Abstract
Leaf composts were studied for their suppressive effects on Pythium ultimum sporangium germination, cottonseed colonization, and the severity of Pythium damping-off of cotton. A focus of the work was to assess the role of fatty-acid-metabolizing microbial communities in disease suppression. Suppressiveness was expressed within the first few hours of seed germination as revealed by reduced P. ultimum sporangium germination, reduced seed colonization, and reduced damping-off in transplant experiments. These reductions were not observed when cottonseeds were sown in a conducive leaf compost. Microbial consortia recovered from the surface of cottonseeds during the first few hours of germination in suppressive compost (suppressive consortia) induced significant levels of damping-off suppression, whereas no suppression was induced by microbial consortia recovered from cottonseeds germinated in conducive compost (conducive consortia). Suppressive consortia rapidly metabolized linoleic acid, whereas conducive consortia did not. Furthermore, populations of fatty-acid-metabolizing bacteria and actinobacteria were higher in suppressive consortia than in conducive consortia. Individual bacterial isolates varied in their ability to metabolize linoleic acid and protect seedlings from damping-off. Results indicate that communities of compost-inhabiting microorganisms colonizing cottonseeds within the first few hours after sowing in a Pythium-suppressive compost play a major role in the suppression of P. ultimum sporangium germination, seed colonization, and damping-off. Results further indicate that fatty acid metabolism by these seed-colonizing bacterial consortia can explain the Pythium suppression observed.
Disease-suppressive soils have been known for over a century and identified from many parts of the world (16). Both naturally occurring and compost-amended soils and container media suppressive to Pythium diseases have been described elsewhere (15). Because of the overwhelming evidence that disease suppression in these soils and composts is due to the activities of microorganisms, such systems provide examples of natural and effective biological control. Such suppressive soils can serve potentially as models for understanding the mechanisms by which complex plant-associated microorganisms interfere with plant pathogenesis. However, the specific microorganisms and mechanisms that give rise to disease suppression in these systems have not been clearly identified.
Previous studies of Pythium-suppressive soils and composts have provided some key observations about the role of resident microbial communities in affecting pathogen responses to plants and subsequent disease development in these systems. For example, present evidence indicates that disease suppression is most likely a property of the microbial community as a whole and not the result of any single species. Single microbial antagonists recovered from suppressive soil (21) or potting mix (3) failed to induce disease suppression at a level observed in the suppressive medium. Similarly, the frequency with which antagonistic microorganisms (antagonistic to Pythium ultimum) can be isolated from some suppressive and conducive composts is no higher from suppressive composts than from conducive composts (E. B. Nelson and C. M. Craft, unpublished observations).
Despite the evidence linking Pythium suppression and disease suppression with compost-inhabiting microorganisms, identification of the specific microorganisms and processes involved in disease suppression has remained elusive. In our work, we have reasoned that clues to the identities of the microorganisms and processes involved in the suppression of Pythium damping-off might be found by looking more closely at the temporal responses of Pythium sporangia to cottonseeds sown in suppressive and conducive composts. Results of such comparative analyses should provide important information about when and how suppressiveness is expressed.
The critical stages and timing of seed infection by P. ultimum as well as the seed exudate molecules responsible for eliciting the germination of P. ultimum sporangia have been documented elsewhere (23). Exogenously dormant sporangia of P. ultimum serve as major survival structures and as primary inocula for plant infections (33). In cotton, long-chain fatty acids are key germination stimulants of P. ultimum sporangia (31), eliciting germination responses within 1.5 to 2 h of sowing cottonseeds (34). Significant seed colonization is observed by 8 to 12 h after sowing (20, 30) with embryo infection occurring by 24 h (20).
Because of the strong link between disease suppression and suppression of pathogen propagule germination, differential responses of P. ultimum sporangia to cottonseeds germinating in suppressive or conducive composts or soils may provide clues as to when disease-suppressive microbial communities are active. For example, by observing P. ultimum propagule germination and seed colonization over time in response to cottonseeds germinating in suppressive or conducive compost, we might be able to identify time points at which Pythium is suppressed and use this information for determining when to sample for Pythium-suppressive microorganisms.
The major objectives of this study were to determine (i) when P. ultimum sporangium germination and seed colonization are suppressed following the sowing of cottonseeds in suppressive composts, (ii) whether microorganisms colonizing cottonseeds at the times when pathogen suppression is expressed contribute to disease suppression, (iii) what are the populations of fatty-acid-metabolizing microorganisms colonizing cottonseeds germinating in suppressive or conducive compost, and (iv) whether these seed-colonizing microorganisms are capable of metabolizing fatty acids and suppressing Pythium damping-off.
MATERIALS AND METHODS
Composts.
Two leaf composts consisting of leaves and twigs from mixed deciduous trees were collected from a composting facility in the village of Endicott, N.Y., in 1989 and 1999 (LC89 and LC99, respectively). LC89 was stored for 10 years at 4°C prior to its use in this study. Disease-suppressive properties of the two composts used in this study had been determined at the time of collection, and they were monitored throughout the study. Suppressiveness or conduciveness of the compost batches was no different than when they were originally collected.
At the commencement of this study in 1999, composts were sieved through a 2-mm-pore-size screen and stored at −20°C. For all laboratory experiments, each compost was mixed with sterile, washed, oven-dried quartz sand with a particle size ranging from 0.5 to 1.0 mm at a rate of 80 mg (dry weight) of compost per cm3 (total volume) of material.
The two composts were similar in their concentrations of major nutrients and organic matter content (Table 1). Minor nutrients and metals such as Mn, B, and Pb were higher in LC99 than in LC89, whereas all other minor nutrients and metals were higher in LC89 than in LC99. Most notable were concentrations of Cu, Fe, and Na that were from 3.4 to 5.6 times higher in LC89 than in LC99.
TABLE 1.
Chemical analyses of suppressive (LC99) and conducive (LC89) leaf composts used in this studya
| Analyte | Value for compost:
|
|
|---|---|---|
| LC89 | LC99 | |
| Major nutrients (%) | ||
| Organic matter | 52.7 | 64.0 |
| Total N | 1.8 | 1.6 |
| NH4-N | 1.4 | 1.6 |
| NO3-N | 0.4 | 0.0 |
| P | 1.7 | 0.3 |
| K | 0.1 | 0.8 |
| Ca | 1.4 | 4.5 |
| Mg | 0.5 | 0.5 |
| Minor nutrients and metals (ppm) | ||
| Mn | 303.1 | 1,007.0 |
| Fe | 31,120.0 | 5,783.0 |
| Cu | 99.0 | 17.7 |
| B | 41.4 | 59.3 |
| Z | 142.2 | 114.9 |
| Mo | 25.1 | 0.3 |
| Al | 3,948.0 | 2,123.0 |
| Na | 1,168.0 | 339.3 |
| Co | 3.9 | 1.8 |
| Cd | <0.1 | <0.1 |
| Cr | 17.2 | 5.1 |
| Ni | 14.1 | 5.6 |
| Pb | 7.2 | 10.3 |
| V | 30.9 | 6.6 |
Samples were analyzed at the Horticulture Nutrient Analysis Laboratory, Cornell University.
Production and germination of P. ultimum sporangia.
P. ultimum isolate P4 was used throughout this study and was maintained at 27°C on a defined mineral salts medium amended with soy lecithin (SM+L) (26, 27). Sporangia produced on this medium mimicked the germination responses observed with sporangia produced on plant tissues and germinated readily in response to cottonseed exudate and linoleic acid, the active component of cottonseed exudate (31).
Inoculum consisted of sporangia on 2-mm-diameter agar disks (∼200 to 400 sporangia/disk). Prior to experiments, the germination of sporangia was tested in response to linoleic acid (positive control) and 10 mM glucose (negative control) to confirm their response behavior.
Plant assays for assessing compost suppressiveness.
Pythium damping-off bioassays to evaluate compost suppressiveness have been described previously (2, 9, 36). In these experiments, 5 cm3 of a compost-sand mixture was placed on the filter paper at the bottom of the cylinder followed by an agar disk containing P. ultimum sporangia. A single surface-disinfested cottonseed was then placed directly onto the agar disk. Prior to use in all experiments, cottonseeds were washed with 1 N NaOH for 10 s to neutralize any residual delinting acids and then rinsed with sterile deionized water. For each bioassay, there were six replicate cylinders per treatment. Controls consisted of inoculated sand and noninoculated sand. The entire assembly was then placed in a clear plastic box and incubated at 25°C with a 16-h photoperiod. After 7 days, each cylinder was rated for seedling emergence and seedling quality. Seedling quality was rated on a scale from 0 to 3 as described previously (36, 37): 0, no seedling emergence; 1, seedling had partially emerged but had failed to develop and might or might not be covered with mycelium; 2, seedling appeared healthy but was less vigorous than the noninoculated control; 3, healthy seedling.
Suppression of sporangium germination and seed colonization.
To examine sporangium germination in response to cottonseeds, agar disks with sporangia were removed from cylinders at 0, 1, 2, 4, 6, 8, and 12 h after sowing. Sporangial disks were rinsed, stained, and examined microscopically. Percent germinated sporangia was determined as described above. Three disks per treatment were examined at each time point.
In separate experiments, seed colonization by P. ultimum was examined by removing cottonseeds from each cylinder at 0, 1, 2, 4, 6, 8, 12, 16, and 24 h after sowing. Seeds were then rinsed with sterile deionized water to remove any sand or compost and plated on a Pythium-selective medium (1). Six cottonseeds from each treatment at each sampling time were plated. After 24 h at 24°C, plates were examined and the percentage of cottonseeds colonized was recorded.
Transplant experiments.
To determine when suppressiveness was expressed in plant bioassays, cottonseeds were removed from cylinders at 0, 2, 4, 6, and 8 h after sowing. Seeds were rinsed with sterile deionized water to remove any adhering compost or sand and then placed directly in contact with an agar disk with P. ultimum sporangia in a new glass cylinder containing 5 cm3 of sterile sand. Bioassay assemblies were incubated and rated as described above.
Microbial isolations.
Microbial consortia were isolated from cottonseeds by removing 10 cottonseeds from each of the two compost treatments and from the sterile sand treatment 4 h after sowing. Seeds were placed in 15 ml of sodium pyrophosphate buffer (NaPP), vortexed for 2 to 3 min, and then removed. The remaining solution was centrifuged for 10 min at 10,000 × g to pellet microorganisms dislodged from the seed surface. The recovered microbial cells were suspended in 5 ml of NaPP for use in bioassays or fatty acid metabolism experiments.
Seed-colonizing microorganisms were isolated and enumerated by plating seed washings on 0.1% Trypticase soy broth agar (1.5% agar) (TSBA) at 0, 1, 2, 4, 6, and 8 h after sowing cottonseeds in either suppressive or conducive compost. At each sampling, cottonseeds were removed and rinsed gently with sterile deionized water. One seed was placed in 1 ml of 0.1% NaPP (pH 7.0). There were three replicate cottonseeds per treatment at each sampling time. Seeds were vortexed for 2 to 3 min, and serial dilutions were prepared from this solution. Dilutions were plated and incubated at 25°C. CFU were enumerated 48 h later. Fungi were not detected from any of the seed washings on TSBA or a number of other culture media tested in preliminary experiments.
Populations of fatty-acid-metabolizing microorganisms were enumerated on a canola oil agar medium (COA) that was made by autoclaving Bushnell-Haas broth (Difco Laboratories) amended with 1.5% agar and then cooling the medium to 55°C. Canola oil was used as a rich source of unsaturated fatty acids on which to culture fatty-acid-metabolizing microorganisms. Filter-sterilized canola oil was added to the cooled medium at a concentration of 0.5% (vol/vol) and emulsified with a sterile Waring blender. After plating, COA plates were incubated at 25°C and CFU were enumerated 48 h later. Bacterial colonies were picked from COA plates and streaked on TSBA to obtain single-cell cultures. Isolates were collected from six different cottonseeds germinated in suppressive compost and eight different cottonseeds germinated in conducive compost. Bacterial colonies isolated from culture plates were stored in 0.1% TSB (Difco Laboratories) containing 15% (vol/vol) glycerol at −80°C. Working cultures were maintained on TSBA slants stored at 4°C.
Disease-suppressive properties of seed-colonizing microorganisms.
Microbial consortia and individual bacterial strains isolated from seed surfaces were tested for their ability to induce suppressiveness to Pythium damping-off of cotton. Cells of the microbial consortia or selected bacterial strains were suspended in 5 ml of NaPP on which five surface-disinfested cottonseeds were placed for 2 to 3 min. Each seed was then placed adjacent to an agar disk with P. ultimum sporangia in cylinders each containing 5 cm3 of sterile sand. Prior to filling of each cylinder with 3 cm3 of sterile sand, 1 ml of the consortium or bacterial suspension was placed on top of each seed. Finally, 2 ml of sterile deionized water was added to each cylinder, and the entire bioassay assembly was incubated and rated as described above.
Fatty acid metabolism among seed-colonizing microorganisms.
Microbial consortia and individual bacterial strains were evaluated as described previously (36, 37) for their ability to metabolize linoleic acid. Sporangium germination was used as an effective biosensor to assess linoleic acid metabolism. This was based on previous studies in which direct relationships were found between linoleic acid concentration and sporangium germination at concentrations of ≤0.2 mg/ml (36). To determine whether sporangium germination inhibitors were produced by microbial consortia, the method of van Dijk and Nelson (37) was followed.
Antibiosis screening.
All bacterial isolates collected from cottonseeds were initially screened for in vitro antibiosis toward P. ultimum on 0.1% TSBA, COA, and seed exudate agar, which consisted of 3% water agar supplemented with cottonseed (Gossypium hirsutum cv. Deltapine 50) exudate prepared and collected as described previously (37). Bacterial cells collected from 24-h cultures grown in 0.1% TSB were washed and resuspended in NaPP. Ten microliters of each bacterial suspension was spotted along the periphery of a plate containing the respective culture medium. Pseudomonas fluorescens strain Pf-5, which is known to produce antibiotics inhibitory to P. ultimum (19), was used as a positive control in this study. A total of five bacterial isolates along with the positive control were inoculated onto each plate. A 3-mm-diameter mycelial disk from a 4-day-old P. ultimum culture was placed in the center of each plate. The plates were incubated at 25°C and examined after 48 h for zones of inhibition.
Classification of bacterial isolates.
Selected bacterial isolates were identified by gas chromatographic-fatty acid methyl ester analysis. Fatty acid methyl esters were prepared and extracted from each isolate according to the protocol provided by the Bacterial Strain-Identification and Mutant Analysis Service, Auburn University, Auburn, Ala. These extracts were then shipped to Auburn University for the gas chromatography. Methyl ester extracts of isolates were identified on the basis of similarity of phospholipid fatty acid profiles with the Sherlock system (MIDI, Inc., Newark, Del.). Isolates with a similarity index of ≥0.2 were assigned to a species. Isolates with a similarity index of <0.2 were assigned to a genus only. Isolates that had multiple matches that differed by <0.1 were assigned to the most appropriate group of taxonomically related bacteria.
Statistical analysis.
Damping-off bioassays were established as a completely randomized design. Bioassays were repeated three times. Seedling stand data were transformed to arcsines of square roots. Both transformed seedling stands and seedling quality were statistically analyzed by analysis of variance. Means were separated by the least significant difference (LSD) test. Percentage data from sporangium germination and seed colonization experiments were transformed (arcsines of square roots) prior to analysis of variance. Means were separated by the LSD test. CFU per seed of populations of either bacteria or actinobacteria at each time point were transformed (log) and then analyzed by t tests. All experiments were performed at least three times with similar results.
RESULTS
Suppression of P. ultimum sporangium germination, seed colonization, and damping-off.
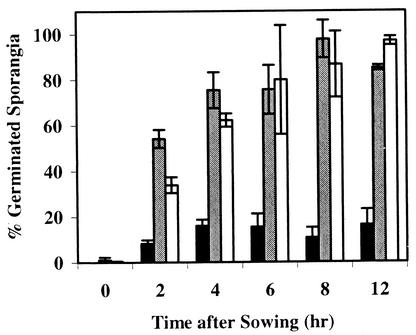
Compared to germination in sterile sand, the germination of P. ultimum sporangia was significantly reduced in response to cottonseeds germinating in LC99 but not in response to cottonseeds germinating in LC89 (Fig. 1). The significant (P = 0.05) reduction in germination of sporangia in LC99 was detected as early as 1 h after sowing cottonseeds. The maximum percentage of germinated sporangia (70 to 100%) in the LC89 and sterile sand was observed between 4 and 12 h after sowing.
FIG. 1.
Germination of P. ultimum sporangia in response to cottonseeds sown in LC99, LC89, and sterile sand during the first 12 h of seed germination. Bars represent the means of two experiments. Error bars represent standard deviations. Black bars, LC99 (suppressive compost); gray bars, LC89 (conducive compost); open bars, sterile sand.
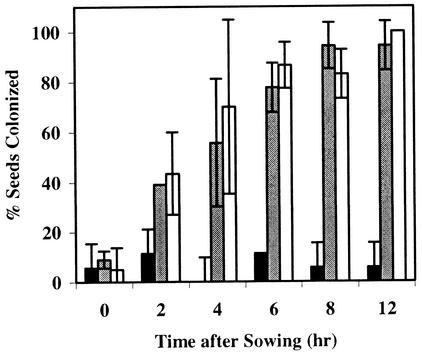
The percentage of cottonseeds colonized by P. ultimum was significantly (P = 0.05) reduced (compared to sterile sand) in LC99 but not LC89 as early as 4 h after sowing (Fig. 2). No seed colonization by P. ultimum was detected at 16 and 24 h after sowing in LC99. The percentage of cottonseeds colonized in LC89 and sterile sand reached a maximum level by 6 h after sowing and remained high for a 24-h period. LC99 was consistently suppressive of Pythium damping-off whereas LC89 was consistently conducive (Table 2).
FIG. 2.
Percentages of cottonseeds colonized by P. ultimum in LC99, LC89, and sterile sand during the first 12 h of seed germination. Bars represent the means of three experiments. Error bars represent standard deviations. Black bars, LC99 (suppressive compost); gray bars, LC89 (conducive compost); open bars, sterile sand.
TABLE 2.
Suppression of P. ultimum damping-off of cotton in leaf compost-sand mixturesc
| Compost | Seedling standa (%) | Seedling qualityb |
|---|---|---|
| Leaf compost (LC89) | 8.5 c | 0.4 b |
| Leaf compost (LC99) | 83.0 a | 2.2 a |
| Inoculated sand control | 16.5 bc | 0.3 b |
| Noninoculated sand control | 83.0 a | 2.2 a |
Cotton seedling stands were calculated 7 days after sowing as a percentage of the number of healthy seedlings (those with a rating of 2 or 3) out of the total number of seeds planted.
Seedling quality was determined 7 days after sowing. See Materials and Methods for details of the rating system.
Means in each column represent results of three separate experiments. Numbers in each column followed by the same letter are not significantly (P = 0.05) different according to the LSD test.
Transplant experiments.
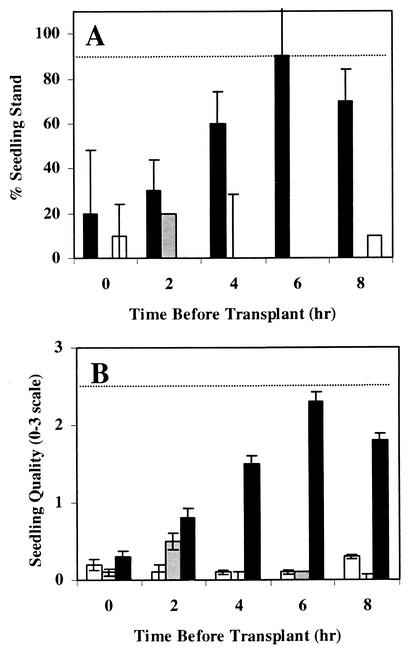
High levels of disease suppression, as indicated by high seedling stands and low disease ratings, were observed when cottonseeds were germinated for 4 to 8 h in LC99 before being transplanted to sand and challenged with P. ultimum (Fig. 3). Little or no suppression was observed with cottonseeds transplanted from LC89 or from sand. Seedling stands and mean disease ratings for cottonseeds germinated in LC99 for 4, 6, and 8 h but not for those germinated in LC89 or sterile sand for the same time period were significantly (P < 0.05) different from those in sterile sand. Seedling stands from cottonseeds transplanted out of LC99 did not differ significantly from seedling stands in noninoculated controls.
FIG. 3.
Influence of germination time in LC99 or LC89 on the damping-off severity of cotton seedlings transplanted into sterile sand and challenged with P. ultimum. (A) Cotton seedling stands were calculated 7 days after sowing as a percentage of the number of healthy seedlings (those with a rating of 2 or 3) out of the total number of cottonseeds transplanted. (B) Mean seedling quality ratings were determined 7 days after transplanting. Ratings were on a scale of 0 to 3 (see Materials and Methods for details of rating system). Means at each time point represent the results of two experiments. Error bars represent standard deviations. Dashed lines represent seedling stands (A) or seedling quality (B) in the noninoculated control. Black bars, LC99 (suppressive compost); gray bars, LC89 (conducive compost); open bars, sterile sand.
Populations of bacteria associated with cottonseeds germinated in composts.
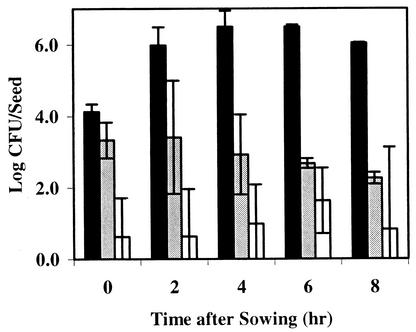
Populations of seed-colonizing microorganisms were enumerated from cottonseeds at various times after sowing by plating seed washings on TSBA (Fig. 4). In preliminary experiments on a variety of culture media, no fungi were detected in our seed washings. In our TSBA plating, only bacterial colonies developed on plates after 48 h of incubation. No significant increases in seed surface bacterial populations were observed on cottonseeds germinated for up to 8 h in conducive compost or sterile sand over populations observed at the time of sowing. Maximum log bacterial populations from cottonseeds germinated in LC89 and sterile sand ranged from 2.26 to 3.33 and 0.63 to 1.63, respectively, over the 8-h sampling period. In contrast, significant increases in bacterial populations were observed on cottonseeds germinated in LC99 over the 8-h sampling period. Log bacterial populations from cottonseeds germinated in LC99 increased rapidly from low levels (4.2) at the time of sowing to 6.50 4 to 6 h later. Population levels between 2 h and at 8 h after sowing in LC99 did not change significantly.
FIG. 4.
Populations of culturable bacteria (on TSBA) from the surface of cottonseeds germinated in suppressive compost (LC99, black bars), conducive compost (LC89, gray bars), or sterile sand (open bars). Bars represent the means of two experiments. Error bars represent standard deviations.
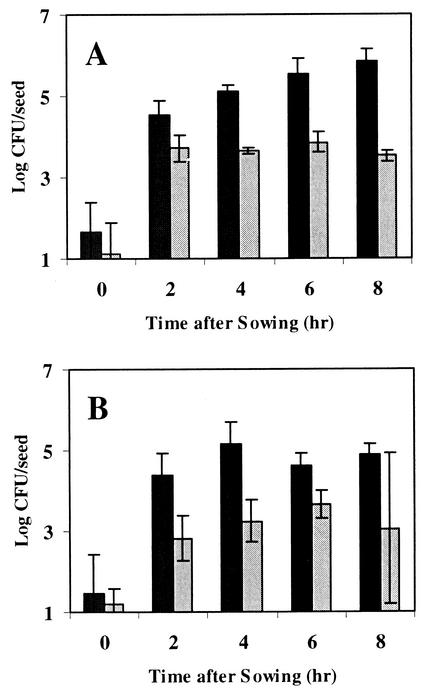
Populations of fatty-acid-metabolizing microorganisms were also enumerated in seed washings taken at various times after sowing by plating on COA (Fig. 5). Bacterial colonies as well as small, filamentous, sporulating colonies were detected on COA from cottonseeds germinated in either compost. Preliminary experiments with antifungal compounds indicated that these small colonies were most likely actinobacteria and not fungi. By 1 h and up until 8 h after sowing, log bacterial populations were significantly (P < 0.05) greater from cottonseeds sown in LC99 (4.7 to 5.6 CFU/seed) than from cottonseeds sown in LC89 (3.6 to 3.8 CFU/seed) (Fig. 5A). Between 2 and 6 h after sowing, log populations of actinobacteria were significantly greater from the surface of cottonseeds germinated in LC99 (4.2 to 4.3 CFU/seed) than from cottonseeds germinated in LC89 (2.3 to 2.5 CFU/seed) (Fig. 5B). By 8 h, however, there were no significant (P = 0.05) differences between populations of actinobacteria from cottonseeds germinated in either compost even though mean population levels were nearly an order of magnitude higher from cottonseeds germinated in LC99 than from those germinated in LC89. Log populations of both bacteria and actinobacteria detected on surface-sterilized cottonseeds alone or from cottonseeds that were germinated in sterile sand were negligible (<1.4 CFU/seed).
FIG. 5.
Populations of fatty-acid-metabolizing bacteria (A) and actinobacteria (B) on the surface of cottonseeds sown in suppressive (black bars) or conducive (gray bars) compost for 8 h. Each was enumerated on Bushnell-Haas medium supplemented with 0.05% canola oil. Bars represent the means of two experiments. Error bars represent standard deviations.
Suppression of damping-off by seed surface microbial consortia.
Microorganisms recovered from the surface of cottonseeds germinated in either LC99 or LC89 for 4 h were tested for their ability to protect cottonseeds from damping-off (Table 3). Seeds treated with microorganisms collected from cottonseeds germinated in LC99 (suppressive microbial consortium) gave rise to significantly (P < 0.05) higher seedling stands and seedling quality than what was obtained in the inoculated control. Seedling quality and percent seedling stands, however, were not improved significantly over the inoculated control when treated with microorganisms from cottonseeds germinated in LC89 (conducive microbial consortium). Seeds from which microbial consortia were removed and cottonseeds treated with the filtered seed washings, regardless of the compost in which they originally germinated, were highly susceptible to damping-off, and no seedlings emerged.
TABLE 3.
Suppression of P. ultimum damping-off of cotton by seed-colonizing microbial consortia from cottonseeds germinated in either LC99 (suppressive) or LC89 (conducive) composta
| Treatmentb | Seedling standc (%) | Seedling qualityd |
|---|---|---|
| Suppressive consortium | 60 b | 1.5 b |
| Conducive consortium | 0 c | 0.2 c |
| Seeds with LC99 consortium removed | 0 c | 0.0 c |
| Seeds with LC89 consortium removed | 0 c | 0.0 c |
| Noninoculated sand control | 85 a | 2.7 a |
| Inoculated sand control | 0 c | 0.2 c |
Means in each column represent results of two separate experiments. Numbers in each column followed by the same letter are not significantly (P = 0.05) different according to the LSD test.
Suppressive consortium refers to the collective group of microorganisms recovered from the surface of cottonseeds germinated in LC99 for 4 h. Conducive consortium refers to the collective group of microorganisms recovered from the surface of cottonseeds germinated in conducive compost for 4 h.
Cotton seedling stands were calculated 7 days after sowing as a percentage of the number of healthy seedlings (those with a rating of 2 or 3) out of the total number of seeds planted.
Seedling quality was determined 7 days after sowing. See Materials and Methods for details of the rating system.
Suppression of damping-off by selected individual seed surface bacterial isolates.
When tested individually, bacterial isolates varied in their efficacy in suppressing Pythium damping-off, regardless of their source (Table 4). Selected strains from cottonseeds germinated in LC99 were generally ineffective in suppressing Pythium damping-off. Seedling stands from cottonseeds treated with various bacterial strains ranged from 0.0 to 23.3%. Only two strains significantly improved seedling stands. Combining all selected strains from cottonseeds germinated in LC99 improved seedling stands over those of the best individual strains alone, but seedling stands were still significantly lower than those in the noninoculated control. Seedling quality was poor, ranging from 0.0 to 1.0.
TABLE 4.
Fatty acid metabolism and biological control of Pythium damping-off among selected bacterial isolates from cottonseeds germinated in suppressive or conducive composta
| Isolate no. | Identificationb | Sporangium germination (%)c | Seedling stands (%)d | Seedling quality (0-3 scale)e |
|---|---|---|---|---|
| LC99 | ||||
| 1-16S | Xanthobacter agilis | 1.5 f | 0.0 e | 0.0 b |
| 1-52S | Enteric bacterium | 2.1 ef | 20.0 c | 0.6 ab |
| 1-36S | Enteric bacterium | 4.1 ef | 0.0 e | 0.6 ab |
| 1-7S | Microbacterium sp. | 8.2 e | 3.3 de | 0.2 ab |
| 1-43S | Paracoccus denitrificans | 10.4 e | 13.3 cd | 0.6 ab |
| 3-15S | Coryneform bacterium | 28.8 d | 0.0 e | 0.0 b |
| 3-44S | Coryneform bacterium | 46.2 c | 0.0 e | 0.0 b |
| 6-4S | Coryneform bacterium | 58.0 bc | 0.0 e | 0.0 b |
| 4-5S | Coryneform bacterium | 59.0 bc | 23.3 c | 0.4 ab |
| 7-7S | Coryneform bacterium | 86.9 a | 13.3 cd | 0.2 ab |
| Combinedf | 40.0 b | 1.0 ab | ||
| LC89 | ||||
| 6-10C | Curtobacterium flaccum- faciens | 6.0 e | 53.3 b | 2.0 a |
| 6-7C | Coryneform bacterium | 6.1 e | 43.3 bc | 1.0 ab |
| 6-5C | Micrococcus luteus | 24.8 d | 6.7 d | 0.0 b |
| 8-8C | Agrobacterium radiobacter | 29.8 d | 0.0 e | 0.0 b |
| 4-2C | Agrobacterium radiobacter | 62.0 b | 6.7 d | 0.0 b |
| 7-13C | No match | 65.4 b | 6.7 d | 0.0 b |
| 4-1C | Paenibacillus macerans | 81.1 a | 36.7 bc | 1.0 ab |
| 5-2C | Agrobacterium radiobacter | 83.9 a | 36.7 bc | 1.0 ab |
| Combinedf | 20.0 c | 0.6 ab | ||
| Noninoculated | 80.0 a | 1.9 a | ||
| No bacteria | 89.0 a | 0.0 e | 0.0 b |
Numbers in each column followed by the same letter are not significantly (P = 0.05) different according to the LSD test.
See Materials and Methods for details of identification.
Linoleic acid (0.35 mg/ml) was incubated with bacterial isolates (5.6 × 106 CFU/ml) for 16 h. Sporangium germination was used to assess linoleic acid metabolism based on previous studies (36).
Seedling stands were determined 7 days after inoculation (25°C).
See Materials and Methods for seedling quality rating details.
Represents an equal mixture of all the individual suppressive or conducive strains.
Selected strains from cottonseeds germinated in LC89 generally were more effective in suppressing Pythium damping-off than were those from cottonseeds germinated in LC99. Seedling stands arising from cottonseeds treated with various bacterial strains from LC89 ranged from 0.0 to 53.3%. Four strains significantly improved seedling stands over the inoculated no-bacteria control. Effective strains did not give rise to stands equivalent to those from noninoculated cottonseeds. Combining all strains from cottonseeds germinated in LC89 induced low but significant levels of damping-off suppression (20% seedling stands). As with the strains from LC89, seedling quality was poor among most strains. Only Curtobacterium flaccumfaciens strain 6-10C gave rise to seedlings with high quality ratings (mean rating, 2.0).
Fatty acid metabolism by seed surface microbial consortia.
In 16 h, suppressive microbial consortia (from LC99) significantly (P = 0.05) reduced the stimulatory activity of linoleic acid to levels capable of inducing <6% germination of P. ultimum sporangia (Table 5). Even when linoleic acid was incubated with a 1:100 dilution of the suspension containing suppressive consortia, the stimulatory activity of linoleic acid was still reduced, resulting in the induction of 30% sporangium germination. Conversely, conducive microbial consortia (from LC89) did not significantly (P = 0.05) reduce the stimulatory activity of linoleic acid from that induced by the nontreated linoleic acid control. Linoleic acid previously exposed to suppressive consortia and added to nontreated linoleic acid solution at equal ratios also induced levels of sporangium germination (91.4%) that were not significantly (P = 0.05) different from those for the nontreated linoleic acid control (83.6%).
TABLE 5.
Linoleic acid metabolism by microbial consortia recovered from the surface of cottonseeds germinated in suppressive or conducive composta
| Treatmentb | Sporangium germinationc (%) |
|---|---|
| Nontreated | 83.6 a |
| Conducive consortium | 80.2 a |
| Suppressive consortium | 5.9 c |
| Suppressive consortium diluted 100×d | 30.2 b |
| Suppressive consortium + linoleic acide | 91.4 a |
Fifty microliters of a 0.4-mg/ml linoleic acid solution was added to 50 μl of a microbial suspension, giving a final linoleic acid concentration of 0.2 mg/ml. Numbers followed by the same letter are not significantly (P = 0.05) different according to the LSD test.
Conducive consortium refers to the collective group of microorganisms recovered from the surface of cottonseeds germinated in conducive compost for 4 h. Suppressive consortium refers to the collective group of microorganisms recovered from the surface of cottonseeds germinated in suppressive compost for 4 h.
Sporangium germination was used to assess linoleic acid metabolism based on previous studies (36).
100× dilution means that the suppressive consortium suspension was diluted in 0.1% NaPP prior to incubation with linoleic acid.
Fifty microliters of a nonincubated 0.4-mg/ml solution of linoleic acid was added to 50 μl of the linoleic acid solution on which the suppressive consortium was incubated (0.2-mg/ml final concentration).
Fatty acid metabolic activity of selected individual seed surface bacteria.
Each of the bacterial isolates collected from cottonseeds germinated for 4 h in LC99 was capable of significantly (P = 0.05) inactivating the stimulatory activity of linoleic acid and thereby reducing sporangium germination compared to that for a nonincubated linoleic acid control. However, only 60% of the bacterial isolates collected from cottonseeds germinated in LC89 were capable of metabolizing linoleic acid. Among those, linoleic acid metabolism varied considerably. Induction of sporangium germination after 16 h of incubation ranged from 0 to 76% (data not shown).
To test if this variability was due to initial differences in cell densities between isolates, 18 bacterial isolates were selected and screened again with cell densities adjusted to 5.6 × 105 cells/ml. Results of these experiments are given in Table 4. Even when cell densities were standardized, bacteria varied in their ability to metabolize linoleic acid. The stimulatory activity of linoleic acid following treatment with bacterial isolates from cottonseeds germinated in LC99 induced germination of sporangia ranging from 1.5 to 86.9%. The stimulatory activity of linoleic acid following treatment with bacterial isolates from cottonseeds germinated in LC89 induced germination of sporangia ranging from 6.0 to 83%.
Given the small sample size, there was no apparent relationship between the bacterial group to which the isolates belong and their ability to reduce the stimulatory activity of linoleic acid. The reduction in the stimulatory activity of linoleic acid by isolates identified as coryneform bacteria was highly variable, with induced sporangium germination ranging from 6.1 to 86.9%. Even linoleic acid treated with isolates identified as Agrobacterium radiobacter induced a wide range of sporangium germination (29.8 to 81.1%). The only linoleic acid treatments that induced consistent levels of sporangium germination (<5%) were those treated with isolates identified as enteric bacteria.
Antibiosis toward P. ultimum by bacteria isolated from cottonseeds germinated in suppressive or conducive compost.
None of the bacterial isolates from cottonseeds, regardless of the compost from which they originated, inhibited P. ultimum mycelial growth on any of the three media tested. Pseudomonas fluorescens strain Pf-5, which was used as a positive control, inhibited P. ultimum mycelial growth only on 0.1% TSBA. No zones of inhibition were detected around this isolate on COA or seed exudate agar.
DISCUSSION
Results from this study indicated that communities of fatty-acid-metabolizing bacteria that colonize cottonseeds during the first few hours of seed germination played a major role in the suppression of P. ultimum sporangium germination and seed colonization, which ultimately resulted in the suppression of damping-off in a Pythium-suppressive compost. Our findings are significant in that they show that (i) suppression was expressed rapidly within the first 8 h after sowing cottonseeds and not days or weeks after sowing and that (ii) suppression was expressed on the seed surface and not necessarily in the bulk compost.
This early expression of suppressiveness is especially important for Pythium diseases, where responses of Pythium propagules to the plant and subsequent disease development are extremely rapid (23). Sporangia of P. ultimum germinate in the spermosphere within 1.5 h, with maximum germination occurring 3 to 4 h after exposure to exudates from germinating seeds (20, 26, 27, 34, 35, 37). Subsequent germ tube growth may exceed 300 μm/h (34). Seeds may be colonized by Pythium spp. as early as 2 to 4 h after planting, with maximum levels of colonization occurring within 12 to 24 h of planting (14, 24, 25, 28-30, 34). The critical importance of this stage of pathogenesis is evident from the observations that, if rapid sporangium germination and early seed colonization are prevented, seeds do not become infected and disease does not develop (36, 37). This further supports our observations that disease suppression must occur quite rapidly and that it will likely be expressed as a conspicuous reduction of sporangium germination and seed colonization.
Our results demonstrated that as early as 1 h after sowing there were significant reductions in sporangium germination in response to cottonseeds germinating in suppressive but not in conducive compost. This reduction was evident up until 12 h after sowing. In contrast, maximum germination of sporangia (∼80%) was achieved in conducive compost and sterile sand by 4 h after sowing. From these observations we reasoned that, if suppression of sporangium germination is due to the activity of compost-inhabiting microorganisms, they must actively suppress sporangium germination in the spermosphere by 1 h after sowing, with the highest level of activity reached by 4 to 6 h after sowing.
Data from transplant experiments supported this theory. Seeds exposed to suppressive compost for 4 to 8 h were protected from damping-off when transplanted into sand and challenged with P. ultimum whereas cottonseeds germinated in conducive compost for up to 8 h were not protected when challenged. Furthermore, cottonseeds were protected from seed rot and damping-off when treated with microbial consortia removed from the surface of cottonseeds germinated in suppressive compost for as little as 4 h, whereas those treated with microbial consortia recovered from cottonseeds germinated in conducive compost were not. These observations, along with our findings that cottonseeds were not protected if they (i) were treated with microbe-free seed washings or (ii) were freed of their surface-colonizing bacteria, all indicate that compost-inhabiting organisms that colonize seed surfaces rapidly within the first few hours after a seed is planted are involved in suppressiveness.
Studies with known microbial antagonists of P. ultimum have shown that most are effective in suppressing damping-off if they are present and metabolically active on the seed surface during the very early stages of seed germination and if they are able to colonize and proliferate rapidly in the spermosphere (10, 11). Based on these observations, we hypothesized that microorganisms capable of reducing sporangium germination were contributing to damping-off suppression in LC99. From studies of the biological control of P. ultimum, we reasoned that the reduction in sporangium germination observed in Pythium-suppressive compost could result from the metabolism of fatty acid stimulants of sporangium germination (36) or the production of sporangium germination inhibitors such as antibiotics (23). Although parasitism of germinating sporangia by seed-colonizing bacteria might be possible, no evidence for parasitism was found upon microscopic observations of nongerminated sporangia recovered from suppressive or conducive composts.
Previously, no water-soluble or volatile inhibitors of sporangium germination could be detected in Pythium-suppressive soil (18) or compost (22). Our results are consistent with these findings. No inhibitors of sporangium germination were detected by bacteria or actinobacteria isolated from the surface of cottonseeds germinated in Pythium-suppressive compost. Furthermore, no inhibitors of mycelial growth were detected in vitro by bacteria isolated from cottonseeds, regardless of whether they originated from suppressive or from conducive compost.
Other factors that may also preclude antibiosis from being a viable mechanism of damping-off suppression at this stage of the preinfection process are (i) the speed at which the germination of P. ultimum sporangia occurs and (ii) the presence or absence of specific precursor molecules required for antibiotic biosynthesis. For antibiotic production to be an effective mechanism contributing to the suppression of sporangium germination within the spermosphere, it must occur within 1.5 to 2 h after sowing and coincide with the germination of sporangia. Since secondary metabolites such as antibiotics are typically produced during the later growth stages of most microorganisms, they may be produced too late to inhibit sporangium germination. Furthermore, the production of antibiotics has been demonstrated elsewhere to be affected by the presence of sugars (13, 19, 32), and therefore concentrations of antibiotics in situ may vary based on the sugar composition of seed exudate present. In this study, even Pseudomonas fluorescens strain Pf-5, which is known to produce pyoluteorin and inhibit mycelial growth of P. ultimum (17), failed to do so on media containing cottonseed exudate as the sole carbon and energy source. This indicates that important molecules required for biosynthesis of this antibiotic may not be present in the spermosphere by 4 h after sowing.
Few studies have been conducted on the microbial ecology of the spermosphere. However, we do know that this habitat offers a rich source of nutrients in an otherwise nutrient-poor environment, allowing microbial populations to flourish and be maintained over time around the seed (7). Because of the contrast between carbon availability before and after seed imbibition and release of exudates, competitive interactions among microorganisms in the spermosphere may develop. However, for competitive interactions to play a role in disease suppression a limiting resource must be shared between the pathogen and microbial antagonist. Such a competition has been demonstrated previously between P. ultimum and the biological control agent Enterobacter cloacae in the spermosphere. Long-chain unsaturated fatty acid components of cottonseed exudate that are required to elicit germination of P. ultimum sporangia are rapidly metabolized by E. cloacae, thereby reducing seed infection and disease incidence (36).
Based on the competitive interaction between E. cloacae and P. ultimum, we focused part of our investigation on the role of seed-colonizing fatty-acid-metabolizing bacteria from suppressive compost in disease suppression. By 4 h after sowing, we detected higher populations of fatty-acid-metabolizing bacteria on cottonseeds from suppressive compost than on cottonseeds from conducive compost or sterile sand. Furthermore, linoleic acid metabolism was also much greater by microbial consortia associated with cottonseeds in suppressive compost than by consortia on cottonseeds from conducive compost or sterile sand. It is not clear from our studies if this difference in linoleic acid metabolism is related to population size or to increased metabolic activity of seed-colonizing fatty-acid-metabolizing bacteria. However, the significant increase in populations of seed-colonizing fatty-acid-metabolizing bacteria observed by 4 h after sowing directly corresponds with the time point at which we observed the greatest levels of disease suppression and reductions in sporangium germination in Pythium-suppressive compost.
Although only 10 bacterial strains were identified from those isolated from cottonseeds in suppressive compost, many of these were classified as coryneform bacteria. This was not surprising, however, because bacterial species within this group, including Corynebacterium spp., Cellulomonas spp., Micrococcus spp., and Microbacterium spp., have been recovered from the rhizosphere (4, 5) and spermosphere (8) of various plants as well as from compost (12). Furthermore, species within these genera have been shown previously to suppress root rot caused by P. ultimum (4) and to metabolize fatty acids (6, 37).
In previous studies of Pythium-suppressive soils and composts, individual antagonists recovered from the soil or compost failed to induce disease suppression at levels observed in the original suppressive soil (21) or compost (4). Our observations of the fatty acid metabolic activity of the suppressive microbial consortium versus that of individual bacterial isolates corroborate these findings. Linoleic acid metabolism among individual bacterial isolates varied considerably, whereas that of the suppressive microbial consortium was high. Furthermore, individual bacterial strains from either suppressive or conducive consortia failed to induce high levels of damping-off suppression, supporting the conclusion that the suppressive consortium functions as a community and is necessary for disease suppression to be expressed. Work is currently under way to characterize these suppressive bacterial communities.
In this study we have presented evidence for the role of seed-colonizing fatty-acid-metabolizing bacteria in cotton damping-off suppression in Pythium-suppressive compost. Based on our knowledge of key characteristics of Pythium damping-off, along with the known mechanisms of microbial antagonism toward Pythium species, we were better able to direct our search for disease-suppressive microbial communities. Although the applicability of this approach to soils suppressive to other pathosystems is not currently known, it may be limited only by our knowledge of (i) temporal and biochemical mechanisms of pathogenesis, (ii) pathogen responses to a host, and (iii) mechanisms of microbial antagonism toward the pathogen under study.
REFERENCES
- 1.Ali-Shtayeh, M. S., L. H. C. Len, and M. W. Dick. 1986. The phenology of Pythium in soil. J. Ecol. 74:823-840. [Google Scholar]
- 2.Ben-Yephet, Y., and E. B. Nelson. 1999. Differential suppression of cucumber damping-off caused by Pythium aphanidermatum, P. irregulare, and P. myriotylum in composts at different temperatures. Plant Dis. 83:356-360. [DOI] [PubMed] [Google Scholar]
- 3.Boehm, M. J., and H. A. J. Hoitink. 1992. Sustenance of microbial activity in potting mixes and its impact on severity of Pythium root rot of poinsettia. Phytopathology 82:259-264. [Google Scholar]
- 4.Boehm, M. J., L. V. Madden, and H. A. J. Hoitink. 1993. Effect of organic matter decomposition level on bacterial species diversity and composition in relation to pythium damping-off severity. Appl. Environ. Microbiol. 59:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broadway, N. M., F. M. Dickinson, and C. Ratledge. 1993. The enzymology of dicarboxylic acid formation by Corynebacterium sp. strain 7E1C grown on n-alkanes. J. Gen. Microbiol. 130:1337-1344. [Google Scholar]
- 6.Broadway, N. M., F. M. Dickinson, and C. Ratledge. 1992. Long-chain acyl-CoA ester intermediates of β-oxidation of mono- and di-carboxylic fatty acids by extracts of Corynebacterium sp. strain 7E1C. J. Biochem. 285:117-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buyer, J. S., D. P. Roberts, and E. Russek-Cohen. 1999. Microbial community structure and function in the spermosphere as affected by soil and seed type. Can. J. Microbiol. 45:138-144. [Google Scholar]
- 8.Cottyn, B., E. Regalado, B. Lanoot, M. DeCleene, T. W. Mew, and J. Swings. 2001. Bacterial populations associated with rice seed in the tropical environment. Phytopathology 91:282-292. [DOI] [PubMed] [Google Scholar]
- 9.Craft, C. M., and E. B. Nelson. 1996. Microbial properties of composts that suppress damping-off and root rot of creeping bentgrass caused by Pythium graminicola. Appl. Environ. Microbiol. 62:1550-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukui, R., M. N. Schroth, M. Hendson, and J. G. Hancock. 1994. Interaction between strains of pseudomonads in sugar beet spermospheres and their relationship to pericarp colonization by Pythium ultimum in soil. Phytopathology 84:1322-1330. [Google Scholar]
- 11.Fukui, R., M. N. Schroth, M. Hendson, J. G. Hancock, and M. K. Firestone. 1994. Growth patterns and metabolic activity of pseudomonads in sugar beet spermospheres: relationship to pericarp colonization by Pythium ultimum. Phytopathology 84:1331-1338. [Google Scholar]
- 12.Goodfellow, M., M. Mordarski, and S. T. Williams (ed.). 1984. The biology of the actinomycetes. Academic Press, London, United Kingdom.
- 13.Gutterson, N., J. S. Ziegle, G. J. Warren, and T. J. Layton. 1988. Genetic determinants for catabolite induction of antibiotic biosynthesis in Pseudomonas fluorescens HV37a. J. Bacteriol. 170:380-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadar, Y., G. E. Harman, A. G. Taylor, and J. M. Norton. 1983. Effects of pregermination of pea and cucumber seeds and of seed treatment with Enterobacter cloacae on rots caused by Pythium spp. Phytopathology 73:1322-1325. [Google Scholar]
- 15.Hoitink, H. A. J., and M. J. Boehm. 1999. Biocontrol within the context of soil microbial communities: a substrate-dependent phenomenon. Annu. Rev. Phytopathol. 37:427-446. [DOI] [PubMed] [Google Scholar]
- 16.Hornby, D. 1983. Suppressive soils. Annu. Rev. Phytopathol. 21:65-85. [Google Scholar]
- 17.Howell, C. R., and R. D. Stipanovic. 1980. Suppression of Pythium ultimum-induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic, pyoluteorin. Phytopathology 70:712-715. [Google Scholar]
- 18.Kao, C. W., and W. H. Ko. 1983. Nature of suppression of Pythium splendens in a pasture soil in South Kohala, Hawaii. Phytopathology 73:1284-1289. [Google Scholar]
- 19.Kraus, J., and J. E. Loper. 1995. Characterization of a genomic region required for production of the antibiotic pyoluteorin by the biological control agent Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 61:849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lifshitz, R., M. T. Windham, and R. Baker. 1986. Mechanism of biological control of preemergence damping-off of pea by seed treatment with Trichoderma spp. Phytopathology 76:720-725. [Google Scholar]
- 21.Lumsden, R. D. 1987. Suppression of damping-off caused by Pythium spp. in soil from the indigenous Mexican chinampa agricultural system. Soil Biol. Biochem. 19:501-508. [Google Scholar]
- 22.Mandelbaum, R., and Y. Hadar. 1990. Effects of available carbon source on microbial activity and suppression of Pythium aphanidermatum in compost and peat container media. Phytopathology 80:794-804. [Google Scholar]
- 23.Martin, F. N., and J. E. Loper. 1999. Soilborne plant diseases caused by Pythium spp.: ecology, epidemiology, and prospects for biological control. Crit. Rev. Plant Sci. 18:111-181. [Google Scholar]
- 24.Nelson, E. B. 1988. Biological control of Pythium seed rot and preemergence damping-off of cotton with Enterobacter cloacae and Erwinia herbicola applied as seed treatments. Plant Dis. 72:140-142. [Google Scholar]
- 25.Nelson, E. B., W. L. Chao, J. M. Norton, G. T. Nash, and G. E. Harman. 1986. Attachment of Enterobacter cloacae to hyphae of Pythium ultimum: possible role in biological control of Pythium pre-emergence damping-off. Phytopathology 76:327-335. [Google Scholar]
- 26.Nelson, E. B., and C. M. Craft. 1989. Comparative germination of culture-produced and plant-produced sporangia of Pythium ultimum in response to soluble seed exudates and exudate components. Phytopathology 79:1009-1013. [Google Scholar]
- 27.Nelson, E. B., and J. S. T. Hsu. 1994. Nutritional factors affecting responses of sporangia of Pythium ultimum to germination stimulants. Phytopathology 84:677-683. [Google Scholar]
- 28.Ogle, H. J., A. M. Stirling, and P. J. Dart. 1995. Some factors affecting the development and biocontrol of cotton seedling disease. Aust. J. Exp. Agric. 35:771-776. [Google Scholar]
- 29.Osburn, R. M., and M. N. Schroth. 1988. Effect of osmopriming sugar beet seed on exudation and subsequent damping-off caused by Pythium ultimum. Phytopathology 78:1246-1250. [Google Scholar]
- 30.Osburn, R. M., M. N. Schroth, J. G. Hancock, and M. Hendson. 1989. Dynamics of sugar beet seed colonization by Pythium ultimum and Pseudomonas species: effects on seed rot and damping-off. Phytopathology 79:709-716. [Google Scholar]
- 31.Ruttledge, T. R., and E. B. Nelson. 1997. Extracted fatty acids from Gossypium hirsutum stimulatory to the seed-rotting fungus, Pythium ultimum. Phytochemistry 46:77-82. [Google Scholar]
- 32.Shanahan, P., D. J. O'Sullivan, P. Simpson, J. D. Glennon, and F. O'Gara. 1992. Isolation of 2,4-diacetylphlorglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol. 58:353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanghellini, M. E. 1974. Spore germination, growth, and survival of Pythium in soil. Proc. Am. Phytopathol. Soc. 1:211-214. [Google Scholar]
- 34.Stanghellini, M. E., and J. G. Hancock. 1971. Radial extent of the bean spermosphere and its relation to the behavior of Pythium ultimum. Phytopathology 61:165-168. [Google Scholar]
- 35.Stanghellini, M. E., and J. G. Hancock. 1971. The sporangium of Pythium ultimum as a survival structure in soil. Phytopathology 61:157-164. [Google Scholar]
- 36.van Dijk, K., and E. B. Nelson. 2000. Fatty acid competition as a mechanism by which Enterobacter cloacae suppresses Pythium ultimum sporangium germination and damping-off. Appl. Environ. Microbiol. 66:5340-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Dijk, K., and E. B. Nelson. 1998. Inactivation of seed exudate stimulants of Pythium ultimum sporangium germination by biocontrol strains of Enterobacter cloacae and other seed-associated bacteria. Soil Biol. Biochem. 30:183-192. [Google Scholar]