Abstract
As part of a major international project for the validation and standardization of PCR for detection of five major food-borne pathogens, four primer sets specific for Salmonella species were evaluated in-house for their analytical accuracy (selectivity and detection limit) in identifying 43 Salmonella spp. and 47 non-Salmonella strains. The most selective primer set was found to be 139-141 (K. Rahn, S. A. De Grandis, R. C. Clarke, S. A. McEwen, J. E. Galán, C. Ginocchio, R. Curtiss III, and C. L. Gyles, Mol. Cell. Probes 6:271-279, 1992), which targets the invA gene. An extended determination of selectivity by using 364 strains showed that the inclusivity was 99.6% and the exclusivity was 100% for the invA primer set. To indicate possible PCR inhibitors derived from the sample DNA, an internal amplification control (IAC), which was coamplified with the invA target gene, was constructed. In the presence of 300 DNA copies of the IAC, the detection probability for primer set 139-141 was found to be 100% when a cell suspension containing 104 CFU/ml was used as the template in the PCR (50 CFU per reaction). The primer set was further validated in an international collaborative study that included 16 participating laboratories. Analysis with 28 coded (“blind”) DNA samples revealed an analytical accuracy of 98%. Thus, a simple PCR assay that is specific for Salmonella spp. and amplifies a chromosomal DNA fragment detected by gel electrophoresis was established through extensive validation and is proposed as an international standard. This study addresses the increasing demand of quality assurance laboratories for standard diagnostic methods and presents findings that can facilitate the international comparison and exchange of epidemiological data.
Salmonella species are recognized as major zoonotic pathogens for animals and humans (14). In many countries, Salmonella is the leading cause of food-borne outbreaks and infections (23, 24). In order to minimize the risk of infection for consumers, microbiological control of the food chain is being increasingly applied. Thus, the availability of reliable, rapid, and internationally accepted test systems for determination of the presence or absence of food-borne pathogens has become increasingly important for the agricultural and food industry, as well as for legislative regulation of food safety.
In vitro amplification of DNA by the PCR has become a powerful tool in microbiological diagnostics. However, due to the lack of international validation and standard protocols, as well as the variable quality of reagents and equipment, the method produces inconsistent results between expert- and end-user laboratories. For example, many primer sets that have been used for the in vitro amplification of Salmonella DNA by PCR differ in their detection limits and accuracy (1, 6, 7, 16). In some cases, the strain collections used for validation do not include all seven known subspecies of Salmonella enterica and Salmonella bongori and lack epidemiologically important isolates. Furthermore, an internal amplification control (IAC) that is necessary to indicate false-negative results caused by PCR inhibitors is rarely included in the final diagnostic test.
A current European research project (12) is focusing on the validation and standardization of PCR for the detection of thermophilic Campylobacter spp., Escherichia coli O157, Yersinia enterocolitica, Listeria monocytogenes, and Salmonella spp. The objective of this project is to facilitate the implementation of diagnostic PCR for both detection and verification of food-borne pathogens by harmonizing, validating, and standardizing PCR methods in Europe (18).
The present study reports a PCR assay for the detection of all Salmonella enterica subspecies and Salmonella bongori which was developed and validated within the European research project. The results of a collaborative study which included 16 participating laboratories that used reference reagents are presented.
MATERIALS AND METHODS
Terms.
In accordance with the MICROVAL protocol (4), several key terms were defined in this study as follows. Selectivity is defined as a measure of the degree of response from target and nontarget microorganisms and comprises inclusivity and exclusivity. Inclusivity is the ability of an alternative method (PCR in this case) to detect the target pathogen from a wide range of strains, and exclusivity is the lack of response from a relevant range of closely related but nontarget strains. Analytical accuracy includes primary validation on purified cell cultures for establishment of selectivity and detection probability (18).
Bacterial reference strains.
The Salmonella strains used for inclusivity tests are listed in Table 1. A total of 242 strains were selected to represent all known subspecies, with an emphasis on the 10 serotypes most frequently isolated from humans in Europe in 1999 and other epidemiologically important serotypes (Ian Fisher, personal communication). Detailed data on the strains are available on the homepage of the FOOD-PCR project (http://www.pcr.dk). Of the 242 strains, 43 were selected for preliminary screening of the primer sets (Table 1). S. enterica serotype Typhimurium phage type DT104 strain 51K61, isolated in 1996 from pig feces (17), was used as the reference strain.
TABLE 1.
Salmonella strains used for inclusivity PCR tests and results of preliminary screening
| Serotype (subspecies) | Serogroup | Total no. of strains | No. of strains selected for prescreening | No. of strains testing positive with primer set:
|
|||
|---|---|---|---|---|---|---|---|
| P1-P2 | S18-S19 | ST11-ST15 | 139-141 | ||||
| Enteritidis (I) | D1 | 60 | 10 | 10 | 9 | 10 | 10 |
| Typhimurium (I) | B | 60 | 10 | 10 | 10 | 10 | 10 |
| Hadar (I) | C2-C3 | 5 | 1 | 1 | 1 | 1 | 1 |
| Virchow (I) | C1 | 5 | 1 | 1 | 1 | 1 | 1 |
| Infantis (I) | C1 | 5 | 1 | 1 | 1 | 1 | 1 |
| Heidelberg (I) | B | 5 | 1 | 1 | 1 | 1 | 1 |
| Newport (I) | C2-C3 | 5 | 1 | 1 | 1 | 1 | 1 |
| Brandenburg (I) | B | 5 | 1 | 1 | 1 | 1 | 1 |
| Saintpaul (I) | B | 20 | 1 | 1 | 1 | 1 | 1 |
| Agona (I) | B | 5 | 1 | 1 | 1 | 1 | 1 |
| Blockley (I) | C2-C3 | 5 | 1 | 1 | 1 | 1 | 1 |
| Bovismorbificans (I) | C2-C3 | 5 | 1 | 1 | 1 | 1 | 1 |
| Bredeney (I) | B | 5 | 1 | 1 | 1 | 1 | 1 |
| Derby (I) | B | 5 | 1 | 1 | 1 | 1 | 1 |
| Dublin (I) | D1 | 5 | 1 | 1 | 1 | 1 | 1 |
| Livingstone (I) | C1 | 5 | 1 | 1 | 1 | 1 | 1 |
| Montevideo (I) | C1 | 5 | 1 | 1 | 1 | 1 | 1 |
| Paratyphi B (I) | B | 5 | 1 | 1 | 1 | 1 | 1 |
| Senftenberg (I) | E4 | 2 | —b | — | — | — | — |
| Litchfield (I) | C2-C3 | 2 | — | — | — | — | — |
| Typhi (I) | D1 | 2 | — | — | — | — | — |
| 42:r:- (II) | T | 2 | 1 | 1 | 1 | 1 | 1 |
| 9,12:z:z39 (II) | D | 1 | 1 | 1 | 1 | 1 | 1 |
| 48:d:z6 (II) | Y | 1 | — | — | — | — | — |
| 42:b:e,n,x,z15 (II) | T | 1 | — | — | — | — | — |
| 30:1,z28,z6 (II) | N | 1 | — | — | — | — | — |
| 21:g,z51:- (IIIa) | L | 1 | 1 | 1 | 1 | 0 | 1 |
| 47:r:- (IIIa) | X | 1 | — | — | — | — | — |
| 18:z4,z32:- (IIIa) | K | 1 | — | — | — | — | — |
| 50:z:z52 (IIIb) | Z | 1 | 1 | 1 | 1 | 1a | 1 |
| 47:1,v:z (IIIb) | X | 1 | — | — | — | — | — |
| 18:i,v:z (IIIb) | K | 1 | — | — | — | — | — |
| 16:z4,z32:- (IV) | I | 1 | 1 | 1 | 1 | 1 | 1 |
| 48:g,z51:- (IV) | Y | 1 | — | — | — | — | — |
| 11:z4,z23:- (IV) | F | 1 | — | — | — | — | — |
| 44:r:- (V) | V | 1 | 1 | 0 | 0 | 1a | 1 |
| 66:z65:- (V) | 1 | — | — | — | — | — | |
| 48:z35:- (V) | Y | 1 | — | — | — | — | — |
| 45:a:e,n,x (VI) | W | 1 | 1 | 1 | 1 | 1 | 1 |
| 1,6,14,25:a:e,n,x (VI) | H | 1 | — | — | — | — | — |
| 41:b:1,7 (VI) | S | 1 | — | — | — | — | — |
| Total | 242 | 43 | 42 | 41 | 42 | 43 | |
Only faint fragments were obtained but included as positive response.
—, not included.
The non-Salmonella strains used for the exclusivity tests are listed in Table 2. The strains were chosen because they are closely related to Salmonella or because they are found in the same environment and grow under the same conditions. A panel of 47 strains was selected for preliminary screening of the primer sets. The remaining 75 strains were selected for further exclusivity tests, as has been recommended previously (13).
TABLE 2.
Salmonella-related strains used for exclusivity PCR tests
| Organism | No. of strains | No. of strains selected for preliminary screening |
|---|---|---|
| Citrobacter brakii | 1 | 0 |
| Citrobacter diversis | 1 | 1 |
| Citrobacter freundii | 19 | 5 |
| Escherichia coli | 16 | 7 |
| Escherichia fergusonii | 1 | 0 |
| Erwinia herbicola | 1 | 0 |
| Enterobacter aerogenes | 4 | 1 |
| Enterobacter agglomerans | 2 | 1 |
| Enterobacter amnigenus | 1 | 0 |
| Enterobacter asbunae | 1 | 0 |
| Enterobacter asburiae | 1 | 0 |
| Enterobacter cloacae | 2 | 1 |
| Enterobacter gergoviae | 2 | 0 |
| Enterobacter sakazakii | 1 | 0 |
| Enterobacter tarda | 1 | 0 |
| Enterobacter taylorae | 2 | 0 |
| Enterococcus faecalis | 3 | 3 |
| Ewingella americana | 1 | 0 |
| Hafnia alvei | 3 | 1 |
| Klebsiella oxytoca | 3 | 1 |
| Klebsiella pneumoniae | 5 | 4 |
| Kluyvera ascorbata | 1 | 0 |
| Listeria inucua | 2 | 2 |
| Listeria ivanovii | 2 | 1 |
| Listeria monocytogenes | 8 | 3 |
| Micrococcus kristinae | 1 | 0 |
| Morganella morganii | 4 | 1 |
| Obesumbacterium proteus | 1 | 0 |
| Proteus agglomerans | 1 | 0 |
| Proteus mirabilis | 4 | 1 |
| Proteus vulgaris | 3 | 2 |
| Providencia heimbachae | 1 | 0 |
| Providencia stuartii | 1 | 0 |
| Pseudomonas aeruginosa | 2 | 0 |
| Pseudomonas alcaligenes | 1 | 0 |
| Rhanella aquatilis | 1 | 0 |
| Serratia marcescens | 3 | 1 |
| Serratia odorifera | 2 | 0 |
| Shigella boydii | 3 | 3 |
| Shigella flexneri | 1 | 1 |
| Shigella sonnei | 1 | 0 |
| Staphylococcus aureus | 4 | 4 |
| Yersinia enterocolitica | 4 | 1 |
| Total | 122 | 47 |
Preparation of DNA samples.
For selectivity tests, Salmonella or non-Salmonella strains were grown aerobically without shaking at 37°C for 16 h in Luria-Bertani medium (20). Viable counts were obtained by plating a dilution made in 0.9% (wt/vol) NaCl solution onto plate count agar (Merck, Darmstadt, Germany) in duplicate and incubating the plates for 24 h at 37°C. The concentration was estimated by calculating the average number of CFU. A 1-ml aliquot of the enriched culture was centrifuged at 10,000 × g in a microcentrifuge tube for 5 min at 4°C. The supernatant was carefully discarded, and the cell pellet was suspended in 300 μl of TE buffer (10 mM Tris-HCl, 0.1 mM EDTA [pH 8.0]). The microcentrifuge tube was incubated for 10 min at 100°C in a water bath and immediately chilled on ice. After centrifugation at 14,000 × g at 4°C for 5 min, the supernatant containing DNA was carefully transferred to a new microcentrifuge tube. A 5-μl aliquot was used as the template DNA for the PCR.
Primer sets and PCR.
The Salmonella-specific primer sets for comparison and the corresponding thermocycler annealing temperatures that were used are presented in Table 3. The primer sets have been published previously and are commonly used in many studies for detection of Salmonella. PCRs were carried out in a GenAmp PCR System 9700 thermocycler (Applied Biosystems, Weiterstadt, Germany). A typical 25-μl PCR mixture contained 0.4 μM concentrations of each primer, 200 μM concentrations of each dNTP (Roche Diagnostics, Mannheim, Germany), 1× PCR buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl), 1.5 mM MgCl2, 0.75 U of Platinum Taq polymerase (Invitrogen, Karlsruhe, Germany), and 5 μl of sample DNA (approximately 106 CFU per reaction tube). The incubation conditions were 95°C for 1 min, followed by 35 or 38 cycles of 95°C for 30 s, 55 to 64°C (depending on the primer set used [Table 3]) for 30 s, and 72°C for 30 s. A final extension of 72°C for 4 min was employed. For selectivity tests, Salmonella DNA was cycled 35 times and non-Salmonella DNA was cycled 38 times in order to detect possible nontarget PCR fragments.
TABLE 3.
Selected primer sets for selectivity tests
| Target | Primer | Sequence | Size of PCR product (bp) | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| oriC | P1 | TTATTAGGATCGCGCCAGGC | 163 | 55 | 25 |
| P2 | AAAGAATAACCGTTGTTCAC | ||||
| ompC | S18 | ACCGCTAACGCTCGCCTGTAT | 159 | 58 | 16 |
| S19 | AGAGGTGGACGGGTTGCTGCCGTT | ||||
| Random fragment | ST11 | AGCCAACCATTGCTAAATTGGCGCA | 429 | 60 | 1 |
| ST15 | GGTAGAAATTCCCAGCGGGTACTG | ||||
| invA | 139 | GTGAAATTATCGCCACGTTCGGGCAA | 284 | 64 | 21 |
| 141 | TCATCGCACCGTCAAAGGAACC |
A 10-μl aliquot of a PCR product was loaded on a 1.8% agarose gel containing 0.5 μg of ethidium bromide/ml and electrophoresed at 6 V/cm for 90 min. Marker X (Roche Diagnostics) was used in the electrophoresis as the molecular weight standard. The gel was documented with a video camera. A positive response was defined as the presence of a visible band at the expected size, while a negative response was defined as the lack of any band at the expected size.
Construction of the internal controls.
The IAC for primer set 139-141 was constructed according to the method described by Ausubel et al. (5), with slight modifications. The method allows the construction of short artificial DNA fragments. Briefly, four, approximately 100-mer oligonucleotides overlapping in 20 bp were hybridized and gaps were filled with Klenow fragment and dNTPs. The product was reamplified using primers malo2-F (GTATTGTTGATTAATGAGATCCG) and malo2-R (ATATTACGCACGGAAACACG), resulting in a 253-bp fragment which was cloned in pGEM-T Easy vector (Promega, Mannheim, Germany). The cloned internal fragment was amplified as described above at an annealing temperature of 55°C with primers M13 forward and M13 reverse located on the vector, resulting in a 498-bp fragment. The PCR product was purified by using a Centrispin 20 column (Princeton Separations, Adelphia, N.J.), and the concentration was spectrometrically determined at 260 nm. The sequence of the IAC, composed of various invA primer sequences between 18 and 26 bp in size and primer sequences (S18-S19) occurring in the ompC gene of Salmonella spp. (16), was confirmed (Fig. 1). The following equation was used to calculate the copy numbers from a known PCR product concentration: weight of PCR fragment (in grams per microliter)/(660 g per mol × the number of base pairs of the PCR fragment) × (6.023 × 1023) = the number of genomic copies per microliter.
FIG. 1.
Cloned artificial 253-bp sequence of the IAC. For the invA PCR assay, a 157-bp IAC was amplified by using primer set 139-141. The underlined italic sequence is the binding sequence of primer 139, and the underlined roman sequence is the binding sequence of primer 141.
Determination of the detection probability.
The probability of detecting Salmonella in a suspension of known concentration in the presence of defined DNA copy numbers of the IAC was determined essentially as described previously (15). Buffered peptone water (Merck) was inoculated with the reference strain 51K61 and incubated for approximately 5 h at 37°C to the exponential phase (approximately 108 CFU/ml). The cell suspension was serially diluted 10-fold in 0.9% (wt/vol) NaCl within the concentration range of 106 to 100 CFU/ml. The dilutions were stored at −20°C until use. The approximate number of CFU per milliliter of cell suspension was determined by plating 100 μl of the dilution containing 104 CFU/ml onto Luria-Bertani agar five times. The average number of CFU from the five plates was used to recalculate the concentration (CFU per milliliter) of the frozen dilutions. Each dilution was thawed and adjusted to approximately 101 to 106 CFU/ml. Five microliters of each dilution was added to five separate PCR tubes and was run in the presence of 300 or 30 copies of IAC as described above. The experiment was repeated five times, resulting in 30 PCRs for each cell concentration. Each PCR gave a positive or negative result at the concentration tested. The detection probability of the PCR assay was obtained by plotting the relative number of positive PCRs observed against the concentration of the cell suspension. A sigmoidal line fitting was performed using the ORIGIN program (version 4.0; Microcal Software, Northampton, Mass.). The determination of the detection probability was repeated by using purified DNA in the concentration range of 105 to 100 genomic DNA copies per PCR. Template DNA for determination of the detection probability was purified from reference strain 51K61 with a Genomic Tip kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. The concentration of the purified DNA was determined by measuring the optical density at 260 nm with a GenQuant photometer (Amersham Pharmacia, Uppsala, Sweden). The number of Salmonella genomic copies of purified DNA was calculated as follows: m = n × (1.013 × 10−21 g/bp), where m is the mass and n is the number of base pairs. The number of kilobase pairs for one serotype Typhimurium genome was previously determined to be 4,951 (19). Consequently, according to the above equation, one Salmonella genome weighs about 5.0 fg. The concentration of DNA (range, 105 to 100 genomic DNA copies/ml) was adjusted in TE buffer.
Interlaboratory study using the primer set 139-141.
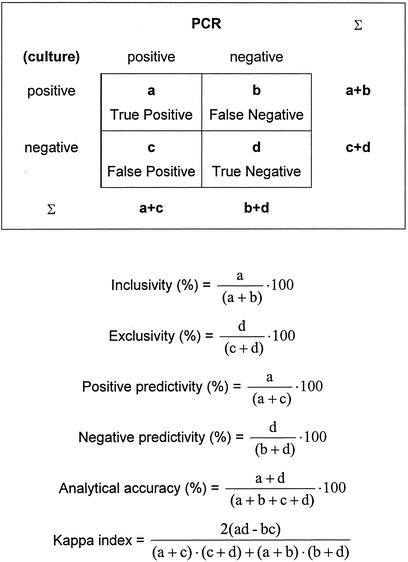
The collaborative study was designed and analyzed according to the basic guidelines of ISO 5725-1 (2) and ISO 5725-2 (3) and the MICROVAL protocol (4). The formulas used for analysis are shown in Fig. 2 and were calculated as described previously (4, 11). The Kappa index, which indicates the strength of the relationship between the row and column variables of a cross tabulation, was calculated as described previously (22). Kappa values of <0.01 indicate no concordance, those between 0.1 and 0.4 indicate weak concordance, those between 0.41 and 0.60 indicate clear concordance, those between 0.61 and 0.80 indicate strong concordance, and those between 0.81 and 1.00 indicate nearly complete concordance.
FIG. 2.
Features of a two-row by two-column contingency table with respect to the reference culture method and the alternative PCR method. The derived formulas used for analysis in the validation study are shown below the table. a, number of samples that are both reference positive and PCR positive (true positive); b, number of samples that are reference negative but PCR positive (false positive); c, number of samples that are reference positive but PCR negative (false negative); d, number of samples that are both reference and PCR negative (true negative).
A total of 16 laboratories (from Austria, Belgium, the Czech Republic, Denmark, England, Finland, France, Germany, Greece, Slovakia, Spain, Sweden, Italy, and The Netherlands) participated in the interlaboratory study. Each laboratory received 28 coded (“blind”) DNA samples, including samples from 12 Salmonella and 16 non-Salmonella strains (see Table 5), one negative (TE buffer) and one positive control DNA, an IAC template (498-bp PCR product, produced as described above), and reagents for use in PCR, including Platinum Taq polymerase (Invitrogen). All DNA samples were prepared according to the thermal cell lysis method as described above. The positive control DNA was purified from reference strain 51K61 with a Genomic Tip kit (Qiagen). Each lab strictly followed a standard operating procedure (available at http://www.pcr.dk). The 25-μl PCR mixture contained 0.4 μM concentrations of primers 139 and 141, 200 μM concentrations of each dNTP (Roche Diagnostics), 1× PCR buffer, 1.5 mM MgCl2, 0.75 U of Platinum Taq polymerase (Invitrogen), 300 copies of the IAC, and 5 μl of sample DNA (approximately 106 CFU per reaction tube). The incubation conditions were 95°C for 1 min, followed by 38 cycles of 95°C for 30 s, 64°C for 30 s, and 72°C for 30 s. A final extension of 72°C for 4 min was employed. The choice of the thermal cycler model was unrestricted. Each set of DNA samples was amplified in three independent experiments. Amplicons were detected after electrophoresis by using a 1.8% agarose gel. The choices of the electrophoresis equipment and buffer were unrestricted. The electric field strength was restricted to 5 V/cm. The calculation took into account all single PCRs from each participating laboratory. The criteria for the acceptance of data generated by the individual laboratories were as follows: (i) both the positive control and negative control were detected correctly, (ii) the 157-bp IAC fragment was visible on the agarose gel from PCRs with non-Salmonella strains, and (iii) missing data, e.g., where a laboratory omitted one of the samples, were excluded.
TABLE 5.
Results of classification of strains used in the interlaboratory study (laboratory 6 excluded)
| Strain and designation | Correct classification of strain (%) |
|---|---|
| Non-Salmonella strains | |
| Citrobacter diversis EU-NS25 | 100.0 |
| Hafnia alvei EU-NS35 | 93.3 |
| Staphylococcus aureus EU-NS22 | 100.0 |
| Citrobacter freundii EU-NS24 | 100.0 |
| Enterobacter cloacae EU-NS32 | 100.0 |
| Enterococcus faecalis EU-NS3 | 100.0 |
| Escherichia coli ATCC 25922 | 97.7 |
| Escherichia coli O128 EU-NS10 | 100.0 |
| Escherichia coli O157 EU-NS11 | 100.0 |
| Klebsiella oxytoca EU-NS18 | 93.3 |
| Klebsiella pneumoniae EU-NS17 | 100.0 |
| Proteus vulgaris EU-NS28 | 100.0 |
| Shigella boydii EU-NS38 | 100.0 |
| Shigella flexneri 2a EU-NS27 | 100.0 |
| Yersinia enterocolitica EU-NS41 | 100.0 |
| Serratia marcescens EU-NS36 | 100.0 |
| Salmonella serotypesa | |
| Hadar 99-601 | 100.0 |
| Enteritidis 00-4 | 90.9 |
| Bredeney 00-215 | 100.0 |
| Dublin 98-443 | 69.7 |
| Blockley 98-1262 | 100.0 |
| Typhimurium 00-510 | 100.0 |
| 42:b:e,n,x,z15 (II) 98-1308 | 90.9 |
| 18:z4,z32:- (IIIa) 99-1556 | 97.0 |
| 47:1,v:z (IIIb) 00-269 | 100.0 |
| 48:g,z51:- (IV) 98-3722 | 100.0 |
| 66:z65:- (V) K-1354 | 97.0 |
| 1,6,14,25:a:e,n,x (VI) 94-2957 | 100.0 |
| Controls | |
| Negative (TE buffer) | 100.0 |
| Positive (serotype Typhimurium 51K61) | 100.0 |
Numerals in parentheses indicate subspecies.
RESULTS
Selectivity.
Table 1 shows the results of the inclusivity tests of the four primer sets tested with a panel of 43 Salmonella strains. All four primer sets amplified a PCR product of the expected size. However, while invA primer set 139-141 identified all 43 strains, the other three primer sets failed to identify one or two Salmonella strains (Table 1). The exclusivity tests with 47 non-Salmonella strains gave unspecific PCR fragments (data not shown). Primer set P1-P2 (oriC) (25) produced multiple, non-target-sized fragments of 0.2 to 3 kb in several non-Salmonella strains. Primer set ST11-ST15 (random genomic fragment) (1) showed faint fragments of similar size from DNA of Citrobacter spp., Yersinia enterocolitica (approximately 400 bp), and Klebsiella oxytocan (approximately 500 bp). Primer set S18-S19 (ompC) (16) produced fragments of similar size from DNA of Proteus vulgaris (approximately 120 bp) and Hafina alvei (approximately 140 bp). Other strains produced several non-target-sized fragments ranging in size from 0.3 to 2 kb. Primer set 139-141 (invA) (21) produced faint non-target-sized fragments from DNA of E. coli (approximately 400 bp) and Citrobacter spp. (approximately 350 bp) strains that could clearly be distinguished from the 284-bp Salmonella target fragment.
Based on these data, the invA primer set 139-141 was selected for extended inclusivity and exclusivity tests. A total of 199 additional Salmonella strains and 75 additional non-Salmonella strains were tested (Table 1 and 2). All Salmonella strains, except one, were identified correctly. The incorrectly identified Salmonella strain belonged to serotype Saintpaul. However, 19 other serotype Saintpaul isolates (collected between 1999 and 2001 from different sources in Germany) were identified correctly. Primer set 139-141 again produced faint non-target-sized fragments from DNA of E. coli (approximately 400 bp) and Citrobacter spp. (approximately 350 bp) strains. The nonspecific fragments were absent in the presence of an IAC (see below).
IAC.
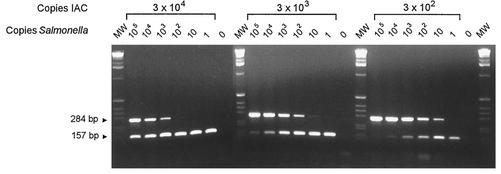
In order to identify false-negative PCR results, an IAC was constructed for primer set 139-141 (Fig. 2). The IAC was coamplified with the target DNA, resulting in a 157-bp amplicon. In the absence of an IAC, the detection limit was determined to be 1 to 10 Salmonella genomic DNA copies (38 cycles of amplification). In the presence of an IAC, the detection limit depended on the initial number of IAC copies (Fig. 3). Since a 10-fold increase in the initial number of IAC copies increased the detection limit of the Salmonella target gene by 10-fold as well, the competition between the two templates could be linear, indicating that the IAC is not preferably amplified. No difference in the detection limit was observed when purified plasmid DNA or PCR product was used as the template for the IAC (data not shown).
FIG. 3.
Coamplification of different copy numbers of the IAC (157-bp fragment) and Salmonella genomic DNA (284-bp fragment) by using primer set 139-141. The initial numbers of Salmonella DNA and IAC copies per reaction tube are indicated above the gels. The sizes of the PCR products are indicated at left. Marker X (Roche Diagnostics) was used as the molecular weight standard (MW). Ten microliters of the 25-μl PCR mixture was loaded per well.
Detection probability.
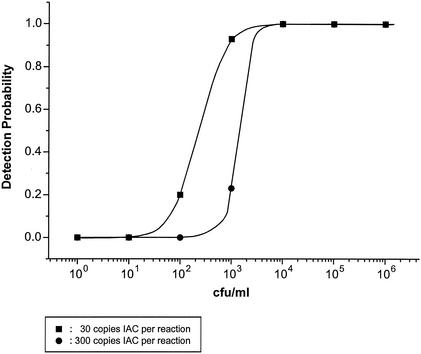
The probability of detecting serotype Typhimurium phage type DT104 reference strain 51K61 was established at different cell and DNA concentrations by using primer set 139-141 in the presence of 30 or 300 copies of IAC. Figure 4 shows that a 10-fold decrease in the initial number of IAC copies increased the detection probability approximately 10-fold. The detection probability for a cell suspension at a concentration of 104 CFU/ml (50 CFU per reaction) was 100% at both 30 and 300 copies of IAC. However, the detection probability of a concentration of 103 CFU/ml was approximately 93% for 30 copies of IAC and 23% for 300 copies of IAC. By using 10 copies of purified Salmonella DNA (50 fg) per reaction as the template, the detection probability was determined to be 100% in the presence of 300 copies of IAC (data not shown). This is a fivefold increase in the detection probability compared with that when cells were used as the template, assuming that 1 genomic copy per cell occurred during the exponential replication of the cells.
FIG. 4.
Detection probability of the Salmonella PCR assay using primer set 139-141 at serially 10-fold-diluted cell concentrations of serotype Typhimurium phage type DT104 reference strain 51K61. The detection probability was established in the presence of 30 or 300 copies of IAC DNA. Five microliters of 10-fold-diluted cell suspensions (100 to 106 CFU/ml) were used as the template in the PCR. The graph shows a sigmoidal fit of data points generated by 30 repetitive PCRs from six independent experiments.
When 30 initial IAC copies were used, the signal intensity of the IAC amplicon varied substantially among identical PCRs, indicating poor reproducibility. Therefore, approximately 300 IAC copies should be used as the template.
Interlaboratory study using primer set 139-141.
The objective of this study was to validate the reproducibility of the analytical accuracy of the described Salmonella PCR assay in a multicenter investigation. Twelve Salmonella and 16 non-Salmonella coded (blind) DNA samples were sent refrigerated to 16 international laboratories. The performance of each participating laboratory is shown in Table 4. Four hundred ninety-five of 516 samples expected to be PCR positive for Salmonella (inclusivity, 96%) and 681 of 688 samples expected to be PCR negative for Salmonella (exclusivity, 99%) were correctly classified by the participating laboratories. The overall analytical accuracy of the study was 98.0%. Laboratory 6 was excluded from the analysis since its Kappa index was significantly lower than those of other laboratories (Table 4). Furthermore, one negative control DNA sample from laboratory 6 was detected as positive, indicating cross contamination of DNA samples at the receiving laboratory. The results of the classification of the strains are shown in Table 5. It was observed that false positives were generally detected as weak signals, indicating carry-over contamination from positive DNA samples. One DNA sample (S. enterica serotype Dublin 98-443) that was expected to be positive was frequently detected as negative (15 of 46 PCRs). This was probably caused by the degradation of the sample during transport or handling during the experiments in some laboratories.
TABLE 4.
Results of the collaborative study validating the reproducibility of the analytical accuracy using primer set 139-141
| Laboratory no. | Inclusivity (%) | Exclusivity (%) | Positive predictivity (%) | Negative predictivity (%) | Analytical accuracy (%) | Kappa indexa |
|---|---|---|---|---|---|---|
| 1 | 100 | 100 | 100 | 100 | 100 | 1.000 |
| 2 | 94 | 100 | 100 | 92 | 96 | 0.926 |
| 3 | 96 | 97 | 98 | 94 | 96 | 0.927 |
| 4 | 100 | 100 | 100 | 100 | 100 | 1.000 |
| 4 | 98 | 92 | 94 | 97 | 95 | 0.903 |
| 6b | 94 | 67 | 65 | 94 | 77 | 0.561 |
| 7 | 100 | 100 | 100 | 100 | 100 | 1.000 |
| 8 | 100 | 100 | 100 | 100 | 100 | 1.000 |
| 9 | 100 | 100 | 100 | 100 | 100 | 1.000 |
| 10 | 91 | 100 | 100 | 88 | 95 | 0.889 |
| 11 | 100 | 100 | 100 | 100 | 100 | 1.000 |
| 12 | 96 | 100 | 100 | 94 | 98 | 0.951 |
| 13 | 94 | 92 | 94 | 92 | 93 | 0.854 |
| 14 | 100 | 100 | 100 | 100 | 100 | 1.000 |
| 15 | 87 | 100 | 100 | 81 | 92 | 0.826 |
| 16 | 100 | 100 | 100 | 100 | 100 | 1.000 |
| All | 97 | 96 | 97 | 96 | 96 | 0.926 |
| All but laboratory 6 | 97 | 99 | 99 | 96 | 98 | 0.952 |
Kappa values of <0.01 indicate no concordance, 0.1 to 0.4 indicate weak concordance, 0.41 to 0.60 indicate clear concordance, 0.61 to 0.80 indicate strong concordance, and 0.81 to 1.00 indicate nearly complete concordance.
Laboratory 6 was excluded from the final statistical analysis due to a markedly lower Kappa index compared with those of the other laboratories.
DISCUSSION
The aim of this study was to validate a PCR assay for the detection of Salmonella spp. which will contribute to an international standard. It was decided that the Salmonella-specific amplicon should be detected by gel electrophoresis in order to offer a simple and economical detection method that is available in most laboratories. We compared four Salmonella-specific primer sets and, in a collaborative study, validated the most selective and sensitive (in terms of detection limit) PCR system for the detection of all S. enterica subspecies and S. bongori. The primer set used for specific amplification of a Salmonella genomic DNA fragment has previously been published by Rahn et al. (21) and amplifies a 284-bp sequence of the invA gene. The invA target gene is located on pathogenicity island 1 of Salmonella spp., which encodes proteins of a type III secretion system (8). The amplification conditions of the PCR assay described here were optimized and differed considerably from those reported by Rahn et al. (21). For instance, the annealing time was shortened from 2 min to 30 s, the extension time was reduced from 3 min to 30 s, and the annealing temperature was made more stringent by being raised from 53 to 64°C. Furthermore, a hotstart Taq polymerase known to reduce amplification of unspecific fragments was used. With these new parameters, the amplification of nonspecific fragments from non-Salmonella strains should have been limited without influencing the inclusivity of the PCR. Some unspecific but faint amplicon products were still detected in some E. coli and Citrobacter freundii isolates when 38 cycles of amplification were performed. However, in the presence of 300 copies of an IAC, which was competitively coamplified with the invA target fragment, the unspecific fragments were totally absent. The addition of an IAC in each PCR tube is necessary if the PCR is to be used as a diagnostic tool. If no IAC is used, we recommend that the number of amplification cycles be decreased to a maximum of 33 to 35. Naturally, the initial DNA copy number of the IAC in the PCR should be kept as low as possible but must be detected reproducibly.
We observed that the invA PCR assay using primer set 139-141 detected, with a high probability, 5 to 50 Salmonella CFU or 10 genomic copies of purified Salmonella DNA per reaction in the presence of 30 to 300 IAC copies, whereas Rahn et al. previously reported a higher detection limit (300 cells per reaction or 27 pg of extracted chromosomal DNA) (21).
Of the 242 Salmonella strains tested, one serotype Saintpaul strain was not detected. We speculate that the invA gene, at least, is absent in this strain. Rahn et al. (21) reported that two serotype Litchfield and two serotype Senftenberg strains were not detected when primer set 139-141 was used. However, it has been shown that the invA gene is essential for the invasion of epithelial cells by Salmonella; consequently, the apparent absence of the invA gene suggests that such strains are not invasive or use alternative invasion mechanisms (9, 10). The two serotype Litchfield and two serotype Senftenberg reference strains that we tested were invA PCR positive. Absence of the invA gene in Salmonella seems to be rare.
The collaborative study showed a high selectivity and reproducibility of the PCR assay among the 16 international laboratories when identical batches of reagents were used. The fact that, in this study, inclusivity (96%) was lower than exclusivity (99%) was probably due to the use of partially degraded sample DNA, especially one sample (serotype Dublin 98-443) expected to be positive that was frequently detected as negative (15 of 46 PCRs). DNA used in the study was prepared by the thermal cell lysis method without subsequent purification of the DNA. Potentially active DNase might be responsible for this degradation.
In conclusion, the invA PCR assay using the primer set 139-141 that was originally published by Rahn et al. (21) demonstrated inclusivity for a wide range of Salmonella serotypes including all subspecies and exclusivity for other species and genera. The assay showed a high (good) detection probability in the presence of 30 or 300 copies of the IAC. The assay was found to be selective and robust by an international collaborative study. The PCR assay will be validated in-house on naturally contaminated samples to investigate the diagnostic accuracy. A second international interlaboratory study is planned to investigate diagnostic accuracy and reproducibility on naturally and artificially contaminated swab samples from pig and poultry rinses.
Acknowledgments
This work was supported by the European Commission (proposal no. QLK1-CT-1999-00226).
We thank Elke Genschow and Nigel Cook for analysis of the collaborative study and Annet N. Jensen for preparation of non-Salmonella DNA. We also appreciate the participation of the following collaborators: P. Rådström (Applied Microbiology, Center for Chemistry and Chemical Engineering, Institute of Technology, Lund University, Lund, Sweden), N. Cook (Central Science Laboratory, York, United Kingdom), M. Wagner (Institute for Milk Hygiene, Milk Technology and Food Science, Vienna, Austria), M. Kuhn (Congen Biotech GmbH, Berlin, Germany), P. T. Tassios (Department of Microbiology, Medical School, University of Athens, Athens, Greece), A. Abdulmawjood (Institute of Veterinary Food Science, Justus-Liebig University Giessen, Giessen, Germany), S. Perelle (Agence Française de Sécurité Sanitaire des Aliments, Maisons Alfort, France), R. Karpiskova (National Institute of Public Health, Brno, Czech Republic), T. Kuchta (Food Research Institute, Bratislava, Slovakia), K. Demnerova (Institute of Chemical Technology, Prague, Czech Republic), T. Aymerich (Institute for Food and Agricultural Research and Technology, Monells, Spain), D. De Medici (Istituto Superiore di Sanità, Rome, Italy), S. Pelkonen (National Veterinary and Food Research Institute, Kuopio, Finland), A. Heuvelink (Inspectorate for Health Protection and Veterinary Public Health, Zutphen, The Netherlands), and B. China (Faculty of Veterinary Medicine, Food Science Department, University of Liege, Liege, Belgium).
REFERENCES
- 1.Aabo, S., O. F. Rasmussen, L. Rossen, P. D. Sorensen, and J. E. Olsen. 1993. Salmonella identification by the polymerase chain reaction. Mol. Cell. Probes 7:171-178. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 1994. Accuracy (trueness and precision) of measurement methods and results. Part 1. General principles and definitions (ISO 5725-1:1994). International Organization for Standardization, Geneva, Switzerland.
- 3.Anonymous. 1994. Accuracy (trueness and precision) of measurement methods and results. Part 2. Basic method for the determination of repeatability and reproducibility of a standard measurement method (ISO 5725-2:1994). International Organization for Standardization, Geneva, Switzerland.
- 4.Anonymous. 2002. Microbiology of food and animal feeding stuffs. Protocol for the validation of alternative methods (EN ISO 16140). European Committee for Standardization, Paris, France.
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Current protocols in molecular biology, p. 8.2.8-8.2.13. John Wiley & Sons, New York, N.Y.
- 6.Bäumler, A. J., F. Heffron, and R. Reissbrodt. 1997. Rapid detection of Salmonella enterica with primers specific for iroB. J. Clin. Microbiol. 35:1224-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, N. D., H. L. Neibergs, D. E. Wallis, R. B. Simpson, E. D. McGruder, and B. M. Hargis. 1994. Genus-specific detection of salmonellae in equine feces by use of the polymerase chain reaction. Am. J. Vet. Res. 55:1049-1054. [PubMed] [Google Scholar]
- 8.Collazo, C. M., and J. E. Galán. 1997. The invasion-associated type-III protein secretion system in Salmonella—a review. Gene 192:51-59. [DOI] [PubMed] [Google Scholar]
- 9.Galán, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galán, J. E., and R. Curtiss III. 1991. Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect. Immun. 59:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner, I. A., and M. Greiner. 1999. Advanced methods for test validation and interpretation in veterinary medicine. Joint cooperation between the Freie Universität Berlin and the University of California, Davis. Freie Universität Berlin, Berlin, Germany.
- 12.Hoorfar, J. 1999. EU seeking to validate and standardize PCR testing of food pathogens. ASM News 65:799. [Google Scholar]
- 13.Hoorfar, J., P. Ahrens, and P. Rådström. 2000. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J. Clin. Microbiol. 38:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphrey, T. 2000. Public-health aspects of Salmonella infection, p. 245-263. In C. Way and A. Way (ed.), Salmonella in domestic animals. CABI Publishing, Oxon, United Kingdom.
- 15.Knutsson, R., Y. Blixt, H. Grage, E. Borch, and P. Rådström. 2001. Evaluation of selective enrichment PCR procedures for Yersinia enterocolitica. Int. J. Food Microbiol. 73:35-46. [DOI] [PubMed] [Google Scholar]
- 16.Kwang, J., E. T. Littledike, and J. E. Keen. 1996. Use of the polymerase chain reaction for Salmonella detection. Lett. Appl. Microbiol. 22:46-51. [DOI] [PubMed] [Google Scholar]
- 17.Malorny, B., A. Schroeter, C. Bunge, B. Hoog, A. Steinbeck, and R. Helmuth. 2001. Evaluation of molecular typing methods for Salmonella enterica serovar Typhimurium DT104 isolated in Germany from healthy pigs. Vet. Res. 32:119-129. [DOI] [PubMed] [Google Scholar]
- 18.Malorny, B., P. T. Tassios, P. Rådström, N. Cook, M. Wagner, and J. Hoorfar. Standardization of diagnostic PCR for the detection of foodborne pathogens. Int. J. Food Microbiol., in press. [DOI] [PubMed]
- 19.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1972. Experiments in molecular genetics, p. 433. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Rahn, K., S. A. De Grandis, R. C. Clarke, S. A. McEwen, J. E. Galán, C. Ginocchio, R. Curtiss III, and C. L. Gyles. 1992. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6:271-279. [DOI] [PubMed] [Google Scholar]
- 22.Sachs, L. 1984. Applied statistics: a handbook of techniques. Springer, Heidelberg, Germany.
- 23.Tirado, C., and K. Schmidt. 2001. WHO surveillance programme for control of foodborne infections and intoxications: results and trends across greater Europe. J. Infect. 43:80-84. [DOI] [PubMed] [Google Scholar]
- 24.Wallace, D. J., T. Van Gilder, S. Shallow, T. Fiorentino, S. D. Segler, K. E. Smith, B. Shiferaw, R. Etzel, W. E. Garthright, F. J. Angulo, et al. 2000. Incidence of foodborne illnesses reported by the foodborne diseases active surveillance network (FoodNet)-1997. J. Food Prot. 63:807-809. [DOI] [PubMed] [Google Scholar]
- 25.Widjojoatmodjo, M. N., A. C. Fluit, R. Torensma, B. H. Keller, and J. Verhoef. 1991. Evaluation of the magnetic immuno PCR assay for rapid detection of Salmonella. Eur. J. Clin. Microbiol. Infect. Dis. 10:935-938. [DOI] [PubMed] [Google Scholar]