Abstract
The total bacterial community of an experimental slow sand filter (SSF) was analyzed by denaturing gradient gel electrophoresis (DGGE) of partial 16S rRNA gene PCR products. One dominant band had sequence homology to Legionella species, indicating that these bacteria were a large component of the SSF bacterial community. Populations within experimental and commercial SSF units were studied by using Legionella-specific PCR primers, and products were studied by DGGE and quantitative PCR analyses. In the experimental SSF unit, the DGGE profiles for sand column, reservoir, storage tank, and headwater tank samples each contained at least one intense band, indicating that a single Legionella strain was predominant in each sample. Greater numbers of DGGE bands of equal intensity were detected in the outflow water sample. Sequence analysis of these PCR products showed that several Legionella species were present and that the organisms exhibited similarity to strains isolated from environmental and clinical samples. Quantitative PCR analysis of the SSF samples showed that from the headwater sample through the sand column, the number of Legionella cells decreased, resulting in a lower number of cells in the outflow water. In the commercial SSF, legionellae were also detected in the sand column samples. Storing prefilter water or locating SSF units within greenhouses, which are often maintained at temperatures that are higher than the ambient temperature, increases the risk of growth of Legionella and should be avoided. Care should also be taken when used filter sand is handled or replaced, and regular monitoring of outflow water would be useful, especially if the water is used for misting or overhead irrigation.
Filtration with slow sand filters (SSF) has a long history of successful use for removing human pathogens during drinking water production (22, 46, 49) but has only recently been introduced into horticultural production systems (62). Contaminated irrigation water has long been recognized as an important source of plant pathogens and can be instrumental in disease spread in commercial horticultural nurseries (9), and SSF filtration shows great promise for control of fungal plant pathogens (62). An SSF relies on physical, chemical, and biological activity for controlling plant pathogens (12, 28, 45, 54, 55, 60, 61). To characterize the microorganisms involved in the suppression of fungal plant pathogens in several agricultural crops, PCR amplification of the 16S rRNA gene has great potential (40, 48, 63). This technique can be used at a general level to detect all bacteria with universal primers, or with more specific primers it can be used to detect specific bacterial groups, such as species. In a study of the total bacterial community in which PCR-denaturing gradient gel electrophoresis (DGGE) analysis of fragments from the 16S rRNA genes was used, a sequence with similarity to the Legionella cherrii sequence was found in the sand column of an experimental SSF unit (L. A. Calvo-Bado, T. R. Pettitt, N. Parsons, G. M. Petch, J. A. W. Morgan, and J. M. Whipps, submitted for publication), and this finding initiated the present study.
Legionella species are aerobic, non-spore-forming, typically flagellated, gram-negative, rod-shaped bacteria (14). They are ubiquitous in aquatic environments and have been found in environmental samples, such as interstitial water (39) and groundwater (41). In humans, Legionella pneumophila can cause Legionnaires' diseases and Pontiac fever (26, 27). In addition, there are other Legionella species that have been implicated in pneumonia which have been isolated from environmental and hospital patient samples (23). There are currently 42 described species representing at least 64 serogroups in the family Legionellaceae and the genus Legionella (7). Legionella species grow in water at temperatures between 20 and 50°C at pHs ranging from 2.0 to 9.5, and optimal growth occurs at 37°C. Legionella species generally occur in low numbers in such aquatic habitats, but under certain environmental conditions the number of these organisms can increase, causing outbreaks of disease. In general, an infection is contracted by inhalation of contaminated water containing Legionella cells in aerosols. Some sources of transmission which have led to outbreaks of disease have been reported to be cooling towers, natural hot spas, evaporative condensers, hot and cold water systems in hotels and hospitals, showers, soil and potting soil, a whirlpool bath, and protozoans (1, 11, 18, 19, 31, 35, 36, 44, 47, 50, 51).
The aim of this study was to characterize the Legionella populations present in an experimental SSF unit during the filtration process and in filters on commercial farms. PCR amplification was used as the main characterization method, as it allowed accurate analysis of the diversity of bacteria present and detected nonculturable bacteria and quantitative PCR allowed the relative abundance of Legionella DNA in samples to be determined.
MATERIALS AND METHODS
SSF unit and samples.
The SSF rig used has been described elsewhere (Calvo-Bado et al., submitted). Briefly, three replicate experimental SSF rigs were constructed, and these rigs were supplied from a common source of untreated water. The filters were made by using 3 m of 160-mm-diameter polyvinyl chloride pipe (Terain Ltd., Hampshire, United Kingdom). The water flow (0.15 m h−1) from each filter was regulated by using a 0.25-in. straight Wade coupling needle valve inserted into the end of a 40-mm-diameter pipe. Each column was constructed with a sand depth of 1 m and a 1.5-m head of water above the sand. In this system, water flow through the column was gravity assisted. Water was pumped to the top of the column (headwater) from a tank containing untreated reservoir water (300 liters) at a continuous rate (1 liter min−1); the untreated reservoir water was obtained from a large storage tank containing water from a reservoir. This storage tank was refilled with water from the reservoir weekly. An overflow pipe was used to maintain the water level, and the water from this pipe was pumped back to the tank containing the headwater via an overflow sump (50 liters). The sand used in all experiments was an autoclaved fine washed plasterer's sand obtained in southwest Hampshire, United Kingdom (New Milton Sand and Ballast Co.) as recommended by Visscher et al. (56) for drinking water filter sand. Sand and water samples were removed via inspection hatches or through a series of ports mounted at 100-mm intervals in the column. In this experiment, two SSF runs were carried out, each with three replicate SSF columns for a minimum of 4 weeks. Each water sample was taken with a replicate. Water samples were obtained from the reservoir, the storage tank, the headwater tank, and the outflow water. Sand samples were taken from surface, the top (1 cm), the middle (50 cm), and the bottom (80 cm) of the sand column. In addition, samples were obtained from a series of commercial sand filters in operation in the United Kingdom horticulture industry; the characteristics of these filters are described in Table 1.
TABLE 1.
Characteristics of the experimental and commercial SSF units
| SSF | Type | Approx surface area (m2) | Location | Temp (°C) |
|---|---|---|---|---|
| SSF-1 | Experimentala | 0.02 | Glasshouse | 10-20 |
| SSF-2 | Commercial | Outdoors | Ambientb | |
| SSF-3 | Commercial | 115 | Glasshouse | 10-20 |
| SSF-4 | Commercial | 100 | Outdoors | Ambientb |
| SSF-5 | Commercial | 1 | Outdoors | Ambientb |
| SSF-6 | Commercial | 115 | Glasshouse | 10-20 |
| SSF-7 | Commercial | 50 | Outdoors | Ambientb |
SSF used in this study.
Temperature range during operation, 2 to 15°C.
DNA isolation.
Briefly, for each water sample, 1 liter of water was taken from the reservoir, the storage tank, the headwater tank, or the outflow. The water samples were filtered (pore size, 0.22 μm; GV-Durapore; Millipore Ltd., Watford, United Kingdom), and the cells were washed off the filter surface with 5 ml of sterile water. Each sample was centrifuged at 13,000 × g for 20 min, and the pellet was saved. Sand samples (0.5 g, wet weight) from different depths in the SSF columns (top layer [1 cm], middle layer [50 cm], and bottom layer [80 cm]) were taken 1, 2, and 4 weeks after the columns were loaded with sand. To each cell pellet or sand sample, 1 ml of extraction buffer (0.12 M Na2HPO4, pH 8.0) and 1/3 volume of 0.1-mm-diameter glass beads (BioSpec Products Inc., Bartlesville, Okla.) were added. The sample was shaken vigorously for 3 min in a mini bead beater (BioSpec Products Inc.). Immediately, 0.1% (wt/vol) sodium dodecyl sulfate was added, and the sample was homogenized and placed on ice for 10 min. One milliliter of phenol (pH 8.0; Sigma, Cambridge, United Kingdom) was added, and the sample was centrifuged at 5,000 × g for 15 min. The supernatant containing the DNA was recovered, and 1 ml of chloroform-isoamyl alcohol (24:1) was added to the DNA suspension. After mixing, the sample was centrifuged at 5,000 × g for 15 min, and the aqueous phase was saved. To this phase, 0.6 volume of isopropanol and 0.1 volume of 5 M NaCl were added. The sample was centrifuged at 13,000 × g for 30 min, and the pellet was washed with 70% (vol/vol) ethanol. The pelleted DNA was air dried for 10 to 15 min and resuspended in 100 μl of sterile water. The DNA was further purified by using a Geneclean spin kit (Bio 101 Inc., Nottingham, United Kingdom) according to the manufacturer's recommendations. The DNA from the sample was finally eluted in 50 μl, stored at −70°C, and analyzed by agarose gel electrophoresis to estimate the yield. Appropriate dilutions were made for PCR amplification.
PCR amplification.
For analysis of the total bacterial community, the V3 region (38) of the 16S rRNA-encoded gene between positions 341 and 534 (Escherichia coli numbering [15]) was amplified by PCR. The primers used were forward primer 341 with 40 GC-rich bases (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCA-3′) and reverse primer 534 (5′-ATTACCGCGGCTGCTGG-3′). For Legionella species, a seminested PCR specific for the bacterial 16S rRNA gene was used (37). Forward primer LEG-225 (5′-AAGATTAGCCTGCGTCCGAT-3′) and reverse primer LEG-858 (5′-GTCAACTTATCGCGTTTGCT-3′) were used for the initial PCR step, and they amplified a 654-bp fragment. For the second PCR step, forward primer LEG-448 (5′-GAGGGTTGATAGGTTAAGAGC-3′) and primer LEG-858 were used, and they amplified a 430-bp fragment. These primers were located at positions 225 to 244 and 880 to 859 (E. coli numbering [15]). For DGGE analysis, PCR amplification with primers LEG-448 GC clamp (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGGAGGGTTGATAGGTTAAGAGC-3′) and LEG-858 was used to obtain a 470-bp fragment. In addition, a GC clamp was also added to reverse primer LEG-858, which was used in combination with LEG-448 to assess variation in the DGGE banding patterns. Routinely, 10 to 50 ng of DNA template, 25 pmol of each primer, a deoxynucleoside triphosphate mixture containing each deoxynucleoside triphosphate at a concentration of 20 mM, 1.25 U of thermostable DNA polymerase, 10× reaction buffer, and 1.5 mM MgCl2 (Advance Biotechnologies, Surrey, United Kingdom) were used in a 100-μl (final volume) reaction mixture. For the initial PCR (with LEG-225 and LEG-858), one cycle of 95°C for 90 s followed by 30 cycles of 95°C for 10 s, 64°C for 60 s, and 72°C for 60 s and a final extension cycle of 72°C for 5 min was performed. For the second step (with LEG-448 and LEG-858), one cycle of 95°C for 90 s followed by 20 cycles of 95°C for 30 s, 66°C for 60 s, and 72°C for 60 s and a final extension cycle of 72°C for 5 min was used. The PCR products were analyzed by agarose gel electrophoresis to determine the yield and purity. For DGGE analysis, the 430-bp fragment was excised from the gel and purified with a QIAquick gel extraction kit (Qiagen, West Sussex, United Kingdom). A further PCR amplification (with LEG-448 GC clamp and LEG-858) in which DNA from the 430-bp purified fragment was used as a target was performed as follows: one cycle of 95°C for 90 s, followed by 35 cycles of 95°C for 30 s, 66°C for 60 s, and 72°C for 60 s and a final extension cycle of 72°C for 10 min.
DGGE.
DGGE analysis was performed by using a DCode mutation detection system (Bio-Rad, Hertfordshire, United Kingdom). Gels containing 8% (wt/vol) acrylamide (ratio of acrylamide to bisacrylamide, 37:1) were formed between 10 and 60% denaturant, with 100% denaturant defined as 7 M urea and 40% (vol/vol) formamide (38). The acrylamide gels were polymerized by adding 0.15% tetramethylethylenediamine and 0.03% ammonium peroxodisulfate (from a 10% [wt/vol] stock solution). Linear denaturant gradients were constructed with a gradient maker (BDH, Leicestershire, United Kingdom) by using a 16-cm-long gel and a 1-mm gel width. Normally, 300 to 500 ng of PCR product was loaded onto each lane of a gel. The gels were electrophoresed at 60 V for 16 h and were maintained at 60°C in 7 liters of 0.5× TAE buffer (40 mM Tris-acetate, 1 mM EDTA; pH 8.0). The gels were stained for 20 min with distilled water containing 25 μl of ethidium bromide (10 mg ml−1) and washed with distilled water for 20 min prior to visualization. A set of seven laboratory strains (Agrobacterium rhizogenes, Arthrobacter polychromogenes, Bacillus subtilis, Burkholderia phenazinium, Paenibacillus amylolyticus, Pseudomonas fluorescens, and Sphingomonas yanoikuyae) was used to construct a standard marker for DGGE analysis.
Quantitative PCR.
Quantitative analysis of Legionella cell numbers based on DNA concentrations in SSF samples was performed by real-time quantitative PCR with SYBR green. Amplification was performed with the ABI Prism 7900HT sequence detection system (Perkin Elmer-Applied Biosystems, Warrington, United Kingdom) and a Quantitect SYBR green PCR kit (Qiagen). The PCR was performed as follows: one cycle of 50°C for 2 min and 95°C for 15 min, followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s. A final step of 95°C with a 2% ramp rate for melting curve analysis was included. Data were collected at each step, as well as the ramp up to 94°C. Each 50-μl reaction mixture contained 25 μl of 2× Quantitect SYBR green PCR master mixture, 0.67 μM primer LEG-448, 0.67 μM primer LEG-858, and 2 μl of DNA. The standard curve was generated by using fivefold dilutions (800 pg to 51.2 fg) of purified (Qiagen) chromosomal DNA from a pure culture of L. pneumophila (a gift from C. Winstanley, Department of Medical Microbiology and Genito-Urinary Infections, University of Liverpool, Liverpool, United Kingdom). Each dilution of standard DNA was measured by quantitative PCR in triplicate. The average of four readings was calculated for each sample: two at a 10−1 dilution and two at a 10−2 dilution for water samples and two at a 10−2 dilution and two at a 10−3 dilution for sand bed samples. Standard curves relating the threshold cycle to the log10 DNA concentration and the DNA concentration of unknown samples were generated with the ABI Prism 7900HT software. The detection threshold was adjusted manually to give the largest R2 value possible for the standard curve and a gradient closest to −3.32, which represents 100% efficiency. The amounts of Legionella DNA present in samples were calculated, and the numbers of cells present were estimated (5).
Cloning and sequencing.
The PCR products were cloned into pGEM-T Easy Vector System I (Promega, Southampton, United Kingdom) by following the manufacturer's instructions. The DNA was electroporated into Escherichia coli DH10B electrocompetent cells (Invitrogen-Life Technologies, Paisley, United Kingdom) by using a field strength of 12.5 V cm−3. The transformed cells were plated onto Luria-Bertani agar (Merck, Leicestershire, United Kingdom) containing 50 μg of ampicillin per ml and 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml. The plates were incubated at 37°C overnight, and white colonies were isolated. To confirm the success of cloning and selection of clones for sequencing, direct PCR amplification from colonies with primers LEG-448 GC clamp and LEG-858 followed by DGGE analysis was carried out. For selected colonies, plasmid DNA was extracted from the cultures with a Qiagen 8 Ultra Plasmid kit (Qiagen). Sequencing reactions with mixtures (20 μl) containing 2 μl of plasmid DNA (250 ng μl−1), 6 μl of an ABI PRISM BigDye terminator cycle sequence Ready Reaction kit (Perkin Elmer-Applied Biosystems), 3.2 μl of the SP6 promoter primer (1 pmol μl−1; Promega, Madison, Wis.), and 8.8 μl of distilled sterile water were performed with a Hybaid PCR MultiBlock system (Hybaid Ltd., Middlesex, United Kingdom). Standard PCR sequencing reaction conditions were used according to the manufacturer's recommendations. The products were analyzed with an ABI PRISM 377 DNA cycle sequencer (Perkin Elmer-Applied Biosystems).
Sequence data analysis.
All sequences were edited and assembled by using the DNAstar SeqMan II sequence analysis package (Lasergene Inc., Madison, Wis.). Sequences were compared to sequences in the Ribosomal Database Project II (http://rdp.cme.msu.edu/html/) (34) and EMBL DNA databases by using Fasta 3 (http://www.ebi.ac.uk/embl/index.html) (52). Multiple sequences were analyzed by using the PHYLIP SEQBOOT, DNADIST, and NEIGHBOR packages as described by Ludwig et al. (33). Multiple data sets were used (n = 1,000), and a number of dendrograms were compared. Results were viewed by using DRAWTREE. The dendrogram shown is representative of the dendrograms obtained.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the EMBL database under accession no. AJ512248 to AJ512274. The accession numbers for the sequences used for comparison in this study are as follows: Legionella anisa, Z32635; Legionella birminghamensis, Z49717; Legionella cherrii, Z49720; Legionella erythra, Z32638; Legionella gresilensis, AF122883; Legionella lytica comb. nov., X66835; Legionella (Tatlockia) maceachernii, AF227161; Legionella parisiensis, U59697; Legionella pneumophila, AF129524; Legionella quinlivanii, Z49733; Legionella wadsworthii, Z49738; strain LLAP-1, U64034; strain LLAP-2, U44909; and strain LLAP-9, U44911.
RESULTS AND DISCUSSION
PCR detection of Legionella.
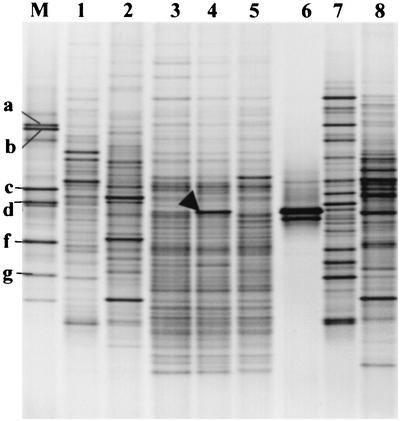
The total bacterial population in sand extracted from the SSF unit was profiled by using universal primers and DGGE profiling. Sequence analysis of visible bands identified a range of bacteria that were present (Calvo-Bado et al., submitted); however, one band showed sequence similarity (95% identity over 194 bp) to L. cherrii (Fig. 1). In one sample this band was a major band representing approximately 5% of the total PCR product; in the other samples this band was detected and indicated that Legionella-like bacteria were present at levels that were higher than expected. Therefore, a detailed analysis of this group of bacteria was initiated with specific PCR primers. The sensitivity and specificity of the seminested PCR described above were tested, and the technique produced results similar to those of Myamoto et al. (37).
FIG. 1.
Ethidium bromide-stained DGGE gel (negative image) showing the 194-bp PCR-amplified fragment of the 16S rRNA genes (V3 region) from the total bacterial communities from the SSF samples. Lane 1, reservoir; lane 2, storage tank; lanes 3 to 5, top layer of three replicate sand columns; lane 6, L. cherrii; lane 7, headwater; lane 8, outflow water; lane M, bacterial marker containing P. fluorescens (a), S. yanoikuyae (b), B. subtilis (c), B. phenazinium (d), P. amylolyticus (e), A. rhizogenes (f), and A. polychromogenes (g). The arrowhead indicates the DGGE band that was initially excised from the total bacterial community, cloned, and sequenced (homology to L. cherrii).
DGGE analysis.
Agarose gel electrophoresis indicated that a single intense 470-bp band was amplified from the gel-purified fragment used for DGGE analysis (with primers LEG-448 GC clamp and LEG-858). One or two dominant DGGE bands were obtained for the reservoir, storage tank, and headwater tank samples, as well as the top, middle, and bottom sand column samples. However, greater numbers of DGGE bands (12 to 15 bands) having the same intensity were detected in the outflow water samples, indicating that there was an increase in the diversity of the Legionella population following passage of the water through the SSF. As band position within a gel reflects a change in the melting properties of a PCR product, no inference concerning the types of changes that took place could be made as the results could reflect changes in sequence composition and also the position of a change. As none of the DGGE bands were dominant in the outflow water, no dominant strain was washed off into the water flowing through the SSF (Fig. 2). Similar DGGE results were obtained for outflow water samples obtained in different months during the experimental run of the SSF unit. As sequence analysis of the bands indicated that the double bands observed had the same sequence, DGGE analysis with primers LEG-448 and LEG-858 GC clamp was carried out. This analysis produced the same results (data not shown); however, a single DGGE band was detected in all the samples except the outflow water samples. As the sequence analysis was carried out with products obtained by using the first primer pair, this gel is shown in Fig. 2.
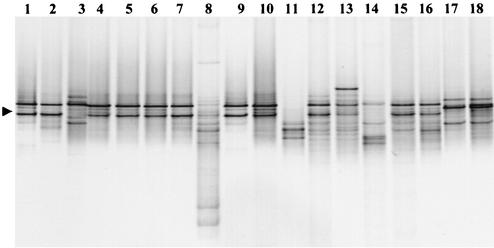
FIG. 2.
Ethidium bromide-stained DGGE gel (negative image) of PCR products amplified by using the Legionella-specific LEG-448 GC clamp and LEG-858 primers for the 16S rRNA genes. Samples were taken from experimental and commercial SSF units. Lanes 2 to 8 show the changes within the Legionella populations in the sand column and water samples in the experimental SSF unit (SSF-1). Lanes 9 to 18 show comparative DGGE banding patterns obtained from the top layer sand column samples from the experimental system (SSF-1) and six commercial farms (SSF-2 to SSF-18). Lanes 1 and 9, L. pneumophila type strain; lane 2, reservoir; lane 3, storage tank; lane 4, top layer; lane 5, middle layer; lane 6, bottom layer; lane 7, headwater; lane 8, outflow water; lane 10, SSF-1; lane 11, SSF-2; lane 12, SSF-3; lane 13, SSF-4; lane 14, SSF-5; lanes 15 and 16, SSF-6 samples a and b; and lanes 17 and 18, SSF-7 samples a and b. Sample a was a sand sample taken from an SSF when it was not in use (winter period), and sample b was a sand sample taken from an SSF when it was in use. The arrowhead indicates the DGGE band from L. pneumophila.
As the SSF unit was an experimental system designed to simulate industrial-size units, samples were obtained from commercial units to confirm the findings obtained with the experimental system. All six commercial filters examined were positive for Legionella and gave PCR products of the appropriate size. DGGE analysis of commercial SSF showed that one or two bands dominated the sand samples (Fig. 2). This indicated that the occurrence of Legionella in samples was not restricted to the experimental system and that this group of bacteria is potentially widespread in the industry. As sand filters provide a surface that is conducive to the growth of Legionella, this result may not be surprising. However, the extent of colonization by a diversity of strains raised concerns about the use of SSF units for providing water that could be used for misting crops or splashed within glasshouse facilities. Although the bands were the correct size, diverse denaturing points for bands were evident. The point at which the bands are denatured can be used to tentatively determine identities with specific primers; however, suitable standards from a variety of strains were not available for this work. Therefore, sequence analysis of bands was used to confirm the identities of bands and to determine similarity to known species.
Sequence analysis of the 470-bp PCR products.
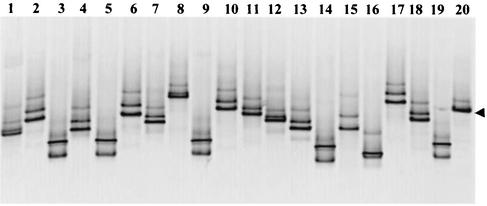
The PCR products from each sample were cloned and reanalyzed by PCR and DGGE analysis. For the outflow water clones, as expected, great variation in the DGGE banding patterns was observed. For the clones obtained from reservoir, storage tank, and headwater tank samples and top, middle, and bottom sand samples, bands with the same migration pattern as the bands detected in the initial samples were obtained. This confirmed the identities of the clones and suggested that one dominant but different clone was present in each sample. When the banding patterns of 40 clones were compared, at least 15 PCR products with different denaturing points were identified in the samples (Fig. 3). Interestingly, each clone produced more than one DGGE band even though sequence analyses of the clones showed that a single clean sequence was present in every clone. This multiple-band pattern could be explained if several melting domains were present in the PCR product. The sequence next to the LEG-448 primer sequence is AT rich (>50% AT) and clamped internally by an 80% GC region. There is also an AT-rich region (70% AT) consisting of 40 bp next to the LEG-858 primer sequence. Therefore, when the clamp was added to the LEG-858 primer, it stabilized the 70% AT-rich region, but when it was added to the LEG-448 primer, a region with several melting domains (denaturing points) was produced. This could have resulted in a multiple-band pattern with one pair of primers and not with the other pair. For the outflow water, bands were separated by DGGE that had as few as 4-bp differences in the 430-bp sequence. This highlights the resolving power of DGGE analysis.
FIG. 3.
Ethidium bromide-stained DGGE gel (negative image) obtained by using PCR products from 19 different clones (lanes 1 to 19) and L. pneumophila type strain (lane 20) amplified with the Legionella-specific LEG-448 GC clamp and LEG-858 primers for the 16S rRNA genes. Samples were obtained from the outflow water from the experimental SSF unit. The arrowhead indicates the DGGE band from L. pneumophila.
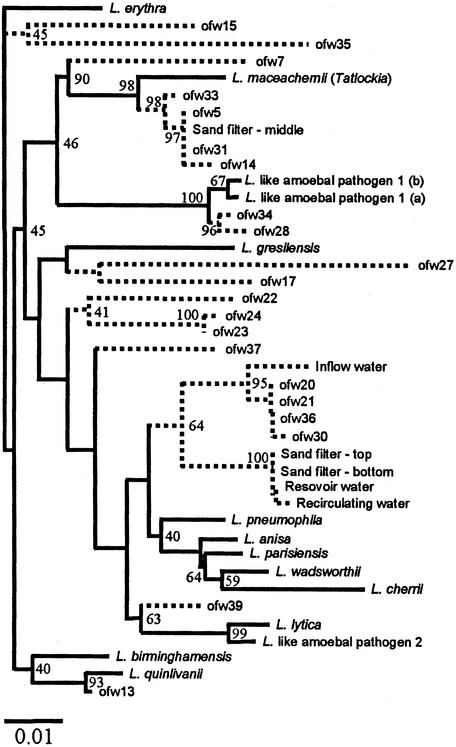
When sequences from the PCR products were compared to database entries, they exhibited the greatest similarity to 16S rRNA-encoding genes from Legionella species (Table 2). DNA sequence similarity to at least 10 different species of the genus Legionella was detected. In general, most of the sequences showed between 95 and 99% sequence identity to this group, and this confirmed that the PCR primers used were detecting bacteria belonging to the genus Legionella. When a phylogenetic analysis of the clones was carried out, this area of the 16S rRNA gene divided known Legionella species (Fig. 4). L. lytica, L. pneumophila, and L. parisiensis grouped together separate from L. birminghamensis, L. erythra, L. quinlivanii, L. maceachernii, and L. micdadei. Within the groups, subgroups containing Legionella-like amoebal sequences were also present. Since only partial sequences (430 bp) could be used, the analysis was limited. However, this type of analysis allows comparisons of sequences to each other, as well as to database entries. Only two of the sequences obtained, clones ofw-27 and ofw-35, showed low sequence similarity to the genus Legionella (92.5 and 93.7%, respectively). Both of these sequences exhibited the greatest similarity to L. erythra, L. rubrilucens, and L. quinlivanii, which were isolated from water (6, 13). However, even lower levels of similarity to bacteria outside the Legionella group (e.g., Coxiella burnettii) were detected. Therefore, these sequences probably represent a distant group of Legionella-like bacteria that are not closely associated with members having known 16S rRNA sequences. In the SSF, L. parisiensis and L. maceachernii-like sequences were found in reservoir, storage tank, and headwater tank samples and top, middle, and bottom sand samples. These Legionella species were likely to be present at low levels in the storage tank or reservoir water as this was the only source for introduction of strains because the system was sterilized prior to operation. It appears that the strains colonized the sand column of the SSF, where one of them may have been dominant in any particular part of the population. L. parisiensis and L. maceachernii were originally isolated from water from a cooling tower and a potable water cistern-home evaporator and appear to be well suited to the SSF environment according to our results (13). Originally, a 194-bp PCR-DGGE fragment corresponding to an L. cherrii-like sequence was identified in the sand column samples of this SSF unit by using universal bacterial primers. However, this study showed that the small PCR fragment corresponded to an L. parisiensis sequence. Analysis of the 16S rRNA-encoded genes for these species showed that L. cherrii and L. parisiensis have 98.2% sequence similarity and are closely related (data not shown). The larger sequence obtained with the Legionella-specific primers indicates that the bacteria present had a 16S rRNA gene more like that of L. parisiensis.
TABLE 2.
Sequence similarity of the PCR products amplified by using the Legionella-specific LEG-448 and LEG-858 primers for the 16S rRNA genes of 27 clones from reservoir, storage tank, headwater, sand column (top, middle, and bottom layers), and outflow water samples from the experimental SSF unit
| SSF clonea | Most closely related Legionella speciesb | % Similarity | Sero- group(s)c |
|---|---|---|---|
| ofw-23 | L. birminghamensis | 96.0 | 1 |
| ofw-24 | L. birminghamensis | 95.8 | 1 |
| ofw-15 | L. erythra | 95.4 | 1 and 2 |
| ofw-35 | L. erythra, L. rubrilucens, L. quinlivanii | 93.7 | 1 and 2 |
| ofw-37 | L. lytica | 96.7 | 1 |
| Middle layer | L. maceachernii | 97.9 | 1 |
| ofw-5 | L. maceachernii | 97.9 | 1 |
| ofw-14 | L. maceachernii | 97.4 | 1 |
| ofw-31 | L. maceachernii | 97.9 | 1 |
| ofw-33 | L. maceachernii | 97.9 | 1 |
| ofw-7 | L. macdadei | 95.1 | 1 |
| Top layer | L. parisiensis | 97.4 | 1 |
| Bottom layer | L. parisiensis | 97.4 | 1 |
| Storage tank | L. parisiensis | 96.0 | 1 |
| Reservoir | L. parisiensis | 97.4 | 1 |
| Headwater | L. parisiensis | 97.2 | 1 |
| ofw-20 | L. parisiensis | 97.4 | 1 |
| ofw-21 | L. parisiensis | 97.4 | 1 |
| ofw-30 | L. parisiensis | 97.2 | 1 |
| ofw-36 | L. parisiensis | 97.4 | 1 |
| ofw-39 | L. parisiensis | 97.4 | 1 |
| ofw-17 | L. pneumophila | 96.5 | 1 |
| ofw-22 | L. pneumophila | 96.0 | 1 |
| ofw-13 | L. quinlivanii | 99.3 | 1 |
| ofw-27 | L. quinlivanii | 92.5 | 1 |
| ofw-28 | Legionella-like amoebal pathogen 1 | 99.0 | |
| ofw-34 | Legionella-like amoebal pathogen 1 | 99.3 |
ofw, outflow water.
Sequences were obtained from the Ribosomal Database Project II (34) and the EMBL DNA database (52). L. birminghamensis, L. macdadei, and L. pneumophila were isolated from hospital patients. L. erythra, L. rubrilucens, L. quinlivanii, and L. maceachernii were isolated from environmental samples. L. lytica and Legionella-like amoebal pathogen 1 were isolated from free-living amoebae. L. parisiensis was isolated from environmental samples and hospital patients.
Serogroups associated with diseases.
FIG. 4.
Phylogenetic tree showing distances based on the 16S rRNA sequences obtained from 27 clones from samples from the reservoir, the storage tank, the headwater tank, the top, middle, and bottom sand column layers, and outflow water from the experimental SSF unit. Sequences of the 16S rRNA genes of Legionella strains used for comparison in this study were retrieved from the EMBL DNA database (52). The 16S rRNA sequences obtained in this work are partial sequences (430 bp), and therefore their positions are indicated by dashed lines. The dendrogram was generated by using the programs DNASIST (maximum likelihood) and NEIGHBOR (neighbor joining). Scale bar = 0.1% estimated sequence divergence. Resampling (bootstrap analysis) was applied to the data, and the values obtained (percentages) are indicated at the nodes.
Diverse sequences were obtained from the outflow water sample, reflecting the diversity of the DGGE bands seen on the gel. Two sequences (ofw-28 and ofw-34) showed high levels of similarity (99.0 and 99.3% identity, respectively) to a Legionella-like amoebal human pathogen, LLAP-1 (43). LLAP-1 was originally isolated in 1981 from the host Acanthamoeba polyphaga from a tank of a portable water well that has been associated with outbreaks of Legionnaires' disease (2). The ofw-37 sequence exhibited 96.7% similarity to sequences of L. lytica, LLAP-2, LLAP-6, and LLAP-9, which were also isolated from Acanthamoeba hosts in water samples (10). Legionella species proliferate inside free-living amoebae in natural environments, including Acanthamoeba spp., Hartmanella spp., and Naegleria spp. (3, 16, 17, 20, 24, 25, 42, 43, 59). These amoebae are potentially pathogenic to humans. Protozoans such as flagellates, ciliates, and amoebae are common in SSF (21, 28). However, a comprehensive investigation of the diversity of protozoans in SSF has not been carried out. PCR primers specific for Acanthamoeba spp. have been developed for preliminary tests with some SSF samples. In all of our samples that were tested, positive results were obtained (data not shown). This is not surprising based on the large number of sequences associated with protozoans and some of the sequences described in this study. One of the potential problems of the association of Legionella and amoebae is that the amoebae can survive environmental temperature extremes and chlorination and produce respirable vesicles containing live L. pneumophila (8). This makes disinfection particularly difficult.
Other sequences with similarity to the sequences of a variety of Legionella species, such as L. micdadei, associated with human pneumonia (30), were evident in the outflow water in our experimental SSF. These results confirm that processing the water through the sand column increased the diversity of strains in the outflow water. Of 27 clones sequenced, only 2 exhibited similarity to strains found in the reservoir, the storage tank, the headwater tank, and the sand column. The rest of them were unique in the outflow water sample. An increase in diversity is not unique to the experimental system used here, as diverse DGGE bands were detected in the outflow water of a commercial unit operated at the Horticulture Research International Efford site (data not shown). Although DGGE bands from this second unit have not been sequenced, all the data which we have for this type of PCR product indicate that only genes similar to those in Legionella sequences are amplified. The increase in diversity in the outflow water may have resulted from removal of a dominant strain from the reservoir water, so that the underlying diverse strains that were present were then detected. Alternatively, the filter may have allowed the growth of less dominant strains, which were then washed off into the outflow water and detected. The sand column in the SSF provides a large surface for colonization by bacteria. High ATP contents, particularly in the top layer of the experimental SSF unit, were detected, and this indicated that a good biofilm had developed (Calvo-Bado et al., submitted). A diverse population could live on this surface, and organisms such as protozoans that harbor Legionella could develop. Little is known about changes in Legionella populations in environmental samples, particularly samples from filtration processes or from recycling of sewage. In the SSF unit described here, Legionella seemed to be ubiquitous, and a variety of strains did well in the system. With such diverse bacteria, including some with sequences with similarity to the sequences of known pathogens, it is likely that the bacteria present included some that were a potential risk to workers in the environment. Since infection studies are not possible, determining similarity to organisms isolated from humans is the best that can be done to address this question.
Quantitative PCR.
In order to estimate the numbers of cells in the SSF samples, the amounts of Legionella DNA in the samples were determined. A quantitative PCR approach was used with an ABI Prism 7900HT sequence detection system. The standard curve for L. pneumophila DNA gave a good R2 value (0.99) and a gradient of −3.61, indicating that the PCR was efficient and reproducible. The presence of contaminants in DNA from environmental samples can cause PCR inhibition, autofluorescence, or quench fluorescence. These effects can be problematic for quantitative PCR. In order to overcome these potential problems, a DNA dilution series of the samples was prepared by using the recommendations of Stults et al. (53), who used this method to accurately quantify DNA from Geobacter spp. in aquifer sediments. The results obtained with Legionella primers showed that 10-fold dilution of DNA resulted in a 10-fold reduction in the quantity of target DNA detected in a manner identical to that observed with the pure L. pneumophila DNA standard. This indicated that there were no major inhibitors in the sample that were diluted out at the lower dilution. The results used were averages of four readings, which were used to calculate the amounts of DNA in the samples. The measurements were transformed into the number of cells by assuming that each Legionella cell contains 4.3 fg of DNA (5). Table 3 shows the sizes of the Legionella populations based on the DNA detected in the SSF samples from the experimental unit and commercial farms by real-time PCR. This experiment showed that only a few Legionella cells were present in the initial reservoir water (3.1 × 103 cells liter of water−1). The concentrations were higher in the storage tank and headwater tank operating prior to filtration (2.3 × 103 to 4.7 × 106 cells liter of water−1). This was probably a result of the conditions used, as storage of water at temperatures greater than 20°C is known to carry a risk of increasing Legionella contamination in horticultural systems (29). However, it was necessary to run the system as described above to maintain a realistically constant head pressure on the SSF. In addition, Legionella has complex nutritional requirements, which include a requirement for large amounts of iron (25). During the experiments, high pH values (pH 7.5 to 8.5) were detected in the water from the headwater tank, and brown-ochre particles were observed in suspension; such particles were also found subsequently on the top surface of the sand column. This could have been mainly due to oxidation of the iron frame holding the pumps in the headwater tank. This source of iron may have resulted in increased Legionella levels in this sample and the top surface of the sand column (6.1 × 105 to 3.9 × 106 cells g of sand−1) (Table 3). Nevertheless, after the water was passed through the sand column, a reduction in the size of the Legionella population was detected (4.4 × 104 to 5.5 × 104 cells liter of water−1). A reduction in the concentration was also observed in the sand itself; the populations were larger in the top layer (2.3 × 105 to 3.9 × 106 cells g of sand−1), and they decreased with depth in the sand column (1.5 × 104 cells g of sand−1). In this way the sand filter itself was beneficial for removal of Legionella. However, in the way that the model system was operated, the increase in the Legionella population prior to filtration resulted in no overall difference between the starting reservoir water levels and the outflow water levels. The results indicate that the prefiltration water and the sand in the SSF pose the greatest risk of Legionella contamination. To determine if the accumulation of Legionella cells in the experimental SSF system was unique and a result of its scale or operation, sand samples from six commercial SSF units were assessed to quantify Legionella DNA. All sand samples were positive, and high DNA concentrations and predicted cell numbers were detected in most of them (Table 3). We therefore anticipate that most SSF are colonized with Legionella species and should be treated with care. A disinfection procedure prior to removal of the sand from the filter is recommended, since the numbers of Legionella cells within the sand are likely to be high. Although the outflow water from the experimental system was shown to contain slightly fewer Legionella cells, it is also recommended that such water should be monitored regularly for Legionella in commercial systems, especially if the water is to be used for misting.
TABLE 3.
Quantitative PCR of the 430-bp PCR product from the experimental and commercial SSF
| Sample | Legionella DNA concn (ng g−1 or ng liter−1) | Estimated % of Legionella DNA in total DNA extracted | No. of cells estimated by real-time PCR (cells g−1 or cells liter−1)
|
|
|---|---|---|---|---|
| Mean | SD | |||
| Reservoir | 0.013 | 0.003 | 3.1 × 103 | 7.8 × 102 |
| IFWa | 0.097 | 0.02 | 2.3 × 104 | 1.3 × 104 |
| IFWa | 0.145 | 0.03 | 3.4 × 104 | 1.4 × 104 |
| IFWa | 0.318 | 0.07 | 7.4 × 104 | 1.3 × 104 |
| Top layer | 0.99 | 0.20 | 2.3 × 105 | 7.7 × 104 |
| T1b | 17.068 | 0.34 | 3.9 × 106 | 2.2 × 106 |
| T2b | 9.212 | 0.18 | 2.1 × 106 | 6.6 × 105 |
| T3b | 2.634 | 0.05 | 6.1 × 105 | 2.1 × 105 |
| Middle layer | 0.268 | 0.05 | 6.2 × 104 | 2.2 × 104 |
| Bottom layer | 0.062 | 0.01 | 1.5 × 104 | 1.0 × 104 |
| Recirculating water | 19.98 | 0.1 | 4.7 × 106 | 2.5 × 106 |
| OFWc | 0.187 | 0.04 | 4.4 × 104 | 1.1 × 104 |
| OFWc | 0.235 | 0.05 | 5.5 × 104 | 1.1 × 104 |
| SSF-2 | 0.635 | 0.051 | 1.7 × 105 | 3.7 × 104 |
| SSF-3 | 0.326 | 0.0004 | 3.4 × 105 | 5.4 × 104 |
| SSF-3 (5 cm down) | 0.566 | 0.006 | 6.8 × 105 | 2.7 × 105 |
| SSF-3 (10 cm down) | 0.119 | 0.00014 | 3.1 × 104 | 1.4 × 104 |
| SSF-3 (15 cm down) | 0.078 | 0.00009 | 6.0 × 104 | 1.6 × 104 |
| SSF-3 (20 cm down) | 0.260 | 0.0003 | 5.5 × 104 | 3.0 × 104 |
| SSF-4 | 0.207 | 0.0003 | 1.3 × 105 | 1.6 × 104 |
| SSF-5 | 0.728 | 0.018 | 3.8 × 105 | 1.4 × 105 |
| SSF-6a | 0.904 | 0.023 | 5.5 × 105 | 8.1 × 104 |
| SSF-6b | 1.227 | 0.031 | 4.5 × 105 | 2.3 × 105 |
| SSF-7a | 0.454 | 0.011 | 2.5 × 105 | 5.1 × 104 |
| SSF-7b | 1.408 | 0.035 | 6.9 × 105 | 7.1 × 105 |
IFW, inflow water. The results obtained for three replicate samples taken during the study are shown.
A sand bed sample was taken in the first 1 cm of the top layer sample in three replicate columns.
OFW, outflow water. The results obtained for two replicate samples taken during the study are shown.
Direct detection of Legionella by PCR circumvents problems associated with the culturability of the organisms on standard microbiological media. Dilution plate counting for Legionella carried out by the Public Health Laboratory Service (Coventry and Warwickshire Hospital, Coventry, United Kingdom) with standard procedures gave values below those obtained from PCR results. A 20-fold difference between the quantitative PCR estimate and the number of CFU was obtained. This difference is consistent with results of other experiments (57). The results of plate analysis are also commonly presented as numbers of CFU for L. pneumophila serogroup I. When specific L. pneumophila mip gene primers (32) were used with SSF samples, no PCR products were obtained. However, as expected, a positive 186-bp PCR fragment was detected by using purified DNA from a pure culture of L. pneumophila (a gift from C. Winstanley). These results indicated that the level of L. pneumophila, if the organism was present, was below the limit of detection (data not shown). Sequence analysis of the PCR products indicated that the strains that were present in the SSF showed similarity to isolates that are termed serogroup I isolates. However, the sequences did not exhibit the greatest similarity to L. pneumophila 16S rRNA. In this way the standard dilution plate count method could detect strains other than L. pneumophila, and such strains are likely to dominate similar samples. The colonies of these organisms may or may not react with the L. pneumophila-specific antiserum. The dilution plate count results should also be interpreted as underestimates of the true numbers of Legionella cells present.
A melting curve analysis of the final PCR products was also carried out at the end of the quantitative PCR; the amount of fluorescence was measured while the temperature of the sample was slowly increased to 95°C. Since SYBR green binds only to double-stranded DNA, a sharp decrease was observed when the product melted. When the rate of the decrease in fluorescence was plotted against temperature, a sharp peak was visible, which represented the melting temperature of the PCR product. The analysis of the melting curve for all samples and standards showed a single peak, indicating the absence of nonspecific products, such as primer dimers. Interestingly, the melting temperature of the products as judged by the position of the peak varied from sample to sample. This indicates that slightly different Legionella species predominated in the different samples. This agrees with the results of DGGE analysis and sequencing, in which products had different melting points and sequences. Melting temperature analysis could provide a useful indicator of the types of legionellae present in samples and complement further analysis.
SSF represent a potentially useful system in which fungal plant pathogens can be suppressed by microbial activity. As these systems are open and uncontrolled, they also represent a source of potentially harmful bacteria. As Legionella species are human pathogens known to be suited to the aquatic environment, it is not surprising that they are detected in the SSF system. It is their high levels and dominance within the total bacterial population in the system that are most surprising and raise health concerns. A regular maintenance and disinfection system is required, along with suitable operating procedures. In addition, monitoring of filtered water (outflow water) should be adopted for these systems. Since the outflow water is used in irrigation systems and for misting plants, this system is an ideal dispersal system for Legionella, and the potential hazards should be taken seriously. With suitable operating procedures in place, SSF systems should not represent a greater risk than the initial water flowing in. Operation of an SSF within a glasshouse at a temperature higher than the ambient temperature allows better colonization by Legionella and results in greater risk. In the 1990s it was widely thought that temperatures greater than 15°C were required for effective SSF function against fungal plant pathogens (4, 58). However, it has been demonstrated that SSF are effective against Phytophthora, Pythium, and Fusarium spp. at operational temperatures down to 2°C (T. R. Pettitt and M. F. Wainwright, unpublished data). Therefore, it may be better to operate horticultural sand filter units outside glasshouses at lower ambient temperatures.
Acknowledgments
This work was funded by Department for Environment, Food and Rural Affairs DEFRA (UK) project HH1751.
We thank C. Winstanley, Department of Medical Microbiology and Genito-Urinary Infections, University of Liverpool, Liverpool, United Kingdom, for supplying L. pneumophila DNA used in this study.
REFERENCES
- 1.Addiss, D. G., J. P. Davis, M. LaVenture, P. J. Wand, M. A. Hutchinson, and R. M. McKinney. 1989. Community-acquired Legionnaires' disease associated with a cooling tower: evidence for longer distance transport of Legionella pneumophila. Am. J. Epidemiol. 130:557-568. [DOI] [PubMed] [Google Scholar]
- 2.Adeleke, A., J. Pruckler, R. Benson, T. Rowbotham, M. Halablab, and B. S. Fields. 1996. Legionella-like amebal pathogens—phylogenetic status and possible role in respiratory disease. Emerg. Infect. Dis. 2:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbaree, J. M., B. S. Fields, J. C. Feeley, G. W. Gorman, and W. T. Martin. 1986. Isolation of protozoa from water associated with legionellosis outbreaks and demonstration of intracellular multiplication of Legionella pneumophila. Appl. Environ. Microbiol. 51:422-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barletta, A. 1995. Isle of delight. Greenhouse Grower 9:28-31. [Google Scholar]
- 5.Bender, L., M. Ott, R. Marre, and J. Hacker. 1990. Genome analysis of Legionella spp. by orthogonal field alternation gel electrophoresis (OFAGE). FEMS Microbiol. Lett. 60:253-257. [DOI] [PubMed] [Google Scholar]
- 6.Benson, R. F., W. L. Thacker, R. P. Walters, P. A. Quinlivan, W. R. Mayberry, D. J. Brenner, and H. W. Wilkinson. 1989. Legionella quinlivanii sp. nov. isolated from water. Curr. Microbiol. 18:195-197. [Google Scholar]
- 7.Benson, R. F., and B. S. Fields. 1998. Classification of the genus Legionella. Semin. Respir. Infect. 13:90-99. [PubMed] [Google Scholar]
- 8.Berk, S. G., R. S. Ting, G. W. Turner, and R. J. Ashburn. 1998. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl. Environ. Microbiol. 64:279-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bewley, W. F., and W. Buddin. 1921. On the fungus flora of glasshouse water supplies in relation to plant disease. Ann. Appl. Biol. 8:10-19. [Google Scholar]
- 10.Birtles, R. J., T. Rowbotham, D. Raoult, and T. Harrison. 1996. Phylogenetic diversity of intra-amoebal legionellae as revealed by 16S rRNA gene sequence comparison. Microbiology 142:3525-3530. [DOI] [PubMed] [Google Scholar]
- 11.Bollin, G. E., J. F. Plouffe, M. F. Para, and B. Hackman. 1985. Aerosols containing Legionella pneumophila generated by shower heads and hot-water faucets. Appl. Environ. Microbiol. 50:1128-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brand, T., and W. Wohanka. 2000. Importance and characterization of the biological component in slow filters. Acta Hortic. (The Hague) 554:313-321. [Google Scholar]
- 13.Brenner, D. J., A. G. Steigerwalt, G. W. Gorman, H. W. Wilkinson, W. F. Bibb, M. Hackel, R. L. Tyndall, J. Campbell, J. C. Feeley, W. L. Thacker, P. Skaliy, W. T. Martin, B. J. Brake, B. S. Fields, H. V. Mceachern, and L. K. Corcoran. 1985. Ten new species of Legionella. Int. J. Syst. Bacteriol. 35:50-59. [Google Scholar]
- 14.Brenner, D. J., J. C. Feeley, and R. E. Weaver. 1984. Family VII. Legionellaceae Brenner, Steigerwalt and McDade 1979:658, p. 279-288. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams and Wilkins, Baltimore, Md.
- 15.Brosius, J., T. L. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organisation and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 16.Brown, M. R. W., and J. Barker. 1994. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140:1253-1259. [DOI] [PubMed] [Google Scholar]
- 17.Brown, M. R. W., and J. Barker. 1999. Unexplored reservoirs of pathogenic bacteria: protozoa and biofilms. Trends Microbiol. 46:45-50. [DOI] [PubMed] [Google Scholar]
- 18.Dondero, T. J., R. C. Rendtorff, G. F. Mallison, R. M. Weeks, J. S. Levy, E. W. Wong, and W. Schaffner. 1980. An outbreak of Legionnaires' disease associated with a contaminated air-conditioning cooling tower. N. Engl. J. Med. 302:365-370. [DOI] [PubMed] [Google Scholar]
- 19.Drozanski, W. 1956. Fatal bacterial infection in soil amoebae. Acta Microbiol. Pol. 5:315-317. [PubMed] [Google Scholar]
- 20.Drozanski, W. 1991. Sarcobium lyticum gen. nov., sp. nov., an obligate intracellular bacterial parasite of small free-living amoebae. Int. J. Syst. Bacteriol. 41:82-87. [Google Scholar]
- 21.Duncan, A. 1988. The ecology of slow sand filters, p. 163-180. In N. J. D. Graham (ed.), Slow sand filtration. Horwood, Chichester, United Kingdom.
- 22.Ellis, K. V. 1986. Slow sand filtration. Crit. Rev. Envion. Control 15:315-354. [Google Scholar]
- 23.Fang, G. D., V. L. Yu, and R. M. Vickers. 1989. Disease due to the Legionellaceae (other than Legionella pneumophila). Historical, microbiological, clinical and epidemiological review. Medicine 68:116-132. [DOI] [PubMed] [Google Scholar]
- 24.Fields, B. S. 1993. Legionella and protozoa; interaction of a pathogen and its natural host, p. 129-136. In J. M. Barbaree, R. F. Breiman, and A. P. Dufour (ed.), Legionella: current status and emerging perspectives. American Society for Microbiology, Washington, D.C.
- 25.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 26.Fraser, D. W., D. C. Deubner, D. L. Hill, and D. K. Gilliam. 1979. Nonpneumonic short-incubation-period legionellosis (Pontiac fever) in men who cleaned a steam turbine condenser. Science 205:690-691. [DOI] [PubMed] [Google Scholar]
- 27.Fraser, D. W., T. R. Tsai, W. Orenstein, W. E. Parkin, H. J. Beecham, R. G. Sharrer, J. Harris, G. F. Mallison, S. M. Martin, J. E. McDade, C. C. Shepard, P. S. Brachman, and the Field Investigation Team. 1977. Legionnaires' disease: description of an epidemic. N. Engl. J. Med. 297:1189-1197. [DOI] [PubMed] [Google Scholar]
- 28.Haarhoff, J., and J. L. Cleasby. 1991. Biological and physical mechanisms in slow sand filtration, p. 19-68. In G. S. Logsdon (ed.), Slow sand filtration, vol. 1. American Society of Civil Engineers, New York, N.Y.
- 29.The Health and Safety Commission-Britain's General Union (HSC-GMB). 2000. Legionnaires' disease: the control of Legionella bacteria in water systems. Approved code of practice and guidance. HSE Books, Sudbury, Suffolk, United Kingdom.
- 30.Hebert, G. A., A. G. Steigerwalt, and D. J. Brenner. 1980. Legionella micdadei species nova: classification of a third species of Legionella associated with human pneumonia. Curr. Microbiol. 3:255-257. [Google Scholar]
- 31.Ishimatsu, S., H. Miyamoto, H. Hori, I. Tanaka, and S. Yoshida. 2001. Sampling and detection of Legionella pneumophila aerosols generated from an industrial cooling tower. Ann. Occup. Hyg. 45:421-427. [PubMed] [Google Scholar]
- 32.Koide, M., A. Saito, N. Kusano, and F. Higa. 1993. Detection of Legionella spp. in cooling tower water by the polymerase chain reaction method. Appl. Environ. Microbiol. 59:1943-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 34.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farri, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The Ribosomal Database Project (The RDP-II). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangione, E. J., R. S. Remis, K. A. Tait, H. B. McGee, G. W. Gorman, B. B. Wentworth, P. A. Baron, A. W. Hightower, J. M. Barbaree, and C. V. Bromme. 1985. An outbreak of Pontiac fever related to whirlpool use, Michigan. JAMA 253:535-539. [PubMed] [Google Scholar]
- 36.Marrie, T. J., D. Raoult, B. La Scola, R. J. Birtles, and E. de Carolis. 2001. Legionella-like and other amoebal pathogens as agents of community-acquired pneumonia. Emerg. Infect. Dis. 7:1026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyamoto, H., H. Yamamoto, K. Arima, J. Fujii, K. Maruta, K. Izu, T. Shiomori, and S. Yoshida. 1997. Development of a new seminested PCR method for detection of Legionella species and its application to surveillance of legionellae in hospital cooling tower water. Appl. Environ. Microbiol. 63:2489-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogino, A., H. Koshikawa, T. Nakahara, and H. Uchiyama. 2001. Succession of microbial communities during a biostimulation process as evaluated by DGGE and clone library analyses. J. Appl. Microbiol. 91:625-635. [DOI] [PubMed] [Google Scholar]
- 40.Postma, J., M. J. E. I. M. Willemsen-de Klein, and J. D. van Elsas. 2000. Effect of the indigenous microflora on the development of root and crown rot caused by Pythium aphanidermatum in cucumber grown on rockwool. Phytopathology 90:125-133. [DOI] [PubMed] [Google Scholar]
- 41.Riffard, S., S. Douglass, T. Brooks, S. Springthorpe, L. G. Filion, and S. A. Sattar. 2001. Occurrence of Legionella in groundwater: an ecological study. Water Sci. Technol. 43:99-102. [PubMed] [Google Scholar]
- 42.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowbotham, T. J. 1983. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J. Clin. Pathol. 36:978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowbotham, T. J. 1993. Legionella-like amoebal pathogens, p. 137-140. In J. M. Barbaree, R. F. Breiman, and A. P. Dufour (ed.), Legionella: current status and emerging perspectives. American Society for Microbiology, Washington, D.C.
- 45.Runia, W. T., J. M. G. P. Michielsen, E. J. van Kuik, and E. A. van Os. 1997. Elimination of root-infecting pathogens in recirculating water by slow sand filtration solutions, p. 395-407. In Proceedings of the 9th International Congress on Soil-less Culture, Jersey, United Kingdom. ISOSC, Wageningen, The Netherlands.
- 46.Shadwell, A. 1899. The London water supply. Longmans, London, United Kingdom.
- 47.Shelton, B. G., W. D. Flanders, and G. K. Morris. 1994. Legionnaires-disease outbreaks and cooling-towers with amplified Legionella concentrations. Curr. Microbiol. 28:359-363. [Google Scholar]
- 48.Shiomi, Y., M. Nishiyama, T. Onizuka, and T. Marumoto. 1999. Comparison of bacterial community structures in the rhizoplane of tomato plants grown in soils suppressive and conducive towards bacterial wilt. Appl. Environ. Microbiol. 65:3996-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinclair, J. 1808. The code of health and longevity. Constable Co., Edinburgh, Scotland.
- 50.Spitalny, K. C., R. L. Vogt, L. A. Orciari, L. E. Witherell, P. Etkind, and L. F. Novick. 1984. Pontiac fever associated with a whirlpool spa. Am. J. Epidemiol. 120:809-817. [DOI] [PubMed] [Google Scholar]
- 51.Steele, T. W., J. Lanser, and N. Sangster. 1990. Isolation of Legionella longbeachae serogroup 1 from potting mixes. Appl. Environ. Microbiol. 56:49-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stoesser, G., W. Barker, A. van den Broek, E. Camon, M. Garcia-Pastor, C. Kanz, T. Kulikova, R. Leinonen, Q. Lin, V. Lombard, R. Lopez, N. Redaschi, P. Stoehr, M. A. Tuli, K. Tzouvara, and R. Vaughan. 2002. The EMBL nucleotide sequence database. Nucleic Acids Res. 30:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stults, J. R., O. Snoeyenbos-West, B. Methe, D. R. Lovley, and D. P. Chandler. 2001. Application of the 5′ fluorogenic exonuclease assay (TaqMan) for quantitative ribosomal DNA and rRNA analysis in sediments. Appl. Environ. Microbiol. 67:2781-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Os, E. A., J. J. Amsing, A. J. van Kuik, and H. Willers. 1999. Slow sand filtration: a potential method for elimination of pathogens and nematodes in recirculating nutrient solution from glasshouse-grown crops. Acta Hortic. (The Hague) 481:519-526. [Google Scholar]
- 55.van Os, E. A., M. A. Bruins, J. van Buuren, D. J. van der Veer, and H. Willers. 1997. Physical and chemical measurements in slow sand filters to disinfect recirculating nutrient solutions, p. 313-327. In Proceedings of the 9th International Congress on Soil-less Culture, Jersey, United Kingdom. ISOSC, Wageningen, The Netherlands.
- 56.Visscher, J. T., R. Paramasivam, A. Raman, and H. A. Heijnen. 1987. Slow sand filtration for community water supply. Planning, design, construction and maintenance. Technical paper no. 24. International Reference Centre for Community Water Supply and Sanitation, The Hague, The Netherlands.
- 57.Wellinghausen, N., C. Frost, and R. Marre. 2001. Detection of Legionella in hospital water samples by quantitative real-time LightCycler PCR. Appl. Environ. Microbiol. 67:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wijchman, G. 1994. Langzame zandfiltratie vint snel terrein. Vakbl. Bloemisterij 11:26-29. [Google Scholar]
- 59.Winiecka-Krusnell, J., and E. Linder. 2001. Bacterial infection of free-living amoebae. Res. Microbiol. 152:613-619. [DOI] [PubMed] [Google Scholar]
- 60.Wohanka, W., and M. Helle. 1997. Suitability of various filter media for slow filtration, p. 551-557. In Proceedings of the 9th International Congress on Soil-less Culture, Jersey, United Kingdom. ISOSC, Wageningen, The Netherlands.
- 61.Wohanka, W., H. Luebke, H. Ahlers, and M. Luebke. 1999. Disinfection of recirculating nutrient solutions by slow sand filtration. Acta Hortic. (The Hague) 481:539-544. [Google Scholar]
- 62.Wohanka, W. 1993. Slow sand filtration and UV radiation, low-cost techniques for disinfection of recirculating nutrient solution or surface water, p. 497-511. In Proceedings of the 8th International Congress on Soil-less Culture, Hunter's Rest, South Africa. ISOSC, Wageningen, The Netherlands.
- 63.Yang, C. H., D. E. Crowley, and J. A. Menge. 2001. 16S rRNA fingerprinting of rhizosphere bacterial communities associated with healthy and Phytophthora infected avocado roots. FEMS Microbiol. Ecol. 35:129-136. [DOI] [PubMed] [Google Scholar]