Abstract
Saprophytic bacteria in cultures of the marine dinoflagellate Alexandrium catenella were removed to assess their effect on growth and paralytic shellfish poisoning toxin production of this dinoflagellate. The actual axenic status was demonstrated by the lack of observable bacteria both immediately after treatment and following extended incubation in the absence of antibiotics. Bacteria were measured by counting CFU and also by epifluorescence microscopy and PCR amplification of bacterial 16S-23S spacer ribosomal DNA to detect noncultivable bacteria. Removal of bacteria did not have any effect on the growth of the dinoflagellate except for the inhibition of A. catenella disintegration after reaching the stationary phase. Toxicity was determined in dinoflagellate cell extracts by different methods: high-performance liquid chromatography (HPLC); an electrophysiological test called the Electrotest, which measures the inhibition of saxitoxin-sensitive Na+ channels expressed in a cell line; and a mouse bioassay, which measures the toxic effect on the whole mammal neuromuscular system. A lower toxicity of the dinoflagellates in axenic culture was observed by these three methods, though the difference was significant only by the mouse bioassay and HPLC methods. Altogether the results indicate that axenic cultures of A. catenella are able to produce toxin, though the total toxicity is probably diminished to about one-fifth of that in nonaxenic cultures.
Marine dinoflagellates of the genus Alexandrium (Halim) Balech include a number of species that produce saxitoxin and its derivatives, known as paralytic shellfish poisoning (PSP) (9, 20, 21). Toxic species of the genus Alexandrium are spread widely throughout many regions of the world (10, 17, 18, 21, 22). Within this genus, members of A. catenella/tamarense/fundyense (known as the tamarensis complex) (18) are among the most toxic species. In Chile, the first documented toxic bloom was reported in 1972 in the Magallanes region (7). The dominant toxic dinoflagellate species was identified by morphological criteria as A. catenella (8). Later, by comparison of rRNA sequence, this strain was grouped with the Asian Southern Pacific A. catenella ribotype (23).
Cultures of A. catenella that were obtained in Chile, like dinoflagellates obtained worldwide, contain a considerable amount of bacteria which probably accompanied the dinoflagellate in the original sample. Hence, as with most cultures, the role of the bacteria in dinoflagellate PSP production is a contentious topic. Bacteria in dinoflagellate cultures might produce saxitoxin autonomously or complement the dinoflagellates in either toxin production or growth. This problem has been thoroughly reviewed by Gallacher and Smith (5). As they state, several publications describing the production of PSPs by dinoflagellates in the absence of bacteria have appeared, but these previous studies lacked unequivocal demonstration of the absence of bacteria in their cultures and/or adequate monitoring of the bacterium-free status throughout their experimentation. Two recent related studies are those by Dantzer and Levin (3) and Doucette and Powell (4). The first set of authors checked the axenic nature of their culture only by showing that bacterial growth did not appear on marine agar, which would detect a very low percentage of the total bacteria found in seawater (19). The second set of authors used observation by epifluorescence microscopy besides bacterial growth to check the axenic status; however, they did not report the frequency of monitoring by microscopy. More recently, Hold et al. (11) determined by high-performance liquid chromatography (HPLC) the ability of two Alexandrium species to produce toxins in bacterium-free cultures. They found that the growth rate and toxin profile of Alexandrium lusitanicum were similar in bacterium-free and control cultures, although the bacterium-free culture generally produced a larger amount of toxins. Bacteria elimination in the other species examined, Alexandrium tamarense, did not affect the growth rates and toxin profile but in some cases decreased the amount of some toxins. However, since most but not all toxins produced by A. tamarense were measured, it was not possible to draw conclusions on the effect on overall toxicity. Here we describe the achievement of axenic cultures of A. catenella and the growth rate and toxicity of this dinoflagellate in the absence of bacteria. In the present study, we provide unequivocal evidence of the absence of bacteria in cultures of A. catenella and measure its effect on toxicity by two bioassays, one measuring the inhibition of saxitoxin-sensitive Na+ channels (24) and the other, the mouse bioassay, measuring the toxic effect on the whole mammal (2, 9). Toxins were also measured analytically by HPLC.
The original clonal cultures of A. catenella contained about 10,000 to 100,000 bacteria per dinoflagellate. These cultures were maintained by growth in f/2 medium (6, 16) at 17°C with 400 microeinsteins/m2, under a 16:8 light-darkness illumination cycle. To obtain axenic cultures, cells from a previously described clonal culture, ACC07 (23), were subjected to sequential washing (after about three duplications) and treatment with gentamicin (0.05 mg/ml) and penicillin G (0.2 mg/ml) (Sigma Chemical Co., St. Louis, Mo.). For washing, 20 to 30 ml of media, containing approximately 104 cells, were filtered through an 11-μm-pore-size nylon mesh attached to the end of a 15-mm-diameter polypropylene cylinder. The cells retained by the mesh were then gently washed by submersion in 20 to 30 ml of fresh sterile medium. After this procedure was repeated, the cells were suspended at a concentration of approximately 500 cells/ml in medium containing either gentamicin or penicillin antibiotic and incubated until they reached a concentration between 2,000 and 10,000 cells/ml. The procedure was repeated several times, alternating each time between the two antibiotics. The filtration procedure avoided damage to the cells caused by centrifugation. Bacterial presence in the cultures was determined by measuring the number of CFU in marine agar (26) (Difco Laboratories, Detroit, Mich.) and by direct or epifluorescence microscopy after staining with acridine orange (12, 14); the procedure for microscopy allowed detection to one bacteria per dinoflagellate. For detection by PCR amplification of bacterial 16S-23S rDNA spacers, DNA was extracted as previously described (23) except that centrifugation of the cells was performed at 10,000 × g for 10 min. Amplification was performed according to the method of Jensen et al. (13). PCR amplification of the D1-D2 fragment of the 28S ribosomal DNA (rDNA) of dinoflagellates, performed as described by Zardoya et al. (25) and modified by Uribe et al. (23), was used as a control of DNA extraction. Toxicity was determined in extracts of cells obtained from cultures at different times of antibiotic treatment and containing decreasing amounts of bacteria. Dinoflagellates were collected at late exponential phase when concentrations reached 10,000 to 20,000 cells/ml as described by Anderson et al. (1). Cell extracts were obtained by addition of 1 ml of 0.1 M HCl and heating at 100°C for 5 min according to the official methods of analysis of the Association of Official Analytical Chemists (2). Three methods were employed: an electrophysiological assay called Electrotest based on the PSP blocking effect on recombinant muscle Na+ ion channels (μ1), stably expressed in HEK293 cells (24); HPLC for measurement of saxitoxin, neosaxitoxin, and gonyautoxins 1 to 4 using purified standards as described previously (15); and the mouse bioassay for total toxicity, following the standard procedure of the Association of Official Analytical Chemists (2). Toxin standards were obtained from the Institute for Marine Bioscience of the National Research Council of Canada.
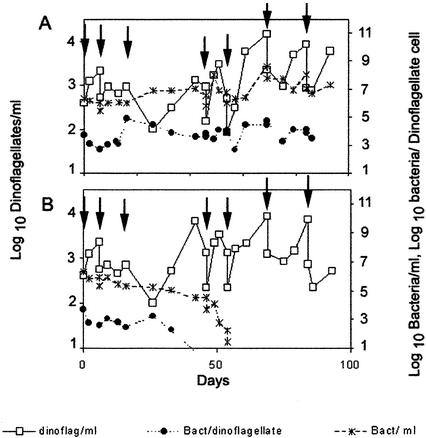
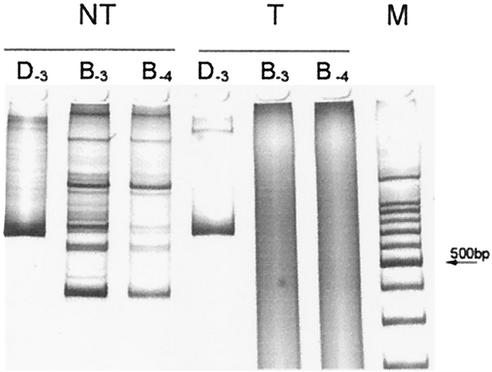
Figure 1 shows the growth of dinoflagellates and bacteria after successive washing and subsequent incubation without (A) and with (B) antibiotic. In both cultures, the number of CFU paralleled, in a ratio of 1 to 10, the number of bacteria counted under the microscope. In the culture without antibiotics, bacteria were partially eliminated by washing, but after about 20 days, they reached a roughly constant number of 1,000 to 10,000 bacteria per dinoflagellate cell. On the other hand, treatment with antibiotics eliminated any observable bacteria by either CFU counting or epifluorescence microscopy after five successive treatments. Removal of bacteria was achieved after five to seven treatments in four independent experiments. The absence of bacteria in the antibiotic-treated culture was checked by detection of bacterial genes (i.e., the region present between the rRNA genes 16S and 23S). DNA extracted from cultures containing bacteria and that extracted from the axenic cultures were used as templates for the amplification of 16S-23S rDNA spacers, with primers common for most of the known bacterial species (13). This assay would allow detection of potential bacteria unobservable by microscopy that tightly adhered to external or internal components of the dinoflagellate cells. The results of this assay are illustrated in Fig. 2 and showed the absence of amplifiable bacterial DNA in the axenic culture, even when using at least 10 times more template DNA than in the bacterium-containing culture. Bacteria were also checked after long-term incubation in the absence of antibiotics. In this assay, it was expected that any bacteria remaining in culture would eventually increase in number, as observed with the culture untreated with antibiotics. None of several antibiotic-treated cultures showed the presence of bacteria after six more subcultures of 1 to 10 dilutions by both epifluorescence microscopy and PCR amplification of the bacterial ribosomal spacer (results not shown).
FIG. 1.
Growth of A. catenella and bacteria in batch cultures after successive washing (A) and after successive washing and antibiotic treatments (B). Dinoflagellate and bacterial cells were determined by direct and epifluorescence microscopy, respectively. Dinoflagellate cell concentration (□), bacterial cell concentration (∗), and number of bacterial cells per dinoflagellate cell (•) are shown. Arrows indicate the points at which washing was conducted. Drops in dinoflagellate cell concentration at these points correspond to dilution after washing.
FIG. 2.
Polyacrylamide gel electrophoresis of the PCR amplification products obtained from nontreated (NT) and antibiotic-treated (T) A. catenella cultures. DNA extracted from dinoflagellate and bacterial cells concentrated by centrifugation was used for amplification of bacterial 16S-23S rDNA spacers to detect the presence of bacteria. 28S rDNA of dinoflagellates was also amplified as a control of DNA extraction. Lanes D correspond to the product after amplification for 28S rDNA from A. catenella (D1-D2 fragment of 699 bp). Lanes B correspond to amplification for 16S-23S bacterial rDNA spacers. Subscripts indicate the DNA dilution (log 10 dilution) employed for amplification. Lane M corresponds to the molecular size ladder marker; the arrow indicates 500 bp.
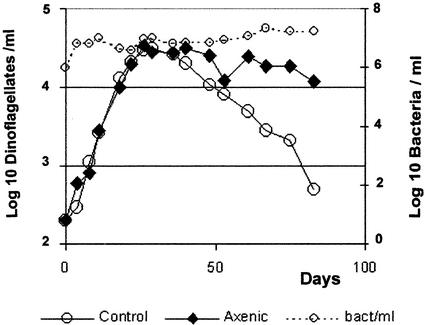
The growth parameters of the untreated culture and the treated axenic cultures are shown in Fig. 3. The growth rates of A. catenella were indistinguishable between the two, with a value of 0.2 day−1, corresponding to an approximate doubling time of 3.3 days. The standard deviation of the growth rate observed in more than 10 growth curves with this isolate was 0.03 day−1. Both cultures reached a stationary phase at a concentration of about 3 × 104 cells/ml. The difference observed between cultures was at the stationary phase, when the number of dinoflagellate cells observed under the microscope decreased much faster in the bacterium-containing culture. This suggests that disintegration of starving or dead A. catenella cells is mainly due to the presence of bacteria in the culture. Furthermore, the growth of bacteria in the untreated culture paralleled the increase of dinoflagellates during the exponential growth phase, maintaining a roughly constant ratio of 3,000 bacteria per dinoflagellate cell. The total number of bacteria remained constant after the dinoflagellates reached the stationary phase; however, they looked significantly larger. These last observations suggest that bacteria in this culture are saprophytic, probably feeding on dinoflagellate by-products. The growth culture parameters, growth rate, and stationary- phase cell concentration were highly reproducible for both dinoflagellates and bacteria.
FIG. 3.
Growth curve of A. catenella in axenic (♦) and control (○) dinoflagellate cultures. The growth of bacteria in the control culture, as determined by microscopy, is shown as a dotted line (○).
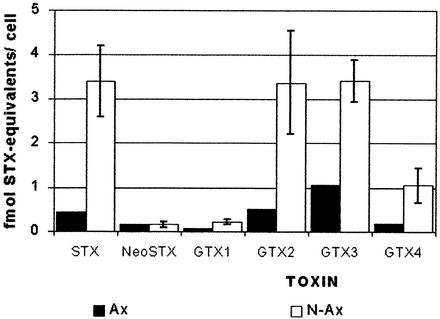
Toxicity was determined in these extracts by the different methods described above. As observed in Table 1, no significant difference was observed between axenic and nonaxenic cultures by the Electrotest assay, but both mouse bioassay and HPLC showed a lower level of toxicity in the axenic cultures. The result of the Student t test for the differences observed were 0.12 for the Electrotest and 0.05 for the mouse assay. Though the decrease observed could be considered significant for the mouse assay and not for the Electrotest, they are not contradictory. The results of the t test indicate that the probability that the difference observed could be due to a different mean toxicity between axenic and nonaxenic cultures is 0.88 in one case and 0.95 in the other. The Student t test could not be applied for the results obtained by HPLC because only one axenic culture could be analyzed. However, the confidence interval range with a 99% level between the values for the axenic and nonaxenic cultures did not overlap, suggesting that the difference observed is significant. Individually, each toxin decreased to a similar extent, except neosaxitoxin, which remained at the same level in axenic and nonaxenic cultures (Fig. 4). Since in every assay toxicity was measured in extracts of pelleted cells, the data reflect exclusively the level of toxin remaining inside the cells, and additional work is required to more completely characterize the effects of bacterial removal on toxin. These figures cannot be extrapolated to toxin production since the extent of excretion is unknown. However, we show here that A. catenella cells remain toxic in bacterium-free culture though this toxicity may be decreased to about one-fifth. We also show that removal of the original accompanying bacteria does not change A. catenella growth properties, except for conferring a higher stability after reaching the stationary phase. If the effect of associated bacteria in field conditions is similar to that observed in this work, they could have a marked effect on the toxicity of the dinoflagellate growing in a bloom. The associated bacteria could also increase dinoflagellate deterioration after the bloom reaches maximum density, having a significant role in clearing the dinoflagellates. However, the effect described here corresponds to that observed in cultures of an isolate of A. catenella associated with bacteria which had been selected during isolation and continuous culturing of the dinoflagellate. These bacteria might represent a particular group from a much more diverse community originally present in the bloom.
TABLE 1.
Toxicity (femtomoles of saxitoxin equivalents/cell) in extracts from A. catenella cultures with varying numbers of associated bacteriaa
| Sampled | No. of bacteria/A. catenella cell | Result
|
||
|---|---|---|---|---|
| Electrotestb | HPLCc | Mouse bioassay | ||
| Untreated 1 | 9,000 | 14.1 (2.8) | ||
| Untreated 2 | 27,000 | 11.4 (2.5) | 9.6 | |
| Treated 1 | 7,500 | 6.0 (0.9) | ||
| Treated 2 | 3,500 | 8.8 (2.2) | 5.8 | |
| Treated 3 | 1,000 | 15.9 (3.0) | 18.0 | |
| Treated 4 | 10 | 7.5 (1.0) | ||
| Treated 5 | 10 | 12.6 (3.9) | 13.0 | 9.5 (1.9) |
| Treated 6 | 10 | 14.2 (2.4) | ||
| Treated ax-1 | Axenic | 7.9 (3.2) | 2.3 (0.5) | |
| Treated ax-2 | Axenic | 5.3 (0.4) | 2.4 | 1.7 (0.3) |
Standard deviations are given in parentheses.
Electrotest measures the inhibition of saxitoxin-sensitive Na+ channels expressed in a cell line.
HPLC values correspond to the sum of the saxitoxin equivalents for saxitoxin, neosaxitoxin, and gonyautoxins 1, 2, 3 and 4.
Samples were obtained by extraction of dinoflagellate collected at late exponential phase when the concentration was 10,000 to 20,000 cells/ml. “Treated” refers to samples collected from cultures at different stages of the successive antibiotic treatment.
FIG. 4.
Toxin levels measured by HPLC in axenic (▪) and nonaxenic (□) A. catenella cultures. STX and NeoSTX correspond to saxitoxin and neo-saxitoxin, respectively. GTX1, -2, -3, and -4 correspond to gonyautoxins 1, 2, 3, and 4, respectively. The values for nonaxenic cultures correspond to the mean of the values measured in each of the cultures containing bacteria appearing in Table 1. The bars represent the standard deviations.
Acknowledgments
We thank Carmen Alcayaga and Benjamin Suárez from Laboratorio Toxinas Marinas, Instituto de Ciencias Biomédicas, University of Chile, for the electrophysiological determinations of PSP toxins, Orialis Villarroel from Instituto de Salud Pública for HPLC analysis, and Illani Atwater for editing. P.U. specially acknowledges the support of B. Suárez as Ph.D. thesis adviser.
This work was supported by a grant from Fondo Nacional para Investigación Científica y Tecnológica, FONDECYT 1990765.
REFERENCES
- 1.Anderson, D. M., D. M. Kulis, J. J. Sullivan, S. Hall, and C. Lee. 1990. Dynamics and physiology of saxitoxin production by the dinoflagellates Alexandrium spp. Mar. Biol. 104:511-524. [Google Scholar]
- 2.Association of Official Analytical Chemists. 1990. Paralytic shellfish poison. Biological method. Final action, p. 881-882, sec 959.08. In K. Hellrich (ed.), Official methods of analysis, 15th ed. Association of Official Analytical Chemists, Arlington, Va.
- 3.Dantzer, W. R., and R. E. Levin. 1997. Bacterial influence on the production of paralytic shellfish toxins by dinoflagellated algae. J. Appl. Microbiol. 83:464-469. [DOI] [PubMed] [Google Scholar]
- 4.Doucette, G. J., and C. L. Powell. 1998. Algal-bacterial interaction: can they determine the PSP-related toxicity of dinoflagellates? p. 406-409. In B. Reguera, J. Blanco, M. L. Fernández, and T. Wyatt (ed.), Harmful algae. Xunta de Galicia and Intergovernmental Oceanographic Commission of UNESCO, Paris, France.
- 5.Gallacher, S., and E. Smith. 1999. Bacteria and paralytic shellfish toxins. Protist 150:245-255. [DOI] [PubMed] [Google Scholar]
- 6.Guillard, R. R. L. 1995. Culture methods, p. 45-62. In G. M. Hallegraeff, D. M. Anderson, and A. D. Cembella (ed.), IOC manuals and guides: manual on harmful marine microalgae. Intergovermental Oceanographic Commission of UNESCO, Paris, France.
- 7.Guzmán, L., and I. Campodónico. 1975. Marea Roja en la región de Magallanes. Publ. Inst. Pat. Ser. Monogr. Punta Arenas (Chile) 9:44. [Google Scholar]
- 8.Guzmán, L., and I. Campodónico. 1978. Mareas Rojas en Chile. Interciencia 3:144-151. [Google Scholar]
- 9.Hall, S., G. Strichartz, E. Moczydlowski, A. Ravindran, and P. B. Reichardt. 1990. The saxitoxins: sources, chemistry, and pharmacology, p. 29-65. In S. Hall and G. Strichartz (ed.), Marine toxins origin, structure and molecular pharmacology. ACS symposium series. American Chemical Society, Washington, D.C.
- 10.Hallegraeff, G. M. 1995. Harmful algal blooms: a global overview, p. 1-22. In G. M. Hallegraeff, D. M. Anderson, and A. D. Cembella (ed.), Manual on harmful marine microalgae. IOC manuals and guides no. 33. Intergovernmental Oceanographic Commission of UNESCO, Paris, France.
- 11.Hold, G. L., E. A. Smith, T. H. Birkberk, and S. Gallacher. 2001. Comparison of paralytic shellfish toxin (PST) production by the dinoflagellates Alexandrium lusitanicum NEPCC 253 and Alexandrium tamarense NEPCC 407 in the presence and absence of bacteria. FEMS Microbiol. Ecol. 36:223-234. [DOI] [PubMed] [Google Scholar]
- 12.Imai, I. 1987. Size distribution, number and biomass of bacteria in intertidal sediments and seawater of Ohmi Bay, Japan. Bull. Jpn. Soc. Microb. Ecol. 2:1-11. [Google Scholar]
- 13.Jensen, M. A., J. A. Webster, and N. Straus. 1993. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl. Environ. Microbiol. 59:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuwae, T., and Y. Hosokawa. 1999. Determination of abundance and biovolume of bacteria in sediments by dual staining with 4′,6-diamino-2-phenylindole and acridine orange: relationship to dispersion treatment and sediment characteristics. Appl. Environ. Microbiol. 65:3407-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oshima, Y. 1995. Post column derivatization liquid chromatographic method for paralytic shellfish toxins. J. AOAC Int. 78:528-532.
- 16.Provasoli, L., J. J. A. McLaughlin, and M. R. Droop. 1957. The development of an artificial media for marine algae. Arch. Mikrobiol. 25:392-428. [DOI] [PubMed] [Google Scholar]
- 17.Richardson, K. 1997. Harmful or exceptional phytoplankton blooms in the marine ecosystem. Adv. Mar. Biol. 31:301-385. [Google Scholar]
- 18.Scholin, C. A., and D. M. Anderson. 1993. Population analysis of toxic and nontoxic Alexandrium species using ribosomal RNA signature sequences, p. 95-102. In T. J. Smayda and Y. Shimizu (ed.), Toxic phytoplankton blooms in the sea. Elsevier Science Publishers, New York, N.Y.
- 19.Schut, F., E. J. de Vries, J. C. Gottschal, B. R. Robertson, W. Harder, R. A. Prins, and D. K. Button. 1993. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shumway, S. E. 1990. A review of the effects of algal blooms on shellfish and aquaculture. J. World Aquacult. Soc. 21:65-104. [Google Scholar]
- 21.Taylor, F. J. R. 1984. Toxic dinoflagellates: taxonomic and biogeographic aspects with emphasis on Protogonyaulax, p. 77-97. In E. P. Ragelis (ed.), Seafood toxins. ACS symposium series 262. American Chemical Society, Washington, D.C.
- 22.Taylor, F. J. R. 1993. The species problem and its impact on harmful phytoplankton studies, p. 81-86. In T. J. Smayda and Y. Shimizu (ed.), Toxic phytoplankton blooms in the sea. Elsevier Science Publishers, New York, N.Y.
- 23.Uribe, P., B. A. Suárez-Isla, and R. T. Espejo. 1999. Ribosomal RNA heterogeneity and identification of toxic dinoflagellate cultures by heteroduplex mobility assay. J. Phycol. 35:884-888. [Google Scholar]
- 24.Vélez, P., J. Sierralta, C. Alcayaga, M. Fonseca, H. Loyola, D. Johns, G. Tomaselli, E. Marbán, and B. Suárez-Isla. 2001. A functional assay for paralytic shellfish toxins that uses recombinant sodium channels. Toxicon 39:929-935. [DOI] [PubMed] [Google Scholar]
- 25.Zardoya, R., E. Costas, V. López-Rodas, A. Garrido-Pertierra, and J. M. Bautista. 1995. Revised dinoflagellate phylogeny inferred from molecular analysis of large-subunit ribosomal RNA gene sequences. J. Mol. Evol. 41:637-645. [DOI] [PubMed] [Google Scholar]
- 26.Zobell, C. E. 1941. Studies on marine bacteria. J. Mar. Res. 4:42-75. [Google Scholar]