recover from the exposure to PQ were examined. SB1 was only marginally affected by exposure to PQ at concentrations as high as 100 μM (Fig. 2), whereas the growth of SB10 was significantly reduced by 10 μM PQ and was reduced by approximately 15% by 1 μM PQ (Fig. 2). Samples which showed inhibition of growth were diluted 10-fold with fresh nutrient broth. These cultures grew to higher densities (Fig. 2).
FIG. 2.
Effects of PQ on bacterial isolates in a liquid environment. The inhibitory effects of PQ on isolates SB1 (A and B) and SB10 (C and D) were examined by measuring the change in OD600. Inocula were added to ZoBell broth amended with PQ, and growth was monitored (A and C). To examine whether PQ was bactericidal or bacteriostatic, subsamples were taken from the initial preparations and diluted 1:10 with fresh ZoBell broth after 60 min (B and D). Open symbols, PQ concentrations between 0.1 and 100 nM; ♦, 1 μM PQ; •, 10 μM PQ; ▾, 100 μM PQ. (For panels B and D the PQ concentrations are the concentrations in the initial inoculum, not in the dilution.)
We tested if the environment in which the target cells resided had an effect on their susceptibility to PQ. Target cells either were placed either on top of solidified agar containing PQ or were embedded within soft agar containing PQ (to simulate a matrix) that was laid on top of hard agar that contained the same concentration of PQ. Lawns of target cells were overlaid onto hard agar containing PQ. In general, isolates were more sensitive to PQ when they were in the matrix than when they were applied on top of the matrix (Table 2).
TABLE 2.
Inhibition of mobility of isolates by PQ
| Treatment
|
Motility of isolatesa
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (min) | PQ concn (μM) | SB1 | SB2 | SB4 | SB6 | SB8 | SB10 | SB11 | SB13 | SB14 | SB15 | SB18 | DC |
| 15 | 100 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| 10 | − | − | + | + | + | − | + | + | + | + | + | − | |
| 1 | − | − | − | − | − | − | − | − | + | − | − | − | |
| Controlb | − | − | − | − | − | − | − | − | − | − | − | − | |
| 60 | 100 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| 10 | ++ | ++ | ++ | + | ++ | − | + | + | + | + | ++ | − | |
| 1 | + | − | + | − | + | − | − | − | + | − | − | − | |
| Controlb | − | − | − | − | − | − | − | − | − | − | − | − | |
Motility was qualitatively scored by dark-field microscopy. −, no noticeable change in motility; +, 25 to 50% decrease in motility; ++, > 50% decrease in motility.
The control contained 0.08% ethanol. PQ concentrations of 1, 10, and 100 nM were also tested but had no effect.
Effect of PQ on physiological properties of bacterial isolates.
The effect of PQ on the motility of sensitive motile isolates was examined by dark-field microscopy. Observations after 15 min of exposure to PQ showed that all isolates were affected at high concentrations (≥10 μM). Longer exposure times decreased the concentration that caused the same degree of inhibition (Table 3). Concentrations down to 100 nM inhibited the most sensitive isolate.
TABLE 3.
Effect of PQ on bacterial isolates in and on a surface matrix
| Treatmenta
|
Growth (normalized to control)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Location | PQ concn | α-Proteobacteria
|
Bacteroidetes
|
γ-Proteobacteria
|
|||||
|
Pseudoalteromonas
|
Vibrio
|
||||||||
| Target 1 | Target 2 | Target 1 | Target 2 | Target 1 | Target 2 | Target 1 | Target 2 | ||
| Surface of matrix | Ethanol controlb | 110 | 98 | 98 | 101 | 88 | 115 | 95 | 86 |
| 1 μM | 153 | 92 | 102 | 113 | 108 | 119 | 117 | 121 | |
| 10 μM | 127 | 70 | 93 | 81 | 93 | 94 | 100 | 94 | |
| 50 μM | 27 | 42 | 32 | 29 | 38 | 30 | 87 | 41 | |
| 100 μM | 18 | 31 | 24 | 27 | 11 | 50 | 62 | 4 | |
| In matrix | Ethanol controlb | 86 | 84 | 94 | 99 | 94 | 95 | 89 | 97 |
| 50 nM | 131 | 70 | 107 | 107 | 100 | 99 | 100 | 104 | |
| 100 nM | 82 | 58 | 97 | 107 | 99 | 98 | 97 | 98 | |
| 1 μM | 61 | 61 | 103 | 104 | 97 | 94 | 95 | 87 | |
| 10 μM | 26 | 54 | 68 | 63 | 65 | 67 | 87 | 81 | |
| 50 μM | 20 | 29 | 62 | 61 | 69 | 67 | 64 | 72 | |
| 100 μM | 26 | 50 | 57 | 50 | 51 | 49 | 38 | 49 | |
Target cells were placed either on or in an agar matrix treated with PQ in a 96-well microtiter plate and incubated overnight. The effects of PQ on two known targets belonging to each of four groups were examined. Growth was measured by determining the density for each treatment and then was normalized to the growth in the blank control, which was defined as 100.
The control contained 0.08% ethanol. The PQ concentrations used ranged from 0.5 nM to 100 μM, and only the results for concentrations which produced significant inhibition are shown.
Both exponential-phase and stationary-phase cultures were preincubated with PQ, and protease and phosphatase activities were measured. No inhibition of activity was detected (Table 4). The BATH-xylene assay revealed no change in the cell surface charge for isolates treated with PQ (data not shown).
TABLE 4.
Inhibition of bacterial ectoenzymatic activities
| PQ concna | Enzyme activitiesb
|
|
|---|---|---|
| Phosphatase | Protease | |
| 0.1 nM | 97 | 107 |
| 1 nM | 102 | 103 |
| 10 nM | 97 | 98 |
| 100 nM | 94 | 120 |
| 1 μM | 102 | ND |
Ethanol controls had values similar to the control values.
The control values were defined as 100.
Effects on phytoplankton and bacterium-phytoplankton interactions.
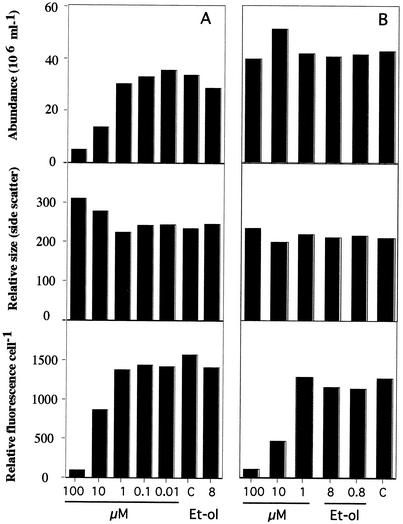
The effects of PQ on two strains of Synechococcus were examined. Growth of a nonmotile isolate, WH7803, was inhibited at a concentration of 10 μM (data not shown). Isolate CC93-01, a motile Synechococcus strain isolated from a sample collected off Scripps Pier, was used to examine the effect of PQ on growth, cell size, phycouribilin content, and motility. Both stationary and freshly inoculated cultures were treated with PQ and incubated for 4 days. There was no effect on the stationary cultures in terms of cell abundance or size, while the level of autofluorescence per cell was dramatically reduced by the 10 and 100 μM PQ treatments (Fig. 3). The freshly inoculated cultures showed no growth in the presence of 100 μM PQ, while in the presence of 10 μM PQ the final cell levels were reduced to 25% of the control levels. As in the stationary culture, autofluorescence was significantly reduced by the 10 and 100 μM PQ treatments. The cell size increased, as detected by side scattering measurements with the flow cytometer, after these two treatments (Fig. 3).
FIG. 3.
Effects of PQ on the growth and physiology of Synechococcus. Stationary (A) and exponentially growing (B) cultures of Synechococcus strain CC93-01 were treated with PQ in SN media and grown for 4 days in low light. Abundance, relative size (side scatter), and cell-specific fluorescence were measured with a flow cytometer. Et-ol, ethanol; C, control.
The effect of PQ on Synechococcus motility was examined with a stationary culture of strain CC93-01 following a 10-min treatment with PQ. Qualitatively, motility was significantly reduced by treatment with ≥10 μM PQ; both the speed at which the cells swam and the percentage of swimming cells were reduced (data not shown).
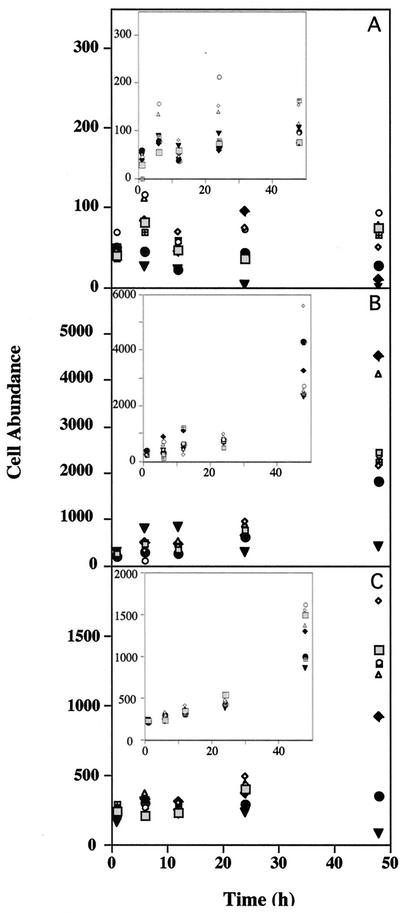
The tests of the inhibitory effects of PQ on diatoms (T. weissflogii, C. simplex, and C. fusiformis) showed that all three species were sensitive to this compound (Fig. 4). The LD50 were 1 to 10 μM, while measurable inhibition occurred at concentrations down to 10 nM. The ethanol control also inhibited diatom growth, but to a lesser extent. Examination of PQ-treated samples showed that there was colonization of all three diatoms by bacteria at PQ concentrations as low as 10 nM. In one set of experiments, cultures treated with sublethal concentrations of PQ had higher levels of mucus than the control cultures had, and they were heavily colonized by bacteria.
FIG. 4.
Effect of PQ on the growth of two diatom species and a phytoplankton-diatom natural assemblage. (A) Filtered (pore size, 63 μm) seawater; (B) C. fusiformis. (C) T. weissflogii. C. fusiformis and T. weissflogii were grown in f/20 media, and all preparations were treated with PQ and grown with a cycle consisting of 12 h of light and 12 h of darkness. Cell density and fluorescence were monitored by flow cytometry. The insets show the results for ethanol controls for each treatment. Grey squares, control (no PQ); open symbols, PQ concentrations between 0.1 and 100 nM; ♦, 1 μM PQ; •, 10 μM PQ; ▴, 100 μM PQ.
Effect of PQ on bacterial community development on a particle.
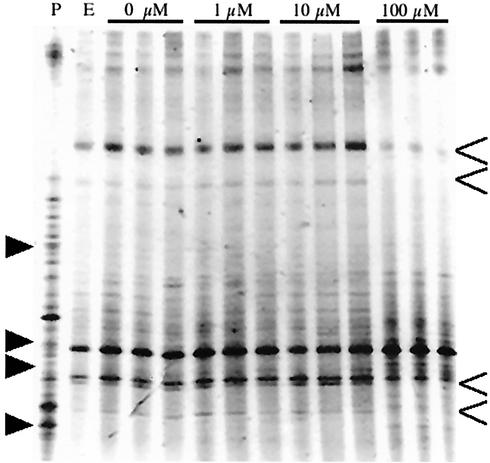
In two colonization experiments with ZoBell agar or agarose amended with PQ, we examined how PQ changed the structure of the bacterial community that colonized model particles. We observed changes in the colonizing bacterial community structures as determined by DGGE in both the short- and long-term experiments. In the 5-h capillary experiment, two phylotypes showed reduced presence at higher concentrations of PQ, while a third phylotype appeared at higher concentrations (data not shown).
Similar results where observed in the 22-h colonization experiment. DGGE analysis revealed an average of 21 ± 2.7 ribotypes in the four treatments, and the means were 22.3, 22.3, 21.6, and 20.3 ribotypes for treatments with 0, 1, 10, and 100 μM PQ, respectively. Changes in ribotypes and bacterial species colonization were recorded only if they were observed in all three replicates. In the preparation treated with 100 μM PQ, four ribotypes decreased significantly or disappeared, while the three ribotypes either increased or appeared (Fig. 5). The bacterial community that developed on the particles was different than the initial community in the water, as observed by the relative differences in band intensity.
FIG. 5.
DGGE analysis of the effect of PQ on colonization of a model particle system. A region of the 16S rRNA gene was targeted for PCR amplification with the 341F primer with a 40-bp GC clamp and the 534R primer. Samples were electrophoresed on a 30 to 55% denaturant DGGE gel and then stained with SYBR Green I. The gel was documented with an UVP Epi-chemi Darkroom charge-coupled device camera system. P, pelagic filtered free-living bacteria; E, ethanol control ZoBell agarose particles and replicates of ZoBell agarose particles treated with PQ. Both types of preparations were examined in triplicate, and a single representative sample was loaded on the gel due to space limitations. The open arrowheads indicate ribotypes that were lost at higher concentrations of PQ, while the solid arrowheads indicate new ribotypes that appeared.
DISCUSSION
Characterization of quinolinols.
Both inhibitory molecules produced by SWAT5 in our study have been described previously. 2-n-Heptyl-4-quinolinol was first isolated and identified from a strain of Pseudomonas aeruginosa (17, 49). Subsequently, 2-n-heptyl-4-quinolinol and PQ were isolated from a marine pseudomonad (50). Whether the ability to produce 2-alkyl-quinolinols is widespread in γ-proteobacteria was not examined, nor is it possible to determine this by examination of the literature, since rediscovered compounds are rarely reported. In this study, we focused on the dominant metabolite, PQ. The bioactivity of this compound has not been reported previously. Two striking characteristics make PQ interesting to study in the context of bacterium-bacterium interactions on particulate organic matter. First, PQ was produced only when SWAT5 was grown on agar or agarose surfaces, which were polysaccharide matrices. Second, PQ is hydrophobic. Our studies showed that it readily diffused within the agar and agarose matrices but not from agar into seawater. Possibly, the hydrophobic lipid tail of PQ interacts with the matrix, thereby keeping it within the matrix. These properties suggest that PQ could be an effective inhibitory product for a particle-attached bacterium, such as SWAT5, for the purpose of antibiosis.
Metabolic and physiological effects of PQ on heterotrophic bacteria.
We inferred from our studies with isolates that the most sensitive mode of action of PQ is inhibition of respiration, since respiration was strongly inhibited at low concentrations (75 nM). The other metabolic parameters examined, DNA and protein syntheses, had much higher MIC of PQ (and may reflect secondary effects of energy depletion). Recovery experiments with isolates suggested that PQ is bacteriostatic, since cultures treated with PQ recovered upon dilution.
PQ had no effect on the ectoenzymatic activity of the target isolates, nor did it alter the cell surface charge of the isolates. It did, however, inhibit motility at higher concentrations, as well as at lower concentrations with longer exposure times. This suggests that a bacterium attached to particle being attacked by PQ may have a chance to leave the particle. After a period of time, the bacterium would no longer be able to leave, and it would become a metabolically inactive package of ectoenzymes that would hydrolyze organic matter but not consume it. This situation may contribute to the previously observed phenomenon of hypersolubization on marine snow, in which high levels of enzymatic activity were observed while the uptake of hydrolysates and bacterial growth rates were low (47).
It has been proposed that seawater contains gel matrices (2) in which bacteria exist and interact with other organisms and organic matter. We examined whether the microenvironment in which a target organism occurs affects its sensitivity to PQ. Isolates within an agar gel matrix were found to be more sensitive than isolates on the surface of the matrix, suggesting either that the cell surface properties are different within the matrix (e.g., different surface proteins are expressed [34]) or that PQ can more readily access the cells.
The general focus in the literature on antibiotics has been on whether antibiotics are bacteriostatic or bactericidal. However, recent studies have revealed more subtle effects of antibiotics at sublethal concentrations. Studies have shown that bacterial adhesion to surfaces was decreased at one-eighth to one-fourth the antibiotic MIC required to inhibit growth (30), while other antibiotics had the opposite effect (43). Nakajima et al. reported on a quinolone that at a concentration less than the MIC induced overexpression of an outer membrane protein (36). While we did not observe changes in ectoenzymatic activity, it has been shown that such activities can be altered at concentrations less than the MIC (8). Furanones from a marine macroalga have been shown to inhibit luminescence and virulence in Vibrio harveyi but not inhibit growth (32). While we did not see such effects in our studies with target bacteria, it is important to consider potential sublethal effects in modeling antibiosis in an ecological context, since subtle physiological changes might alter bacterium-particulate organic matter interactions and their consequences.
In addition to the sublethal effects of antibiotics, other classes of molecules can regulate bacterial interactions and their effects upon particles. Quorum-sensing molecules, such as acylated homoserine lactones, are population density-sensing molecules that have been found to have an array of effects upon bacterial gene expression and thus phenotypes at the intraspecies level (for a review see reference14). Even more recently, other quorum-sensing molecules have been shown to be effective at the interspecies level (4). Indeed, quorum sensing and antibiosis are not necessarily mutually exclusive. A quorum-sensing molecule which is structurally similar to PQ, 2-heptyl-3-hydroxy-4-quinolone, has been reported. This molecule induced virulence of P. aeruginosa, while 2-heptyl-4-hydroxy-quinoline-N-oxide, which is even more similar to PQ, had no effect (40). It has been suggested that quorum sensing might also regulate bacterial species on particles (26). More recently, it has been demonstrated that bacterial isolates from marine snow produce acylated homoserine lactones (15).
Effect of PQ on bacterial species composition on particles.
Using PQ and defined particle matrices as a model, we examined our hypothesis that antibiosis occurring on organic particles in the water column allows particle specialists to regulate bacterial community structure. In our colonization experiment, we presented polysaccharide surfaces enriched with ZoBell 2216E nutrients to bacterial assemblages for colonization. Increased concentrations of PQ resulted in reduction or disappearance of some phylotypes, suggesting that PQ exerts an influence on the colonization and growth of different species on a particle. Particle specialists, such as SWAT5, may produce compounds that inhibit the physiology of other bacterial species in a variety of ways. They might inhibit colonization by altering behavior or gene expression or by inhibiting the growth of bacteria present on particles, thereby reducing the biochemical influence that particular bacteria may exert enzymatically on a particle's organic matter. This shift of bacterial species can have important consequences in the flux of organic matter (see below). Antibiosis may also contribute to patchiness of bacterial species distribution at the microscale in the pelagic ocean (27).
PQ inhibited 27% of the bacterial isolates, as determined by the disk diffusion assay, and it inhibited a similar percentage of ribotypes in our DGGE analysis. We were unable to load enough material to sequence the ribotypes from this experiment as three ribotypes dominated the PCR products and most bands were not visible without the use of a digital imaging system. Larger quantities of PCR products would have overloaded the gel.
Effect of PQ on autotrophic members of the food web.
PQ inhibited the growth of Synechococcus and altered other aspects of its physiology. It also inhibited the growth of T. weissflogii, C. simplex, and C. fusiformis, suggesting that PQ may be used by SWAT5 to attack the phytoplankton. A series of reports on algicidal bacteria and their inhibitory products (23, 28, 31) support the hypothesis that bacteria can regulate the development of algal blooms (13). Two modes of algicidal activity are direct contact and the release of extracellular compounds. Bacteria belonging to the Cytophaga-Flavobacter-Bacteriodes group are the most common direct-contact algicidal bacteria (52), while γ-proteobacteria release substances and directly attack algal cells (19, 51, 52). Thus, our findings concerning the inhibition of diatoms and Synechococcus by the PQ produced by an Alteromonas sp. are consistent with previous reports. These findings support the notion that bacteria in the pelagic realm are not passive receivers of dissolved organic matter, and they shed light on how bacterial interactions with phytoplankton can have important implications in carbon cycling and the biogeochemistry of the ocean (2).
Our study suggests that PQ influences bacterium-phytoplankton interactions in several ways. In one experiment, sublethal concentrations of PQ did not affect phytoplankton growth or fluorescence, but microscopy revealed that phytoplankton produced exopolymers profusely and were colonized by bacteria. In a second set of experiments with natural assemblages collected at a different time of the year, these effects were not observed. When they occur, such shifts in bacterium-phytoplankton interactions could have important ramifications in the cycling of nutrients and carbon fluxes. For instance, observed shifts in bacterium-diatom interactions could influence dissolved organic matter production.
In summary, PQ produced by SWAT5 could potentially alter bacterial species composition, bacterial metabolic and physiological parameters, and bacterial interactions with organic matter in the ocean. We suggest that the effects of bacterial antibiotics on bacterium-bacterium and bacterium-phytoplankton interactions might have important ramifications for food web structure and biogeochemical dynamics of the pelagic marine ecosystem.
Acknowledgments
This work was supported by NSF grant OCE-9819603 and NIAID grant A146600 to F. Azam and by NSF grant CHE-9816169 and NIH grant CA-49084 to D. J. Faulkner.
We thank Bianca Brahamsha for assistance with the Synechococcus experiments. We thank two anonymous reviewers for their recommendations.
Footnotes
In memory of D. John Faulkner.
REFERENCES
- 1.Alldredge, A. L., U. Passow, and B. E. Logan. 1993. The abundance and significance of a class of large, transparent organic particles in the ocean. Deep Sea Res. 40:1131-1140. [Google Scholar]
- 2.Azam, F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694-696. [Google Scholar]
- 3.Azam, F., and R. E. Hodson. 1981. Multiphasic kinetics for d-glucose uptake by assemblages of natural marine bacteria. Mar. Ecol. Prog. Ser. 6:213-222. [Google Scholar]
- 4.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bidle, K. D., and F. Azam. 1999. Accelerated dissolution of diatom silica by marine bacterial assemblages. Nature 397:508-512. [Google Scholar]
- 6.Bidle, K. D., and M. Fletcher. 1995. Comparison of free-living and particle-associated bacterial communities in the Chesapeake Bay by stable low-molecular-weight RNA analysis. Appl. Environ. Microbiol. 61:944-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkholder, P., R. Pfister, and F. Leitz. 1966. Production of a pyrrole antibiotic by a marine bacterium. Appl. Microbiol. 14:649-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cipriani, P., M. Trancassini, M. R. Chiossi, D. DeVito, and F. Filadoro. 1998. Antibiotics at sub-inhibitory concentrations and virulence factors of Pseudomonas aeruginosa. Microbiologica 21:285-288. [PubMed] [Google Scholar]
- 9.DeLong, E. F., D. G. Franks, and A. L. Alldredge. 1993. Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol. Oceanogr. 38:924-934. [Google Scholar]
- 10.Fenical, W. 1993. Chemical studies of marine bacteria—developing a new resource. Chem. Rev. 93:1673-1683. [Google Scholar]
- 11.Friedrich, U., M. Schallenberg, and C. Holliger. 1999. Pelagic bacteria-particle interactions and community-specific growth rates in four lakes along a trophic gradient. Microb. Ecol. 37:49-61. [DOI] [PubMed] [Google Scholar]
- 12.Fuhrman, J. A., and F. Azam. 1980. Bacterioplankton secondary production estimates for coastal waters of British Columbia, Antarctica, and California. Appl. Environ. Microbiol. 39:1085-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukami, K., T. Nishijima, H. Murata, S. Doi, and Y. Hata. 1991. Distribution of bacteria influential on the development and the decay of Gymnodinium nagasakiense red tide and their effects on algal growth. Nippon Suisan Gakkaishi-Bull. Jpn. Soc. Sci. Fish. 57:2321-2326. [Google Scholar]
- 14.Fuqua, C., and E. P. Greenberg. 1998. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr. Opin. Microbiol. 1:183-189. [DOI] [PubMed] [Google Scholar]
- 15.Gram, L., H. P. Grossart, A. Schlingoff, and T. Kiørboe. 2002. Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68:4111-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 29-60. In W. H. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Publishing Corp., New York, N.Y.
- 17.Hay, E. E., I. C. Wells, P. A. Katzman, C. K. Cain, F. A. Jacobs, S. A. Thayer, and E. A. Doisy. 1945. Antibiotic substances produced by Pseudomonas aeruginosa. J. Biol. Chem. 15:725-749. [PubMed] [Google Scholar]
- 18.Hoppe, H.-G. 1983. Significance of exoenzymatic activities in the ecology of brackish water: measurements by means of methylumbelliferyl-substrates. Mar. Ecol. Prog. Ser. 11:299-308. [Google Scholar]
- 19.Imai, I., Y. Ishida, K. Sakaguchi, and Y. Hata. 1995. Algicidal marine bacteria isolated from northern Hiroshima Bay, Japan. Fish. Sci. 61:628-636. [Google Scholar]
- 20.Jensen, P., and W. Fenical. 2000. Marine microorganisms and drug discovery: current status and future potential, p. 6-29. In N. Fusetani (ed.), Drugs from the sea. Karger, Basel, Switzerland.
- 21.Jensen, P. R., and W. Fenical. 1994. Strategies for the discovery of secondary metabolites from marine bacteria—ecological perspectives. Annu. Rev. Microbiol. 48:559-584. [DOI] [PubMed] [Google Scholar]
- 22.Kirchman, D., E. K'Nees, and R. Hodson. 1985. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49:599-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, S. O., J. Kato, N. Takiguchi, A. Kuroda, T. Ikeda, A. Mitsutani, and H. Ohtake. 2000. Involvement of an extracellular protease in algicidal activity of the marine bacterium Pseudoalteromonas sp. strain A28. Appl. Environ. Microbiol. 66:4334-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemos, M. L., C. P. Dopazo, A. E. Toranzo, and J. L. Barja. 1991. Competitive dominance of antibiotic-producing marine bacteria in mixed cultures. J. Appl. Bacteriol. 71:228-232. [DOI] [PubMed] [Google Scholar]
- 25.Long, R. A., and F. Azam. 1996. Abundant protein-containing particles in the sea. Aquat. Microb. Ecol. 10:213-221. [Google Scholar]
- 26.Long, R. A., and F. Azam. 2001. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol. 67:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long, R. A., and F. Azam. 2001. Microscale patchiness of bacterioplankton assemblage richness in seawater. Aquat. Microb. Ecol. 26:103-113. [Google Scholar]
- 28.Lovejoy, C., J. P. Bowman, and G. M. Hallegraeff. 1998. Algicidal effects of a novel marine Pseudoalteromonas isolate (class Proteobacteria, gamma subdivision) on harmful algal bloom species of the genera Chattonella, Gymnodinium, and Heterosigma. Appl. Environ. Microbiol. 64:2806-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovell, F. M. 1966. The structure of a bromine rich marine antibiotic. J. Am. Chem. Soc. 88:4510-4511. [Google Scholar]
- 30.Majtan, V., and L. Majtanova. 1998. The effect of subinhibitory concentrations of quinolones on cell surface hydrophobicity of Pseudomonas aeruginosa. Biologia 53:713-717. [Google Scholar]
- 31.Manage, P. M., Z. Kawabata, and S. Nakano. 2000. Algicidal effect of the bacterium Alcaligenes denitrificans on Microcystis spp. Aquat. Microb. Ecol. 22:111-117. [Google Scholar]
- 32.Manefield, M., L. Harris, S. A. Rice, R. De Nys, and S. Kjelleberg. 2000. Inhibition of luminescence and virulence in the black tiger prawn (Penaeus monodon) pathogen Vibrio harveyi by intercellular signal antagonists. Appl. Environ. Microbiol. 66:2079-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez, J., D. C. Smith, G. F. Steward, and F. Azam. 1996. Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat. Microb. Ecol. 10:223-230. [Google Scholar]
- 34.Montgomery, M. T., and D. L. Kirchman. 1994. Induction of chitin-binding proteins during the specific attachment of the marine bacterium Vibrio harveyi to chitin. Appl. Environ. Microbiol. 60:4284-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakajima, A., M. Hoshikawa, and T. Nakae. 1998. Antibiotic stress induces a large amount of outer membrane protein in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 165:261-265. [DOI] [PubMed] [Google Scholar]
- 37.Nissen, H., P. Nissen, and F. Azam. 1984. Multiphasic uptake of d-glucose by an oligotrophic marine bacterium. Mar. Ecol. Prog. Ser. 16:155-160. [Google Scholar]
- 38.Noble, P. A., K. D. Bidle, and M. Fletcher. 1997. Natural microbial community compositions compared by a back-propagating neural network and cluster analysis of 5S rRNA. Appl. Environ. Microbiol. 63:1762-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okami, Y. 1986. Marine microorganisms as a source of bioactive agents. Microb. Ecol. 12:65-78. [DOI] [PubMed] [Google Scholar]
- 40.Pesci, E. C., J. B. J. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pietra, F. 1997. Secondary metabolites from marine microorganisms: bacteria, protozoa, algae and fungi. Achievements and prospects. Nat. Prod. Rep. 14:453-464. [DOI] [PubMed] [Google Scholar]
- 42.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 43.Ramadan, M. A., A. F. Tawfik, T. A. Elkersh, and A. M. Shibl. 1995. In vitro activity of subinhibitory concentrations of quinolones on urea-splitting bacteria—effect on urease activity and on cell surface hydrophobicity. J. Infect. Dis. 171:483-486. [DOI] [PubMed] [Google Scholar]
- 44.Rath, J., K. Y. Wu, G. J. Herndl, and E. F. DeLong. 1998. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat. Microb. Ecol. 14:261-269. [Google Scholar]
- 45.Rosenberg, M., D. Gutnick, and E. Rosenberg. 1980. Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 9:29-33. [Google Scholar]
- 46.Smith, D. C., and F. Azam. 1992. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 6:107-114. [Google Scholar]
- 47.Smith, D. C., M. Simon, A. L. Alldredge, and F. Azam. 1992. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359:139-142. [Google Scholar]
- 48.Smith, D. C., G. F. Steward, R. A. Long, and F. Azam. 1995. Bacterial mediation of carbon fluxes during a diatom bloom in a mesocosm. Deep Sea Res. Part II 42:75-97. [Google Scholar]
- 49.Wells, I. C. 1952. Antibiotic substances produced by Pseudomonas aeruginosa. Syntheses of PYO Ib, PYO Ic, PYO III. J. Biol. Chem. 196:331-340. [PubMed] [Google Scholar]
- 50.Wratten, S. J., M. S. Wolfe, R. J. Andersen, and D. J. Faulkner. 1977. Antibiotic metabolites from a marine pseudomonad. Antimicrob. Agents Chemother. 11:411-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshinaga, I., T. Kawai, and Y. Ishida. 1997. Analysis of algicidal ranges of the bacteria killing the marine dinoflagellate Gymnodinium mikimotoi isolated from Tanabe Bay, Wakayama Pref, Japan. Fish. Sci. 63:94-98. [Google Scholar]
- 52.Yoshinaga, I., M. C. Kim, N. Katanozaka, I. Imai, A. Uchida, and Y. Ishida. 1998. Population structure of algicidal marine bacteria targeting the red tide forming alga Heterosigma akashiwo (Raphidophyceae), determined by restriction fragment length polymorphism analysis of the bacterial 16S ribosomal RNA genes. Mar. Ecol. Prog. Ser. 170:33-44. [Google Scholar]