Abstract
The aim of this study was to evaluate how the in situ exposure of a Danish subsurface aquifer to phenoxy acid herbicides at low concentrations (<40 μg l−1) changes the microbial community composition. Sediment and groundwater samples were collected inside and outside the herbicide-exposed area and were analyzed for the presence of general microbial populations, Pseudomonas bacteria, and specific phenoxy acid degraders. Both culture-dependent and culture-independent methods were applied. The abundance of microbial phenoxy acid degraders (100 to 104 g−1 sediment) was determined by most probable number assays, and their presence was only detected in herbicide-exposed sediments. Similarly, PCR analysis showed that the 2,4-dichlorophenoxyacetic acid degradation pathway genes tfdA and tfdB (102 to 103 gene copies g−1 sediment) were only detected in sediments from contaminated areas of the aquifer. PCR-restriction fragment length polymorphism measurements demonstrated the presence of different populations of tfd genes, suggesting that the in situ herbicide degradation was caused by the activity of a heterogeneous population of phenoxy acid degraders. The number of Pseudomonas bacteria measured by either PCR or plating on selective agar media was higher in sediments subjected to high levels of phenoxy acid. Furthermore, high numbers of CFU compared to direct counting of 4′,6-diamidino-2-phenylindole-stained cells in the microscope suggested an increased culturability of the indigenous microbial communities from acclimated sediments. The findings of this study demonstrate that continuous exposure to low herbicide concentrations can markedly change the bacterial community composition of a subsurface aquifer.
During the past decade an increasing number of herbicides has been detected in groundwater aquifers (15). These include the phenoxy acids, which have been widely used for more than 50 years, such as mecoprop [2-(2-methyl-4-chlorophenoxy)propionic acid], dichlorprop [2-(2,4-dichlorophenoxy)propionic acid], and 2,4-D (2,4-dichlorophenoxyacetic acid). Groundwater is an important drinking water resource, and studies exploring the potential of the indigenous microbial communities to degrade herbicides and the impact of these chemicals on microbial community structure may contribute to our understanding of how and when microorganisms indigenous to aquifers acclimate to xenobiotic compounds.
Microbial acclimation is commonly defined as the processes taking place during the time period between entry of a chemical and the evidence of its detectable loss (2). With acclimated microbial communities this period is typically shortened following repeated exposure to the same chemical compound (2). Microbial acclimation is generally ascribed to the enrichment of specific degraders, enzyme induction, and genetic adaptation, but the presence of toxins, predation by protozoa, and diauxie also influence the process (2).
It is only in the past few years that microbial acclimation to herbicides in subsurface aquifer environments has been investigated (1, 24, 37). Most of these studies involved laboratory incubation and primarily focused on the activity and rate of herbicide degradation. None, however, dealt with the effects of herbicides on the indigenous subsurface microbial communities. In contrast, numerous studies have examined the effects of herbicides on surface soil microbial communities, e.g., microbial biomass and the abundance of specific degraders and catabolic genes (12, 19, 22, 36). Subsurface aquifers constitute environments that are physically, chemically, and biologically very different from surface soils. They have reduced concentrations and availability of oxygen, carbon, and inorganic nutrients and a 102 to 106 times lower bacterial density (17). Results from surface soil environments, thus, cannot readily be extrapolated to subsurface aquifer conditions. Furthermore, in agricultural fields herbicides are typically applied in concentrations in the milligrams per kilogram of soil range, and surface soil microbial communities may thus be naturally exposed to high contaminant concentrations. In contrast, herbicide concentrations in groundwater are low: typical concentrations measured from landfills are 10 to 250 μg l−1 (26, 43) (equivalent to 2 to 40 μg kg−1 of sediment), whereas herbicide concentrations in groundwater polluted due to agricultural use most often are <1 μg l−1 (15). To get a realistic picture of how herbicides affect subsurface microbial communities, studies need to be carried out at low contaminant concentrations. Besides, information retrieved from such studies is necessary in order to model the fate of real-world contaminant plumes properly and for suggesting possible actions for remediation of polluted groundwater.
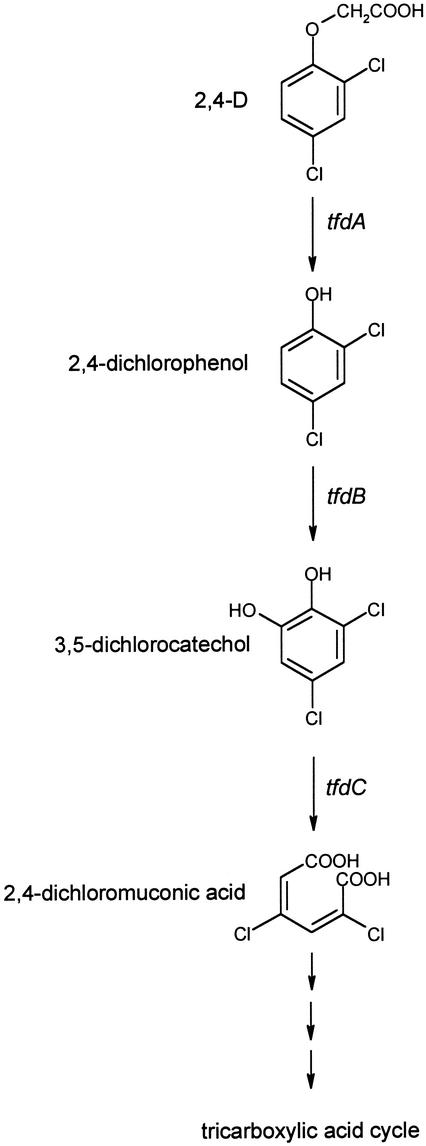
In a natural gradient field injection experiment, a part of the Vejen aquifer (Denmark) was continuously exposed to herbicide concentrations of <40 μg l−1 (8) (equivalent to <7 μg kg−1 of sediment). After an initial lag phase of 80 to 100 days, fast degradation of mecoprop and dichlorprop was observed and herbicide concentrations decreased to below detection levels within the first meter from the injection wells (8). This indicated that the microbial community acclimated to the contamination. This was verified by laboratory experiments showing that sediments close to the injection wells had the highest potential for phenoxy acid herbicide degradation (38). The aim of the present study was to evaluate how the in situ herbicide exposure of the Vejen aquifer affected the indigenous microbial community composition. This was done by measuring the total microbial population density and the abundance of Pseudomonas bacteria and phenoxy acid degraders. Furthermore, the frequency and the composition of the 2,4-D degradation pathway genes tfdA and tfdB (14, 33) (Fig. 1) were estimated.
FIG. 1.
Illustration of the 2,4-D degradation pathway in R. eutropha JMP134, the archetype 2,4-D degrader (13). The catabolic genes tfdA, tfdB, and tfdC were originally discovered in strain JMP134 and encode a 2,4-D/2-oxoglutarate dioxygenase, a chlorophenol hydroxylase, and a chlorocatechol 1,2-dioxygenase, respectively (14, 33). Other tfd genes are involved in the further transformation to products of the tricarboxylic acid cycle (14).
MATERIALS AND METHODS
Field site.
Sediment and groundwater were collected from an unconfined shallow aerobic aquifer located at Vejen, Denmark (5, 8). Part of the aquifer was previously exposed to herbicides during a natural gradient field injection experiment (8). A mixture of the herbicides mecoprop, dichlorprop, DNOC (2-methyl-4,6-dinitrophenol), bentazone [3-(1-methylethyl)-1H-2,1,3-benzothiadiazin-4(3H)-one 2,2-dioxide], isoproturon {N,N-dimethyl-N′-[4-(1-methylethyl)phenyl]urea}, and 2,4-dichlorbenzamide (BAM; a degradation product of dichlobenil) was continuously injected into the aquifer for a period of 216 days, creating a contaminant plume. Bromide was included as a nonreactive tracer. The concentration of each herbicide was approximately 40 μg l−1 immediately down-gradient of the injection wells. Migration of bromide and the herbicides was monitored by using a dense network of multilevel samplers (8).
At the time of sampling (40 days after termination of the field injection), herbicides were no longer present in the aquifer due to advective flow and degradation (8). The field exposures of mecoprop and dichlorprop were expressed as the total amount (in micrograms) of each compound that passed through each sampling point during the whole injection period (38).
Sampling.
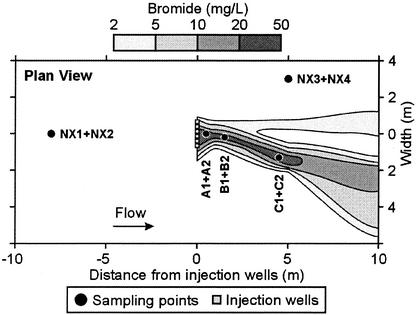
Sediment and groundwater were sampled from the herbicide-exposed area (samples A1, A2, B1, B2, C1, and C2) and from nonexposed areas outside the contaminant plume (NX1 to -4) (Fig. 2). Herbicide-exposed aquifer sediment was collected as three intact cores at different distances from the injection wells (0.5 m for A sites, 1.5 m for B sites, and 4.5 m for C sites). Two subsamples from each core were obtained, an upper sample (number 1) and a lower sample (number 2). The distance between the upper and lower samples was approximately 0.5 m. The nonexposed samples were collected similarly at upper (NX1 and NX3) and lower (NX2 and NX4) depths. All sediment cores were collected with a stainless steel piston sampler from the saturated zone, 5 to 6.25 m below the surface, and were stored (4°C) until partitioning. Two centimeters of each core end and the outermost 0.5 cm were aseptically removed by using a paring device modified from that described by Wilson et al. (41). Individual sediment samples were homogenized thoroughly and were stored at 4°C for a maximum of 9 days until setup of laboratory experiments. Sediments for DNA analysis were stored at −20°C. Groundwater was sampled adjacent to each sediment core by using a peristaltic pump and were stored at 4°C.
FIG. 2.
Field plot of the Vejen aquifer. Plan view showing the bromide tracer plume and the location of aquifer sampling points inside (A1, A2, B1, B2, C1, and C2) and outside (NX1 to -4) the herbicide-exposed area. The groundwater velocity is approximately 0.5 m day−1 (8), and the temperature is 10°C (38). At sampling (40 days after the field injection had been terminated), bromide as well as herbicides were no longer present within the monitoring network (8).
Bacterial strains and culture conditions.
Pseudomonas putida KT2442 was grown in Luria-Bertani medium (28). The Ralstonia eutropha strains JMP134 and AEO106 (a plasmid-cured derivative of R. eutropha JMP134) were grown in minimal medium (10) supplemented with 2 mM 2,4-D (>98%; Merck, Darmstadt, Germany) and 0.2% fructose, respectively. All bacterial strains were grown at 30°C.
Microbial populations.
Microbial population sizes were estimated by analyzing three replicate subsamples of each sediment sample. One gram of sediment was diluted in 9.0 ml of sterile NaCl (0.9% [wt/vol]) and vortexed for 1 min, and 10-fold dilutions were prepared. Direct counting of bacteria was performed by using epifluorescence microscopy (Olympus BX50; Hamburg, Germany) after staining with 4′,6-diamidino-2-phenylindole (DAPI) (2 μg ml−1) for 10 min in darkness. Three replicates of each subsample were enumerated. The number of culturable heterotrophs was estimated by plating on water agar (15.0 g l−1 of BiTek agar [Difco, Detroit, Mich.] in Milli-Q water). The number of culturable Pseudomonas bacteria was estimated by plating on the Pseudomonas-specific (18, 20) Gould's S1 agar medium (18). All agar media were supplemented with nystatin (50 μg ml−1) to prevent fungal growth. Each subsample was plated in triplicate and was incubated at 20°C for 21 days (water agar) and 7 days (Gould's S1) before enumeration of CFU.
The number of mecoprop, dichlorprop, and 2,4-D degraders was estimated by a most probable number (MPN) assay relying on herbicide degradation. Two milliliters of a minimal medium (10) containing 2.5-mg liter−1 concentrations of mecoprop (>99%; Riedel-de Haën, Seelze, Germany), dichlorprop (>99%; Riedel-de Haën), or 2,4-D (Merck) was added to 20-ml glass vials. The vials were then supplied with 200-μl aliquots of each sediment dilution (10−1 to 10−5), set up in five replicates, and incubated at 20°C for 3 months. Samples were filter sterilized (0.2 μm) and stored at 4°C until high-performance liquid chromatography analyses were made for remaining concentrations of herbicides as previously described (11). A similar setup was used to determine the abundance of mecoprop- and 2,4-D-mineralizing bacteria, though these vials also included 14C-labeled herbicides and a NaOH trap to collect 14CO2 as described previously (11). MPN samples were scored as positive if more than 25% of the herbicides were degraded or mineralized.
DNA extraction.
Whole-community DNA was extracted from three subsamples of each sediment sample by a combination of bead beating and freeze-thaw procedures. Bead beating was applied on 500 mg of sediment by using the FastDNA Spin Kit for Soil (BIO 101, Vista, Calif.) followed by two freeze-thaw cycles (1 h at −80°C, 30 min at 37°C). DNA was purified by the use of a GeneClean procedure (BIO 101), and the resulting samples were stored at −20°C.
Detection of Bacteria- and Pseudomonas-specific 16S rDNA.
The DNA primers PRBA338f and PRUN518r (27) were used to amplify general bacterial 16S ribosomal DNA (rDNA) segments, while PSMGf (7) and 785r (3) were used to amplify Pseudomonas-specific 16S rDNA segments. The PSMG primer has been shown to be specific to Pseudomonas (7, 21, 35). PCR mixtures (50 μl) consisted of 1× GeneAmp PCR Buffer (Perkin-Elmer, Applied Biosystem, Foster City, Calif.), 100 μM concentrations of each deoxynucleoside triphosphate (Perkin-Elmer), 0.5 μM concentrations of each primer (Gibco-BRL Custom Primers; Life Technologies, Paisley, Scotland), and 1.25 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer). Five-microliter aliquots of the DNA extracts were added to the reaction mixtures, and PCR was performed with the following program: 10 min at 95°C; 35 cycles of 30 s at 95°C, 30 s at 55°C (Bacteria) or 59°C (Pseudomonas), and 1 min at 72°C; 6 min at 72°C. DNA extracts (1 μl) of P. putida KT2442 and R. eutropha JMP134 cultures were used as positive and negative controls, respectively, for Pseudomonas-specific PCR. The abundance of 16S rDNA genes in DNA samples was estimated by MPN-PCR by analyzing three replicates of each sample dilution (100 to 10−4).
Detection of tfdA, tfdB, and tfdC genes.
PCR amplification of the 2,4-D degradation genes tfdA, tfdB (39), and tfdC (9) was performed with degenerate primers. PCR of tfdA- and tfdB-homologous genes was performed by using a program slightly modified from the methods described by Vallaeys et al. (39): 6 min at 94°C; 40 cycles of 45 s at 94°C, 30 s at 59°C (tfdA) or 49°C (tfdB), and 2 min at 72°C; 6 min at 72°C. PCR of tfdC-homologous genes was performed as described by Cavalca et al. (9). For all tfd PCRs, each tube contained a total of 50 μl of reaction mixture consisting of 1× GeneAmp PCR Buffer (Perkin-Elmer), 200 μM concentrations of each deoxynucleoside triphosphate (Perkin-Elmer), 1.0 μM concentrations of each primer (DNA Technology A/S, Aarhus, Denmark), and 2.5 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer). Five-microliter aliquots of the DNA extracts were added to the reaction mixtures, and PCR was performed as described above. DNA extracts (1 μl) of 2,4-D-grown R. eutropha JMP134 and fructose-grown R. eutropha AEO106 cultures were used as positive and negative controls for the presence of tfd genes. The abundance of tfd genes in DNA samples was estimated by MPN-PCR by analyzing three replicates of each sample dilution (100 to 10−2).
The populations of tfd genes obtained from individual aquifer sediments were analyzed by restriction fragment length polymorphism (RFLP) by digestion of PCR products with HaeIII and CfoI (an isoschizomer of HhaI) (Roche Molecular Biochemicals, Mannheim, Germany) followed by separation on 3.5% agarose gels. The molecular sizes of the restriction fragments were calculated by using a standard curve derived from the 100- to 600-bp bands of a 100-bp molecular size marker.
Statistical analyses.
All measurements of microbial population density and gene abundance are expressed per gram (dry weight) of sediment. The statistical significance of differences was determined by using a Student's t test at the 0.05 significance level. The level of linear correlation between data was expressed by Pearson correlation coefficients (r).
RESULTS AND DISCUSSION
General bacterial populations.
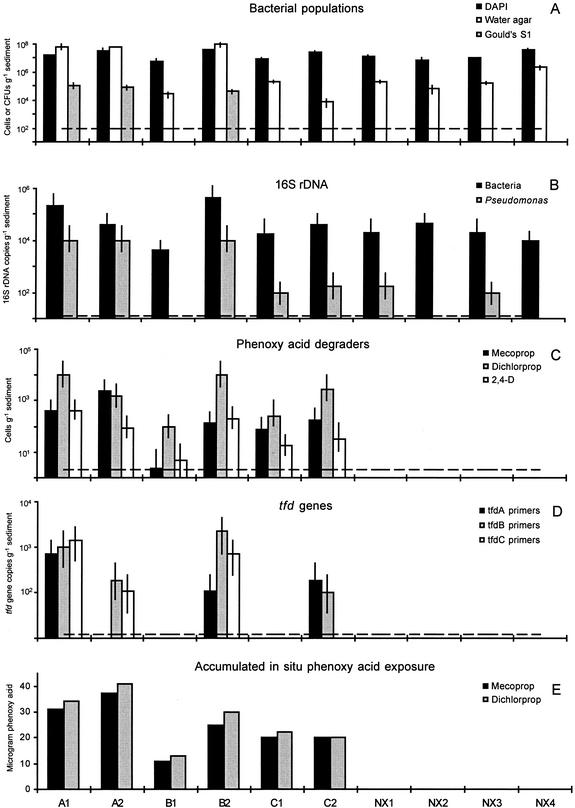
We found a total bacterial density of approximately 107 DAPI-stained cells g−1 of sediment (Fig. 3A), with no significant differences between samples (P > 0.05). In contrast, the population density measured as CFU on water agar was significantly higher in sediments A1, A2, and B2 (approximately 107 CFU g−1) than in any other sediment (approximately 103 to 105 CFU g−1) (Fig. 3A). These three sediments have previously been shown to have a much higher potential for degradation of phenoxy acid herbicides than the nonexposed sediments (NX1 to -4) (38). The higher CFU numbers found in the highly acclimated sediments A1, A2, and B2 suggest an increased culturability of the microbial communities and not an increase in the total bacterial population density. Similar to the total bacterial counts, MPN-PCR of Bacteria-specific 16S rDNA gene segments showed an equal distribution of DNA (approximately 104 to 105 gene copies g−1) (Fig. 3B). Only in B1 were significantly lower levels (P < 0.05) of 16S rDNA gene abundance recorded, and only in B2 were significantly higher levels (P < 0.05) of 16S rDNA gene abundance recorded. The MPN-PCR assay gave a lower estimate of bacterial populations compared to those from the classical microbiological methods. The true copy number, however, is probably several orders of magnitude higher: the calculation of MPNs is based on the capability of one gene copy to yield a positive reaction, and this is rarely enough to produce a visible PCR product (31). Furthermore, complete extraction of all bacterial DNA from the aquifer sediments may not be possible.
FIG. 3.
Effect of in situ herbicide exposure on microbial community composition in the Vejen aquifer. A1, A2, B1, B2, C1, and C2 indicate sediment samples collected within the herbicide-exposed area of the aquifer, while NX1 to -4 indicate samples collected outside the contaminant plume. Field samples were collected in the saturated zone at two depths (1 = upper, 2 = lower) (see also Materials and Methods). (A) Bacterial population density estimated by DAPI staining (cells per gram of sediment) and by plating on water and Gould's S1 agar (CFU per gram of sediment). Each data point represents the means ± standard deviations of nine replicate measurements. The broken line indicates the detection limit (102 CFU g−1) for culturable estimates. (B) Abundance of Bacteria- and Pseudomonas-specific 16S rDNA gene segments determined by MPN-PCR. Each data point represents the mean ± 95% confidence interval of three replicate measurements. The broken line indicates the detection limit (40 gene copies g−1, assuming that one gene copy can give rise to a PCR product). (C) Population density of phenoxy acid degraders as measured by an MPN assay relying on herbicide degradation. Each data point represents the mean ± 95% confidence interval of five replicate measurements. The broken line indicates the detection limit (2 cells g−1). (D) Abundance of tfd genes determined by MPN-PCR. Each data point represents the mean ± 95% confidence interval of three replicate measurements. The broken line indicates the detection limit (40 gene copies g−1, assuming that one gene copy can give rise to a PCR product). (E) Accumulated exposure levels of mecoprop and dichlorprop in the Vejen aquifer during the in situ natural field injection (reprinted from reference 38 with permission of the publisher).
Pseudomonas populations.
It is generally known that Pseudomonas bacteria are able to degrade a wide range of xenobiotic compounds (29). In the Vejen aquifer, culturable Pseudomonas organisms were only detected in the acclimated sediments A1, A2, and B2 (approximately 105 CFU g−1) (Fig. 3A). Nevertheless, PCR analysis demonstrated that Pseudomonas-specific gene segments were present in both herbicide-exposed (A1, A2, B2, C1, and C2) and -nonexposed (NX1 and NX3) sediments (Fig. 3B). The abundance, though, was significantly higher in A1, A2, and B2 than in any other sediment (P < 0.05). Densities recorded as CFU on Gould's S1 agar and Pseudomonas-specific 16S rDNA gene segments were linearly correlated (r = 0.92), and data indicated that the microbial communities of the acclimated sediments A1, A2, and B2 were enriched in bacteria phylogenetically belonging to Pseudomonas. These results might suggest that Pseudomonas bacteria had a possible role in the phenoxy acid degradation in the Vejen aquifer. Therefore, we tested approximately 50 Pseudomonas isolates for the ability to mineralize 14C-labeled mecoprop and 2,4-D, but only one bacterial morphotype had this capacity (data not shown). This suggests that only a few of the enriched Pseudomonas bacteria had a potential direct impact on the in situ herbicide degradation, although the involvement of nonculturable species or the dependence on codegrading microorganisms cannot be excluded. Nor can the possible existence of dichlorprop-degrading bacteria not detected by this assay be excluded.
Phenoxy acid degraders.
Mecoprop, dichlorprop, and 2,4-D degraders were found in all herbicide-exposed sediments (Fig. 3C). In contrast, at the nonexposed sites the number of specific degraders was below the detection limit (2 cells g−1). In general, the abundance of degraders was highest in sediments A1, A2, B2, and C2 (approximately 102 to 104 degraders g−1), whereas B1 harbored the significantly lowest number of phenoxy acid degraders (approximately 100 to 102 g−1) (P < 0.05). In correspondence with this, the mass of field-injected mecoprop and dichlorprop could at most explain increases in population densities of 2 × 105 cells g−1 sediment, assuming (i) a yield coefficient of 0.3 g of carbon in produced biomass per gram of carbon of degraded herbicide (38), (ii) a weight of 1.72 × 10−13 g of carbon per cell (4), and (iii) a homogenous distribution of produced biomass in the plume (2 m3). The calculations included the first meter from the injection where the major part of the phenoxy acids was degraded (8). We found that the estimated density of mecoprop and 2,4-D degraders was identical irrespective of whether the MPN assay was performed by high-performance liquid chromatography or mineralization analyses (data not shown). The phenoxy acid degraders of the Vejen aquifer thus had the potential to degrade the herbicides completely to CO2.
The Vejen aquifer had not been exposed to 2,4-D during the field injection, but the number of 2,4-D degraders corresponded well with the number of mecoprop and dichlorprop degraders (Fig. 3C). Most bacterial phenoxy acid degraders are able to transform several phenoxy acids. For example, mixed and pure bacterial cultures enriched on mecoprop typically show a broad substrate range, including dichlorprop, 2,4-D, and MCPA [2-(4-chloro-2-methylphenoxy)acetic acid] (23, 34, 42). However, isolates enriched on 2,4-D typically do not degrade phenoxypropionic acids (13, 40).
Increases in the density of specific degraders have also been observed in situ in subsurface environments and aquifers contaminated with fuel compounds (6, 25, 32), but they have been observed at much higher contaminant concentrations than the herbicide concentrations in the Vejen aquifer. Also, several studies have demonstrated an increased population density (104 to 108 g−1) of 2,4-D degraders following exposure to 2,4-D (in milligrams per kilogram levels) in laboratory microcosms of surface soil (12, 19). However, no studies have reported on increases in the density of specific microbial degraders following in situ exposure to low herbicide concentrations (<40 μg l−1) in groundwater aquifers.
tfd genes.
PCR amplification with tfd-specific primers was only seen in the herbicide-exposed sediments (Fig. 3D). tfdA-homologous genes were found in sediments A1, B2, and C2 (Fig. 3D), and RFLP analysis revealed a difference between the populations found in C2 and those in A1 and B2 (data not shown). tfdB-homologous genes were observed in sediments A1, A2, B2, and C2 (Fig. 3D). Here, RFLP patterns of the populations in B2 were different from those in the other three sediments (data not shown). The abundance of tfd gene copies was approximately 102 to 103 g−1 of sediment (Fig. 3D), which is lower than was estimated by the culture-dependent assay for measuring phenoxy acid degraders (Fig. 3C). However, as in the case of the MPN-PCR analysis of Bacteria-specific 16S rDNA gene segments, the copy number is probably several orders of magnitude higher. Although resulting in different abundance levels, the culture-dependent and the culture-independent MPN assays generally correlated: tfdA versus mecoprop (r = 0.97), dichlorprop (r = 0.76), and 2,4-D (r = 0.91) degraders; tfdB versus mecoprop (r = 0.50), dichlorprop (r = 0.91), and 2,4-D (r = 0.70) degraders. The lack of tfd genes in sediments B1 and C1 is probably due to a lower population density of phenoxy acid degraders in these samples. It cannot be excluded, however, that the sequence of the tfd genes in these sediments differs from those of the archetype 2,4-D degrader, R. eutropha JMP134. Several bacterial 2,4-D degraders have been reported not to have any homology with the tfd genes of strain JMP134 (9, 16, 39). This could also explain the lack of tfdA genes in sediment A2. Also, presently only tfd gene sequences from culturable 2,4-D degraders are known.
Amplification with tfdC-specific primers resulted in PCR products in sediments A1, A2, and B2 (Fig. 3D), but these were significantly shorter than the product of strain JMP134 (data not shown). Further characterization of the tfdC PCR products by cloning and sequencing followed by a search for known DNA sequences at the GenBank database (http://www.ncbi.nlm.nih.gov/) revealed no homology to known tfdC genes or other DNA sequences (data not shown). At the protein level, however, significantly high homology with bacterial ferredoxin NADP reductases was found (data not shown).
The tfd genes are widespread among bacterial 2,4-D degraders (9, 16, 39), and the enrichment of these catabolic genes following application of 2,4-D to surface soil has previously been observed (19, 22). The detection of tfdA and tfdB genes in acclimated microbial communities of the Vejen aquifer suggests, however, that tfd genes are possibly also responsible for the transformation of phenoxypropionic acid compounds. This is in agreement with the recent finding of tfdA-homologous genes in three bacterial mecoprop degraders (30).
In general, the PCR analysis of tfd genes complemented the MPN measurements of culturable phenoxy acid degraders, although it failed to detect their presence in sediments B1 and C1. It may thus be concluded that the culture-dependent MPN assay is a better tool than PCR in the evaluation of in situ microbial acclimation. However, if nonculturable contaminant degraders need to be detected, the application of molecular analyses is necessary. Also, these analyses can give a more refined picture of the presence and distribution of the catabolic genes responsible for the observed in situ degradation.
The abundance of herbicide degraders as well as the presence of tfd genes correlated with the phenoxy acid field exposure (Fig. 3E). Furthermore, it has previously been shown that the sediments exposed to the highest amount of phenoxy acids also were the most acclimated to mecoprop, dichlorprop, and 2,4-D (38).
Concluding remarks.
Continuous exposure to low herbicide concentrations (< 40 μg l−1) in the Vejen aquifer changed the microbial community composition toward a higher bacterial culturability, increased abundance of Pseudomonas bacteria, and increased abundance of phenoxy acid degraders as well as tfd genes. The most pronounced effects were observed with sediments A1, A2, and B2, which had been exposed to the highest levels of phenoxy acids in the field. Data suggest that the degradation of mecoprop and dichlorprop measured in situ (8) was attributable to acclimated microbial communities (38) enriched by a heterogeneous population of phenoxy acid degraders carrying tfd genes. To our knowledge, this is the first study to show that in situ exposure of subsurface aquifers to low herbicide concentrations can markedly change the indigenous microbial community composition, resulting in acclimated microbial communities. This information contributes to our understanding of the complex processes taking place in polluted aquifers and may be used to decide what actions should be taken for the remediation of polluted groundwater environments. Whether the continuous exposure to even smaller herbicide concentrations coming from agricultural use also will result in acclimated microbial communities is still to be investigated.
Acknowledgments
This work was supported by the Danish Environmental Research Programme, Pesticides and Groundwater, and by the Technical University of Denmark.
We gratefully acknowledge the assistance of Spire Maja Kiersgaard and Rasmus Rune Hansen with the field sampling and of Spire Maja Kiersgaard and Pia Bach Jacobsen with the high-performance liquid chromatography analyses and measurements of DAPI-stained bacteria. We also thank Roger Garrett for access to the MegaBACE 1000 sequencer.
REFERENCES
- 1.Albrechtsen, H.-J., M. S. Mills, J. Aamand, and P. L. Bjerg. 2001. Degradation of herbicides in shallow Danish aquifers: an integrated laboratory and field study. Pest Manag. Sci. 57:341-350. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, M. (ed.). 1999. Acclimation, p. 17-40. In Biodegradation and bioremediation. Academic Press, San Diego, Calif.
- 3.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balkwill, D. L., F. R. Leach, J. T. Wilson, J. F. McNabb, and D. C. White. 1988. Equivalence of microbial biomass measures based on membrane lipid and cell wall components, ATP and direct counts in subsurface aquifer sediments. Microb. Ecol. 16:73-84. [DOI] [PubMed] [Google Scholar]
- 5.Bjerg, P. L., K. Hinsby, T. H. Christensen, and P. Gravesen. 1992. Spatial variability of hydraulic conductivity of an unconfined sandy aquifer determined by a mini slug test. J. Hydrol. 136:107-122. [Google Scholar]
- 6.Braddock, J. F., and K. A. McCarthy. 1996. Hydrologic and microbiological factors affecting persistence and migration of petroleum hydrocarbons spilled in a continuous-permafrost region. Environ. Sci. Technol. 30:2626-2633. [Google Scholar]
- 7.Braun-Howland, E. B., P. A. Vescio, and S. A. Nierzwicki-Bauer. 1993. Use of a simplified cell blot technique and 16S rRNA-directed probes for identification of common environmental isolates. Appl. Environ. Microbiol. 59:3219-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broholm, M. M., K. Rügge, N. Tuxen, A. L. Højberg, H. Mosbæk, and P. L. Bjerg. 2001. Fate of herbicides in a shallow aerobic aquifer: a continuous field injection experiment (Vejen, Denmark). Water Resour. Res. 37:3163-3176. [Google Scholar]
- 9.Cavalca, L., A. Hartmann, N. Rouard, and G. Soulas. 1999. Diversity of tfdC genes: distribution and polymorphism among 2,4-dichlorophenoxyacetic acid degrading soil bacteria. FEMS Microbiol. Ecol. 29:45-58. [Google Scholar]
- 10.de Lipthay, J. R., T. Barkay, J. Vekova, and S. J. Sørensen. 1999. Utilization of phenoxyacetic acid, by strains using either the ortho or meta cleavage of catechol during phenol degradation, after conjugal transfer of tfdA, the gene encoding a 2,4-dichlorophenoxyacetic acid/2-oxoglutarate dioxygenase. Appl. Microbiol. Biotechnol. 51:207-214. [DOI] [PubMed] [Google Scholar]
- 11.de Lipthay, J. R., J. Aamand, and T. Barkay. 2002. Expression of tfdA genes in aquatic microbial communities during acclimation to 2,4-dichlorophenoxyacetic acid. FEMS Microbiol. Ecol. 40:205-214. [DOI] [PubMed] [Google Scholar]
- 12.DiGiovanni, G. D., J. W. Neilson, I. L. Pepper, and N. A. Sinclair. 1996. Gene transfer of Alcaligenes eutrophus JMP134 plasmid pJP4 to indigenous soil recipients. Appl. Environ. Microbiol. 62:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Don, R. H., and J. M. Pemberton. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Don, R. H., A. J. Weightman, H.-J. Knackmuss, and K. N. Timmis. 1985. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4). J. Bacteriol. 161:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.European Environment Agency. 1999. Groundwater quality and quantity in Europe—environmental assessment report no. 3. European Environment Agency, Copenhagen, Denmark.
- 16.Fulthorpe, R. R., C. McGowan, O. V. Maltseva, W. E. Holben, and J. M. Tiedje. 1995. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl. Environ. Microbiol. 61:3274-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghiorse, W. C., and J. T. Wilson. 1988. Microbial ecology of the terrestrial subsurface. Adv. Appl. Microbiol. 33:107-172. [DOI] [PubMed] [Google Scholar]
- 18.Gould, W. D., C. Hagedorn, T. R. Bardinelli, and R. M. Zablotowicz. 1985. New selective media for enumeration and recovery of fluorescent pseudomonads from various habitats. Appl. Environ. Microbiol. 49:28-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holben, W. E., B. M. Schroeter, V. G. M. Calabrese, R. H. Olsen, J. K. Kukor, V. O. Biederbeck, A. E. Smith, and J. M. Tiedje. 1992. Gene probe analysis of soil microbial populations selected by amendment with 2,4-dichlorophenoxyacetic acid. Appl. Environ. Microbiol. 58:3941-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnsen, K., and P. Nielsen. 1999. Diversity of Pseudomonas strains isolated with King's B and Gould's S1 agar determined by repetitive extragenic palindromic-polymerase chain reaction, 16S rDNA sequencing and Fourier transform infrared spectroscopy characterisation. FEMS Microbiol. Lett. 173:155-162. [DOI] [PubMed] [Google Scholar]
- 21.Johnsen, K., Ø. Enger, C. S. Jacobsen, L. Thirup, and V. Torsvik. 1999. Quantitative selective PCR of 16S ribosomal DNA correlates well with selective agar plating in describing population dynamics of indigenous Pseudomonas spp. in soil hot spots. Appl. Environ. Microbiol. 65:1786-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ka, J. O., W. E. Holben, and J. M. Tiedje. 1994. Use of gene probes to aid in recovery and identification of functionally dominant 2,4-dichlorophenoxyacetic acid-degrading populations in soil. Appl. Environ. Microbiol. 60:1116-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilpi, S. 1989. Degradation of some phenoxy acid herbicides by mixed cultures of bacteria isolated from soil treated with 2-(2-methyl-4-chloro)phenoxypropionic acid. Microb. Ecol. 6:261-270. [DOI] [PubMed] [Google Scholar]
- 24.Larsen, L., S. R. Sørensen, and J. Aamand. 2000. Mecoprop, isoproturon, and atrazine in and above a sandy aquifer: vertical distribution of mineralization potential. Environ. Sci. Technol. 34:2426-2430. [Google Scholar]
- 25.Long, S. C., C. M. Aelion, D. C. Dobbins, and F. K. Pfaender. 1995. A comparison of microbial community characteristics among petroleum-contaminated and uncontaminated subsurface soils. Microb. Ecol. 30:297-307. [DOI] [PubMed] [Google Scholar]
- 26.Lyngkilde, J., and T. H. Christensen. 1992. Fate of organic contaminants in the redox zones of a landfill leachate pollution plume (Vejen, Denmark). J. Contam. Hydrol. 10:291-307. [Google Scholar]
- 27.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Sayler, G. S., S. W. Hooper, A. C. Layton, and J. M. H. King. 1990. Catabolic plasmids of environmental and ecological significance. Microb. Ecol. 19:1-20. [DOI] [PubMed] [Google Scholar]
- 30.Smejkal, C. W., T. Vallaeys, S. K. Burton, and H. M. Lappin-Scott. 2001. Substrate specificity of chlorophenoxyalkanoic acid-degrading bacteria is not dependent upon phylogenetically related tfdA gene types. Biol. Fertil. Soils 33:507-513. [Google Scholar]
- 31.Sobecky, P. A., T. J. Mincer, M. C. Chang, and D. R. Helinsky. 1997. Plasmids isolated from marine sediment microbial communities contain replication and incompatibility regions unrelated to those of known plasmid groups. Appl. Environ. Microbiol. 63:888-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song, H.-G., and R. Bartha. 1990. Effects of jet fuel spills on the microbial community of soil. Appl. Environ. Microbiol. 56:646-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Streber, W. R., K. N. Timmis, and M. H. Zenk. 1987. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J. Bacteriol. 169:2950-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tett, V. A., A. J. Willetts, and H. M. Lappin-Scott. 1994. Enantioselective degradation of the herbicide mecoprop [2-(2-methyl-4-chlorophenoxy)propionic acid] by mixed and pure bacterial cultures. FEMS Microbiol. Ecol. 14:191-200. [Google Scholar]
- 35.Thirup, L., K. Johnsen, and A. Winding. 2001. Succession of indigenous Pseudomonas spp. and actinomycetes on barley roots affected by the antagonistic strain Pseudomonas fluorescens DR54 and the fungicide imazalil. Appl. Environ. Microbiol. 67:1147-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Topp, E., T. Vallaeys, and G. Soulas. 1997. Pesticides: microbial degradation and effects on microorganisms, p. 547-575. In J. D. van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker, New York, N.Y.
- 37.Tuxen, N., P. L. Tuchsen, K. Rügge, H.-J. Albrechtsen, and P. L. Bjerg. 2000. Fate of seven pesticides in aquifer materials using aerobic column experiments. Chemosphere 41:1485-1494. [DOI] [PubMed] [Google Scholar]
- 38.Tuxen, N., J. R. de Lipthay, H.-J. Albrechtsen, J. Aamand, and P. L. Bjerg. 2002. Effect of exposure history on microbial herbicide degradation in an aerobic aquifer affected by a point source. Environ. Sci. Technol. 36:2205-2212. [DOI] [PubMed] [Google Scholar]
- 39.Vallaeys, T., R. R. Fulthorpe, A. M. Wright, and G. Soulas. 1996. The metabolic pathway of 2,4-dichlorophenoxyacetic acid degradation involves different families of tfdA and tfdB genes according to PCR-RFLP analysis. FEMS Microbiol. Ecol. 20:163-172. [Google Scholar]
- 40.Vallaeys, T., L. Albino, G. Soulas, A. D. Wright, and A. J. Weightman. 1998. Isolation and characterization of a stable 2,4-dichlorophenoxyacetic acid degrading bacterium, Variovorax paradoxus, using chemostat culture. Biotechnol. Lett. 20:1073-1076. [Google Scholar]
- 41.Wilson, J. T., J. F. McNabb, D. L. Balkwill, and W. C. Ghiorse. 1983. Enumeration and characterization of bacteria indigenous to a shallow water-table aquifer. Ground Water 2:134-142. [Google Scholar]
- 42.Zipper, C., K. Nickel, W. Angst, and H.-P. Kohler. 1996. Complete microbial degradation of both enantiomers of the chiral herbicide mecoprop [(RS)-2-(4-chloro-2-methylphenoxy)propionic acid] in an enantioselective manner by Sphingomonas herbicidovorans sp. nov. Appl. Environ. Microbiol. 62:4318-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zipper, C., M. J.-F. Suter, S. B. Haderlein, M. Gruhl, and H.-P. Kohler. 1998. Changes in the enantiomeric ratio of (R)- to (S)-mecoprop indicate in situ biodegradation of this chiral herbicide in a polluted aquifer. Environ. Sci. Technol. 32:2070-2076. [Google Scholar]