Abstract
The seven botulinum neurotoxins (BoNTs) are zinc metalloproteases that cleave neuronal proteins involved in neurotransmitter release and are among the most toxic natural products known. High-throughput BoNT assays are needed for use in antibotulinum drug discovery and to characterize BoNT protease activities. Compared to other proteases, BoNTs exhibit unusually stringent substrate requirements with respect to amino acid sequences and polypeptide lengths. Nonetheless, we have devised a strategy for development of fluorigenic BoNT protease assays, based on earlier structure-function studies, that has proven successful for three of the seven serotypes: A, B, and F. In synthetic peptide substrates, the P1 and P3′ residues were substituted with 2,4-dinitrophenyl-lysine and S-(N-[4-methyl-7-dimethylamino-coumarin-3-yl]-carboxamidomethyl)-cysteine, respectively. By monitoring the BoNT-catalyzed increase in fluorescence over time, initial hydrolysis rates could be obtained in 1 to 2 min when BoNT concentrations were 60 ng/ml (about 1 nM) or higher. Each BoNT cleaved its fluorigenic substrate at the same location as in the neuronal target protein, and kinetic constants indicated that the substrates were selective and efficient. The fluorigenic assay for BoNT B was used to characterize a new competitive inhibitor of BoNT B protease activity with a Ki value of 4 μM. In addition to real-time activity measurements, toxin concentration determinations, and kinetic studies, the BoNT substrates described herein may be directly incorporated into automated high-throughput assay systems to screen large numbers of compounds for potential antibotulinum drugs.
The structurally related protein neurotoxins produced by Clostridium botulinum (BoNTs) and Clostridium tetani are among the most potent toxins known (10). Seven serotypes of BoNT, types A through G, and one serotype of tetanus toxin have been reported (9). Each consists of a light chain (Mr, ∼50,000) and a heavy chain (Mr, ∼100,000) covalently linked by a single disulfide bond. Receptor-binding and translocation domains on the heavy chain deliver the light chain into the cytosol of the neuronal cell, where the zinc metalloprotease activity of the light chain cleaves and inactivates proteins involved in the mechanism of neurotransmitter release. For example, BoNT A cleaves the Mr ∼25,000 synaptosomal-associated protein (SNAP-25) between residues Q197 and R198. BoNTs B and F both cleave a neuronal vesicle-associated membrane protein (VAMP), but at different locations: residues Q76 and F77 for BoNT B and residues Q58 and K59 for BoNT F. Because the combined functionalities of the heavy and light chains are required for in vivo biological activity, the individual subunits are not toxic (15, 16).
Botulinum toxins have proven to be useful research tools for investigating the process of neurotransmitter release and are highly effective drugs for treating certain muscle dysfunctions and other diseases in humans (8, 10, 15). Therefore, BoNT protease activity has become the subject of intense research with regard to substrate requirements, catalytic mechanisms, and the development of inhibitors that might reverse BoNT-induced paralysis (1, 3, 5, 17, 18, 20, 22). Progress in these areas would be expedited by the availability of assays that provide real-time measurements of BoNT protease activity and are readily adapted for use in automated high-throughput systems. To meet these requirements, we developed soluble fluorigenic substrates for the protease activities of BoNT serotypes A, B, and F. Activity measurements and determinations of kinetic constants showed that the substrates are highly sensitive and suitable for use with very low concentrations of BoNT. An example of a practical application was the characterization of a new competitive inhibitor of BoNT B protease activity (developed for this report) by using the BoNT B fluorigenic substrate. The new fluorigenic assays supplement the capabilities of our previously-reported high-throughput solid-phase BoNT assays (19).
MATERIALS AND METHODS
Peptide synthesis.
The peptide synthesizer used in this study was model 431A from Perkin Elmer-Applied Biosystems, Foster City, Calif. We used reagents and protocols obtained from the synthesizer manufacturer. All peptides were N-terminal acetylated and had carboxamide at the C termini. Peptides were purified by reverse-phase high-pressure liquid chromatography (HPLC) with gradients of acetonitrile in 0.1% trifluoroacetic acid (TFA). HPLC equipment was obtained from Waters Corp., Milford, Mass. Reverse-phase HPLC columns (Hi-Pore RP318 and RP304) were obtained from Bio-Rad Laboratories, Hercules Calif.
The fluorescence donor-acceptor pair was introduced during peptide synthesis by incorporating cysteine and Nɛ-(2,4-dinitrophenyl)-lysine (dnpK), respectively, at the desired locations. The fluorophore was then introduced by reacting the sulfhydryl group of the cysteine residue with 3-iodoacetamido-4-methyl-7-dimethylamino-coumarin (DACIA; Molecular Probes, Inc., Eugene, Oreg.) as follows. Solutions of 2 to 5 mM peptide and 10 mM DACIA were prepared in methyl sulfoxide. Equal volumes of each were mixed, and N, N-diisopropylethylamine was added to approximately 30 mM. All of the peptide reacted with the reagent in 30 min at ambient temperature. Excess reagent was then discharged by adding mercaptoethanol to 0.05 M. After another 20 to 30 min, the reaction mixture was diluted with 8 volumes of water, and the pH was adjusted to ∼2 with TFA. The product was concentrated by solid-phase extraction (C18 Sep-Pak; Waters Corp.) and purified by reverse-phase HPLC. The reaction product of DACIA and the cysteine sulfhydryl group is abbreviated as “daciaC.”
Extinction coefficients of the substrates.
Three solutions of 5 mM DACIA were prepared in methyl sulfoxide from three separate weighings of reagent. To enhance solubility in water, DACIA was reacted with mercaptoethanol in methyl sulfoxide as described above. Aliquots of each solution were diluted to 30 μM in 40 mM HEPES-0.05% Tween (pH 7.3), and absorbance spectra were obtained in an LKB Ultrospec III spectrophotometer (1-cm light path). Samples of known concentrations were also analyzed by reverse-phase HPLC to obtain factors (peak areas per mole) for use in calculating substrate and hydrolysis-product concentrations from HPLC runs. Identical procedures were done with three solutions of dnpK. Extinction coefficients and area factors of DACIA and dnpK were summed to obtain values for the intact substrates. The results are the averages of triplicate determinations ± standard deviations.
Excitation and emission spectra of intact and cleaved substrates.
Fluorescence spectra were obtained with a fluorimeter from Photon Technology International, Lawrenceville N.J., with “ultramicro” cuvettes (70-μl minimum sample volume, 3-mm path length; Hellma, Inc., Plainview, N.Y.). The buffer was 40 mM HEPES-0.05% Tween (pH 7.3). Fluorescence was expressed in arbitrary units. To compare the fluorescence of intact substrate to that of the hydrolyzed products, aliquots were incubated at ambient temperature for 1 h with 50 μg of trypsin (BoNT A and BoNT F substrates) or chymotrypsin (BoNT B substrate) per ml. Complete digestion was confirmed by HPLC analyses (not shown).
Assays of BoNT protease activities.
BoNT A, B, and F were obtained from Metabiologics, Madison, Wis. Recombinant light chains of BoNT A and B (A-Lc and B-Lc, respectively) were kindly provided by Michael Byrne (2) and Melody Jensen, Toxinology and Aerobiology Division, U.S. Army Medical Research Institute of Infectious Diseases.
In HPLC-based assays, initial hydrolysis rates were determined as described previously (17, 18). Briefly, assay mixtures (30 μl) contained 40 mM HEPES-0.05% Tween (pH 7.3) and various concentrations of substrate and BoNT or recombinant light chain. Holotoxin was first preincubated for 30 min with 10 mM dithiothreitol (DTT) and 0.5 mM ZnCl2 and then assayed in the presence of 5 mM DTT and 0.25 mM ZnCl2 (17). However, it was not necessary to preincubate recombinant light chain or to add DTT or ZnCl2 to light-chain assays (data not shown). The temperature was 37°C. Assays were stopped by addition of TFA to 0.5% and then analyzed by reverse-phase HPLC. Concentrations of substrates and hydrolysis products were calculated from peak areas.
Kinetic constants were obtained from plots of initial rates versus seven concentrations of substrate. Results were calculated from nonlinear regression analyses by using the program Enzfitter (Biosoft, Cambridge, United Kingdom). The values are the averages of three independent determinations ± standard deviation. The Ki value for the BoNT B inhibitor was first calculated from a Dixon plot by using the equation Ki = Km/[(slope)(Vmax)(S)], where S is the substrate concentration (21). The Ki value and the type of inhibition (competitive, noncompetitive, etc.) were then determined by calculation of substrate Vmax and apparent Km (Kmapp), as described above, in the presence of three different inhibitor concentrations. Ki values for each of the three assays in the presence of inhibitor were calculated with the equation Ki = [I]/[(Kmapp/Km) − 1], where [I] is the inhibitor concentration (21).
In fluorigenic assays, buffer (40 mM HEPES-0.05% Tween [pH 7.3]) and substrate were mixed in a volume of 80 μl and placed in a fluorimeter cuvette. The reaction was started by adding light chain or BoNT. Fluorescence was monitored at 1 point per s. The excitation and emission wavelengths were 398 and 485 nm, respectively. The temperature was maintained at 25 to 27°C.
Correlation of fluorescence change per unit of time with hydrolysis rate.
Solutions containing various concentrations of substrate and light chain were incubated for different periods of time in the fluorimeter. In each case, after the last fluorescence reading was taken, the reaction was quickly stopped by adding TFA to about 0.5%. The extent of light-chain-catalyzed hydrolysis was then determined by HPLC. Initial substrate concentrations from 5 to 40 μM and light-chain concentrations from 0.05 to 2 μg/ml were employed, yielding product concentrations from 0.6 to 22 μM.
RESULTS AND DISCUSSION
Structures of fluorigenic substrates.
The fluorigenic substrate for BoNT A protease activity (Fl-A) was based on a peptide representing residues 187 to 203 of SNAP-25 (SNAP 187-203), which was a substrate for the protease activity of BoNT A (17). Because the efficiency of energy transfer is inversely proportional to distance between fluorescence donor and acceptor (13), we wanted to place the two as close as possible in the peptide sequence. When modifying SNAP 187-203 to obtain a fluorigenic substrate, we based the potential locations of the donor and acceptor moieties on known catalytic properties and substrate binding requirements of BoNT A. For example, P1 glutamine and/or P3′ threonine could be replaced in the substrate by certain other residues without loss of function, but peptides with substitutions at P2 asparagine, P1′ arginine, or P2′ alanine were not cleaved (18). Therefore, we chose P1 glutamine and P3′ threonine to be replaced with dnpK and daciaC, respectively. In addition, we found that replacing the two lysine residues in the substrate (equivalent to K189 and K201 in SNAP-25) with arginines provided a 1.5-fold increase in the hydrolysis rate (data not shown). Therefore, Fl-A had the following sequence: SNRTRIDEAN[dnpK]RA[daciaC]RML.
This peptide was cleaved by A-Lc between the dnpK and R residues, with a concomitant increase in fluorescence, and proved to be an efficient fluorigenic substrate for the protease activity of BoNT A (see sections below). However, a peptide with the opposite orientation of dnpK and daciaC was not.
To obtain a fluorigenic substrate for BoNT B (Fl-B), the strategy we employed was analogous to that for BoNT A. Fl-B included residues 60 to 94 of human VAMP-2 (22), with P1 glutamine and P3′ threonine replaced by dnpK and daciaC, respectively. The sequence was LSELDDRADALQAGAS[dnpK]FE[daciaC]SAAKLKRKYWWKNLK.
Finally, the fluorigenic substrate for BoNT F (Fl-F) was based on VAMP residues 37 to 75, which was an effective substrate for the protease activity of BoNT F (19): AQVDEVVDIMRVNVDKVLERD[dnpK]KL[daciaC]ELDDRADALQAGAS.
All of the fluorigenic substrates were soluble up to at least 0.3 mM; Fl-A was soluble at 1 mM. Solutions were stable to repeated freeze-thaw cycles, and exposure to ambient laboratory temperature and lighting for up to 60 min had no adverse effects (data not shown).
Fluorescence properties of the fluorigenic substrates.
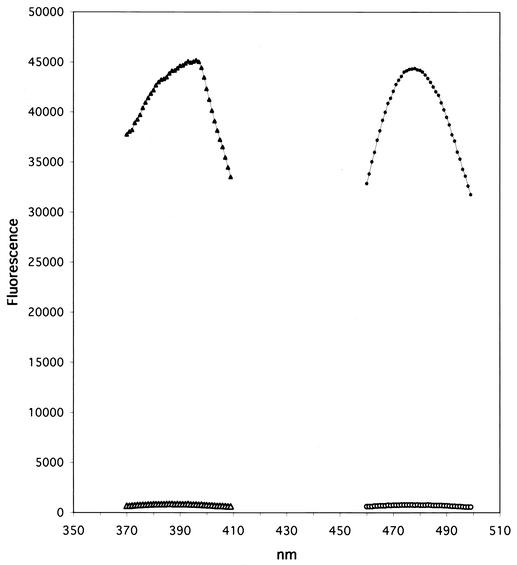
The excitation and emission spectra of intact and hydrolyzed Fl-A are shown in Fig. 1. Intact Fl-A had less than 2% of the fluorescence exhibited by the hydrolysis product; i.e., more than 98% of the available fluorescence was quenched in the unhydrolyzed substrate. This result compares favorably with fluorigenic substrates for many other proteases (3, 9). Excitation and emission maxima were 398 and 485 nm, respectively. Fl-B and Fl-F with the same donor-acceptor pair placed the same distance apart in the sequences gave identical results.
FIG. 1.
Excitation and emission spectra of intact and cleaved Fl-A. Open triangles, excitation scan of intact Fl-A; solid triangles, excitation scan of cleaved Fl-A; open circles, emission scan of intact Fl-A; solid circles, emission scan of cleaved Fl-A.
Determining concentrations of substrates and cleavage products.
Because DACIA and dnpK both had substantial absorbance at 365 nm, we chose this wavelength for calculations of fluorigenic substrate and hydrolysis product concentrations. The molar extinction coefficients for DACIA and dnpK at 365 nm were 18,700 ± 1,160 and 17,800 ± 360, respectively. At this wavelength, contributions from other amino acid side chains were negligible. Therefore, the molar extinction coefficient of the substrates at 365 nm was 36,500. In the widely used Waters model 2487 detector, with the HPLC column effluent monitored at 365 nm, the peak area per nanomole of substrate injected was 2,300 ± 145 mV s.
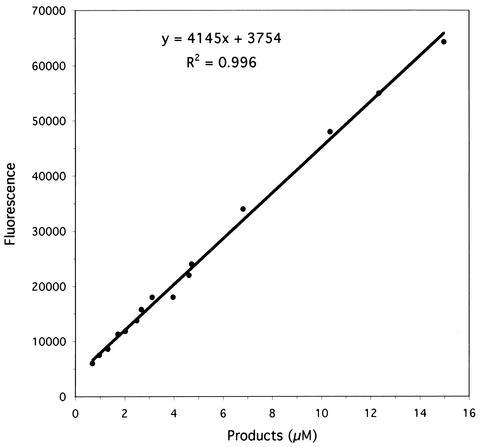
To correlate fluorescence change per unit time with hydrolysis rate, fluorescence values were plotted against the concentrations of hydrolysis products, catalyzed by light chain, and quantitated by HPLC. This approach more accurately reflected actual assay conditions than did synthesis of individual cleavage products and measurement of their fluorescence properties in the absence of intact substrate. The results for product concentrations up to 15 μM with Fl-A cleaved by A-Lc are shown in Fig. 2. The slope of the line, 4,145, represents the fluorescence exhibited by 1.0 μM product and was used to calculate hydrolysis rates in moles per unit of time. The correlation between fluorescence and molarity of product was essentially linear up to about 15 μM when initial substrate concentrations were 20 μM or less, but was nonlinear at higher product and/or initial substrate concentrations. Because both DACIA and dnpK have substantial absorbance at the excitation wavelength, the observed nonlinearity was due to the inner filter effect (13). Such limitations are common to fluorigenic substrates (11). The results for Fl-B and Fl-F were not significantly different from those in Fig. 2.
FIG. 2.
Plot of fluorescence versus concentrations of products from hydrolysis of Fl-A catalyzed by A-Lc.
Mass spectrometry of hydrolysis products recovered from HPLC analyses revealed that each fluorigenic substrate was cleaved by the respective enzyme only between the two residues corresponding to the cleavage site in the natural sequence substrate. For example, Fl-A was cleaved by A-Lc at the dnpK-arginine bond, analogous to residues 197 and 198 of SNAP-25. In all cases, there were no other cleavage products detected (data not shown).
Fluorigenic assays.
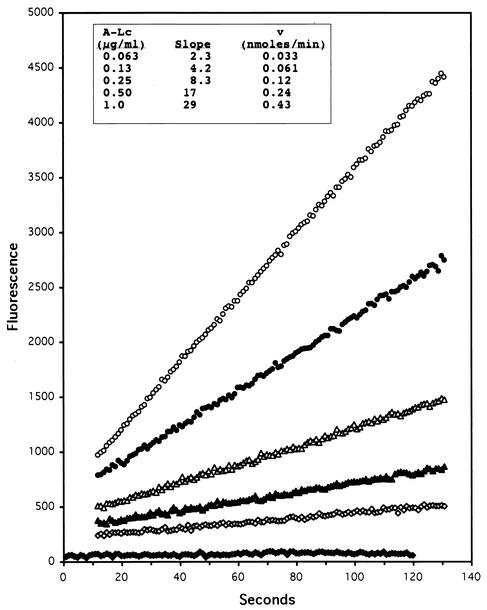
Representative fluorigenic assays of Fl-A hydrolyzed by A-Lc are depicted in Fig. 3. The hydrolysis rates tabulated in Fig. 3 (inset) are averages of triplicate determinations, and standard deviations were <±10% in all cases. For routine assays, initial hydrolysis rates could be obtained in 1 to 2 min. With A-Lc concentrations between 1 and 60 ng/ml, longer assay times of up to 1 h were employed (data not shown). The substrate was stable under these conditions, because the fluorescence readings in the absence of light chain (blanks) did not change significantly during assays. Therefore, it was not necessary to correct assay slopes due to changes in the blanks.
FIG. 3.
Hydrolysis of 10 μM Fl-A catalyzed by various concentrations of A-Lc: no A-Lc, solid diamonds; 0.063 μg/ml, open diamonds; 0.13 μg/ml, solid triangles; 0.25 μg/ml, open triangles; 0.50 μg/ml, solid circles; 1.0 μg/ml, open circles. (Inset) Hydrolysis rate versus concentration of A-Lc.
Similar results were obtained when Fl-B was cleaved by B-Lc, but hydrolysis rates were higher than those in Fig. 3. For example, with 10 μM Fl-B and 1 μg of B-Lc per ml, the rate was 0.88 ± 0.016 nmol/min.
Because an active recombinant BoNT F light chain was not available, Fl-F was assayed with the holotoxin. With 10 μM substrate and 1 μg of BoNT F per ml (equivalent to 0.3 μg of light chain per ml), the hydrolysis rate was 0.075 nmol/min.
The kinetic constants of peptide substrates for BoNT and light-chain protease activities, as well as those of their respective fluorigenic derivatives, are summarized in Table 1. Compared to published results for BoNT A and B holotoxins (17, 22), the kcat was higher and the Km was lower when unmodified peptide substrates were assayed with the respective recombinant light chains. The largest differences were found when SNAP 187-203 was hydrolyzed by A-Lc. In that case, the kcat was almost fivefold higher and the Km was almost fivefold lower than the kinetic constants for the same substrate hydrolyzed by BoNT A. For hydrolysis of VAMP 60-94 by BoNT B and B-Lc, kcat values were similar for both (24 and 38 s−1, respectively), but the Km was about fourfold lower when B-Lc was the protease. These differences could be due to several factors. For example, the protease activity of BoNT must be preactivated by incubating holotoxin with a reducing agent and low concentrations of zinc. However, concentrations of both must be carefully optimized, because these agents can also diminish BoNT protease activity (6, 17, 22). Furthermore, BoNT heavy chain might block access of the substrate to the active site on the light chain (12), which could result in higher Km values when substrates were hydrolyzed by holotoxins instead of recombinant light chains. Therefore, the protease activities of BoNT holotoxins were compromised by the presence of potential inhibitors. In assays with recombinant light chain, the heavy chain was absent, and it was not necessary to include reducing agent or zinc. Consequently, optimum activities were observed.
TABLE 1.
Kinetic constants of substrates for BoNT protease activities
| Substrate (reference) | Enzyme | kcat (s−1) | Km (mM) | kcat/Km [(M−1 s−1) × 103] |
|---|---|---|---|---|
| SNAP 187-203 (17) | BoNT A | 4.7 ± 0.5 | 5.0 ± 0.5 | 0.94 |
| SNAP 187-203 | A-Lc | 23 ± 1 | 1.1 ± 0.1 | 21 |
| F1-A | A-Lc | 7.2 ± 0.4 | 0.096 ± 0.01 | 7.5 |
| VAMP 60-94 (22) | BoNT B | 24 ± 0.9 | 0.33 ± 0.03 | 72 |
| VAMP 60-94 | B-Lc | 38 ± 1 | 0.081 ± 0.004 | 470 |
| F1-B | B-Lc | 4.7 ± 0.2 | 0.027 ± 0.002 | 170 |
| VAMP 37-75 | BoNT F | 17 ± 1 | 1.1 ± 0.1 | 15 |
| F1-F | BoNT F | NDa | ND | ND |
ND, not determined.
The fluorigenic substrates Fl-A and Fl-B had lower kcat and Km values than their respective unmodified substrates. A reduction in kcat was not surprising, because a relatively large residue with an aromatic group (dnpK) replaced the P1 glutamine on the N-terminal side of the cleavage site, and a large fluorescent moiety with a conjugated ring structure (daciaC) replaced the P3′ threonine. Similarly, decreased Km values have been reported for many BoNT substrate modifications (17, 20, 22), including a previously described fluorigenic substrate for BoNT B (3). Nonetheless, selectivity constants (kcat/Km in Table 1) for Fl-A and Fl-B showed that they were practical and efficient substrates for BoNT A and B protease activities, respectively. Finally, cleavage of such extensively modified substrates by BoNT supports our earlier work, which showed that many binding subsites on BoNT are not highly specific for a particular amino acid side chain, and substrate discrimination occurs mainly at the catalytic stage of the reaction rather than at the initial binding stage (18, 19).
Characterization of a BoNT B inhibitor by using the fluorigenic assay.
In earlier work with the BoNT A peptide substrate SNAP 187-203, replacement of P1 glutamine with [d]cysteine yielded an inhibitor of BoNT A protease activity with a Ki of 2 μM (20). We conjectured that substituting [d]cysteine for the equivalent residue, glutamine 76, in VAMP 60-94 would result in an inhibitor of BoNT B. This peptide (Q76[d]C) was synthesized and added to fluorigenic assays of B-Lc with 10 μM Fl-B as the substrate (Fig. 4). Sixteen different concentrations of Q76[d]C were employed, and initial rates were obtained in 2-min assays. It was clear that Q76[d]C is a good inhibitor of BoNT B protease activity. The Ki value, calculated from the Dixon plot (Fig. 4, inset), was 3 μM.
FIG. 4.
Rate of cleavage of 10 μM Fl-B catalyzed by 1 μg of B-Lc per ml in the presence of various Q76[d]C concentrations. (Inset) Dixon plot of the data.
To identify the type of inhibition and confirm Ki, kinetic constants of Fl-B were determined in the presence of three concentrations of Q76[d]C. The results are shown in Table 2. Q76[d]C had no effect on kcat, but Kmapp increased with Q76[d]C concentration. Therefore, Q76[d]C was a competitive inhibitor of B-Lc protease activity. The average Ki was 4 ± 0.7 μM, in good agreement with the estimate from the Dixon plot.
TABLE 2.
Kinetic constants of F1-B and VAMP 60-94 in the presence of the inhibitor Q76[D]C
| Substrate | Q76[D]C concn (μM) | kcat (s−1) | Kmapp (mM) |
|---|---|---|---|
| F1-B | 0 | 4.7 | 0.027 |
| 2 | 4.5 | 0.038 | |
| 5 | 5.0 | 0.064 | |
| 15 | 4.4 | 0.10 | |
| VAMP 60-94 | 0 | 38 | 0.081 |
| 2 | 39 | 0.11 | |
| 5 | 35 | 0.15 | |
| 15 | 39 | 0.33 |
In principle, Ki is an intrinsic property of the inhibitor and is independent of the substrate against which the inhibitor is tested. However, because of the extensive structural changes employed when VAMP 60-94 was modified to obtain the fluorigenic substrate Fl-B, we also tested Q76[d]C as an inhibitor of BoNT B protease activity with unmodified VAMP 60-94 as the substrate. Again, inhibition was competitive (Table 2), and the Ki was 5 ± 0.8 μM, confirming results obtained when Fl-B was the substrate. Based on all three determinations, the average Ki was 4 ± 1 μM.
HPLC analyses confirmed that Q76[d]C was not cleaved by B-Lc under assay conditions. In contrast, Q76[l]C was slowly hydrolyzed by B-Lc and was a relatively weak inhibitor (Ki > 0.1 mM) (data not shown).
We propose that inhibition of B-Lc by Q76[d]C is due to binding of the active-site zinc by the [d]cysteine sulfhydryl group, but this has not yet been confirmed. Nonetheless, because Q76[d]C is a substrate analog and is not cleaved by B-Lc, we anticipate that this competitive inhibitor will prove useful for investigations of BoNT B interactions with substrate, through X-ray analyses of BoNT B-Q76[d]C cocrystals. Additional studies of the mode of binding and structural modifications to improve Ki are in progress.
Comparisons to other fluorescence-based BoNT protease assays.
Earlier work in our laboratory resulted in fluorescence-based solid-phase assays for the protease activities of BoNTs A, B, D, and F (19). Substrate was labeled at the N terminus with fluorescein and then covalently bound at the C terminus to the walls of multiwell plates. BoNT protease activity cleaved the substrate and thereby released fluorescence into solution, which was then measured. The solid-phase assays were economical, reproducible, stable to prolonged storage, sensitive, and afforded true high-throughput capability. Furthermore, they could be used with samples that interfered with direct fluorescence measurements due to turbidity or quenching. However, the solid-phase assays were not compatible with matrix-bound samples, such as combinatorial libraries on polymer beads. In that case, decreased BoNT proteolysis could result simply from binding to compounds on the matrix or to the matrix itself, rather than direct inhibition of protease activity. In addition, solid-phase assays are not suitable for BoNT kinetic studies, because the effective concentration of an immobilized substrate cannot be determined. We developed the fluorigenic substrates described in this report to address these needs and to complement the capabilities of the solid-phase assays.
Another fluorigenic substrate for BoNT B, based on VAMP 60-94, was published (3), but the type and placement of fluorescence donor and acceptor differed from those described herein. Pyrenylalanine (fluorescence donor) was substituted for P3 alanine, and 4-nitrophenylalanine (acceptor) was substituted for P1′ phenylalanine. This approach would not be suitable for a BoNT A fluorigenic substrate, because P1′ arginine cannot be replaced without loss of substrate function (18, 20). Furthermore, production of their substrate first required the stereospecific synthesis of N-(fluorenylmethoxycarbonyl)-3-(pyrene-1-yl)-[l]alanine, because the latter was not commercially available. In contrast, all components needed for synthesizing the substrates in this report were commercially available. Compared to unmodified substrate, their fluorigenic substrate had a higher Vmax and lower Km, allowing assays with very low concentrations of toxin. In addition to routine assays, the authors also proposed use of their substrate to detect the presence of BoNT in food or serum.
We did not determine the lower limits of detection for our substrates, because they were developed primarily for rapid, real-time determinations of BoNT protease activities and for high-throughput assay systems. In these contexts, recombinant light chains are the enzymes of choice, because they are nontoxic, have high specific activities (Table 1), and production methods typically exhibit higher yields with fewer purification steps than those involving BoNT holotoxins (2, 14). BoNT light-chain protease activities do not require preactivation or addition of reducing agents or zinc to assays. However, if necessary, holotoxins may be used with the fluorigenic substrates, as exemplified by cleavage of Fl-F by BoNT F. Therefore, fluorigenic assays could be used for determinations of toxin concentrations in samples intended for research or for therapeutic applications, and in some situations, they could replace the mouse lethality bioassay.
Because fluorescence of the intact substrate was stable and relatively low, very small BoNT-catalyzed changes were accurately measured. Initial rates of hydrolysis could be obtained in 1 to 2 min, even when low enzyme concentrations were employed, and a relatively small quantity of enzyme could be used for many assays. For example, 1 mg of purified A-Lc, in 100-μl assays at 0.25 μg/ml would be sufficient for approximately 40,000 assays.
Fluorigenic substrates have been described for many proteases (3, 4, 7, 11, 23). However, clostridial neurotoxins exhibit unusually stringent substrate requirements with respect to polypeptide length and amino acid sequence (6, 17, 18). Modifications often result in loss of functionality. Therefore, selecting a fluorescence donor-acceptor pair and placing it in the sequence are not obvious decisions. In this report, substituting dnpK for P1 glutamine and daciaC for P3′ threonine gave efficient fluorigenic substrates for the protease activities of BoNT A, B, and F. Application of this and other approaches to the development of such substrates for the remaining BoNT serotypes (C, D, E, and G) is in progress.
REFERENCES
- 1.Adler, M., J. D. Nicholson, F. Cornille, and B. E. Hackley. 1998. Efficacy of a novel metalloprotease inhibitor on botulinum neurotoxin B activity. FEBS Lett. 429:234-238. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, A. S., M. P. Byrne, M. Jensen, H. B. Hines, E. Brueggemann, and L. A. Smith. 2001. Enzymatic autocatalysis of botulinum A neurotoxin light chain. J. Protein Chem. 20:221-231. [DOI] [PubMed] [Google Scholar]
- 3.Anne, C., F. Cornille, C. Lenoir, and B. P. Roques. 2001. High-throughput fluorigenic assay for determination of botulinum type B neurotoxin protease assay. Anal. Biochem. 291:253-261. [DOI] [PubMed] [Google Scholar]
- 4.Carmel, A., and A. Yaron. 1978. An intramolecularly quenched fluorescent tripeptide as a fluorigenic substrate of angiotensin-I-converting enzyme and of bacterial dipeptidyl carboxypeptidase. Eur. J. Biochem. 87:265-273. [DOI] [PubMed] [Google Scholar]
- 5.Eswaramoorthy, S., D. Kumaran, and S. Swaminathan. 2002. A novel mechanism for Clostridium botulinum neurotoxin inhibition. Biochemistry 41:9795-9802. [DOI] [PubMed] [Google Scholar]
- 6.Foran, P., C. C. Shone, and J. O. Dolly. 1994. Differences in the protease activities of tetanus and botulinum B toxins revealed by the cleavage of vesicle-associated membrane protein and various sized fragments. Biochemistry 33:15365-15374. [DOI] [PubMed] [Google Scholar]
- 7.Hawthorne, S. J., P. Harriott, J. Lim, A. J. Turner, B. Walker, and C. H. Williams. 1997. Evaluation of some fluorigenic substrates for continuous assay of aminopeptidase P. Anal. Biochem. 253:13-17. [DOI] [PubMed] [Google Scholar]
- 8.Heckman, M., A. O. Ceballos-Bauman, and G. Plewig. 2001. Botlinum toxin A for axillary hyperhidrosis (excessive sweating). N. Engl. J. Med. 344:488-493. [DOI] [PubMed] [Google Scholar]
- 9.Humeau, Y., F. Doussau, N. J. Grant, and B. Poulain. 2000. How botulinum and tetanus neurotoxins block neurotransmitter release. Biochemie 82:427-446. [DOI] [PubMed] [Google Scholar]
- 10.Kessler, K. R., and R. Benecke. 1997. Botulinum toxin: from poison to remedy. Neurotoxicology 18:761-770. [PubMed] [Google Scholar]
- 11.Knight, C. G., F. Willenbrock, and G. Murphy. 1992. A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett. 296:263-266. [DOI] [PubMed] [Google Scholar]
- 12.Lacy, D. B., W. Tepp, A. C. Cohen, B. R. DasGupta, and R. C. Stevens. 1998. Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nat. Struct. Biol. 5:898-902. [DOI] [PubMed] [Google Scholar]
- 13.Lakowicz, J. R. 1986. Principles of fluorescence spectroscopy. Plenum Press, New York, N.Y.
- 14.Li, L., and B. R. Singh. 1999. High-level expression, purification, and characterization of recombinant type A botulinum neurotoxin light chain. Protein Expres. Purif. 17:339-344. [DOI] [PubMed] [Google Scholar]
- 15.Rossetto, O., M. Seveso, P. Caccin, G. Schiavo, and C. Montecucco. 2001. Tetanus and botulinum neurotoxins: turning bad guys into good by research. Toxicon 39:27-41. [DOI] [PubMed] [Google Scholar]
- 16.Schiavo, G., M. Matteoli, and C. Montecucco. 2000. Neurotoxins affecting neuroexocytosis. Physiol. Rev. 80:717-766. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt, J. J., and K. A. Bostian. 1995. Proteolysis of synthetic peptides by type A botulinum neurotoxin. J. Protein Chem. 14:703-708. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt, J. J., and K. A. Bostian. 1997. Endoproteinase activity of type A botulinum neurotoxin: substrate requirements and activation by serum albumin. J. Protein Chem. 16:19-26. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt, J. J., R. G. Stafford, and C. B. Millard. 2001. High-throughput assays for botulinum neurotoxin proteolytic activity: serotypes A, B, D, and F. Anal. Biochem. 296:130-137. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt, J. J., R. G. Stafford, and K. A. Bostian. 1998. Type A botulinum neurotoxin proteolytic activity: development of competitive inhibitors and implications for substrate specificity at the S1′ binding subsite. FEBS Lett. 435:61-64. [DOI] [PubMed] [Google Scholar]
- 21.Segel, I. H. 1975. Enzyme kinetics. Wiley, New York, N.Y.
- 22.Shone, C. C., and A. Roberts. 1994. Peptide substrate specificity and properties of the zinc-endopeptidase activity of botulinum type B neurotoxin. Eur. J. Biochem. 225:263-270. [DOI] [PubMed] [Google Scholar]
- 23.Zhong, W., and S. J. Benkovic. 1998. Development of an internally quenched fluorescent substrate for Escherichia coli leader peptidase. Anal. Biochem. 255:66-73. [DOI] [PubMed] [Google Scholar]