Abstract
While plasmids from lactic acid bacteria possess many traits that are of industrial value, their exploitation is often frustrated by an inability to conduct food-grade engineering of native plasmids or to readily screen for their transfer. Here we describe a system that uses a RepA+ temperature-sensitive helper plasmid and a RepA− cloning vector to overcome these problems while maintaining the food-grade status of the native plasmid. This strategy was used to precisely delete ltnA1 alone, or in conjunction with ltnA2 (encoding the structural proteins of the lantibiotic lacticin 3147), from the native 60.2-kb plasmid pMRC01 and to select for the transfer of pMRC01 between Lactococcus lactis strains.
The gram-positive bacterium Lactococcus lactis has many plasmid-carried industrially significant traits, including lactose utilization, bacteriocin production, bacteriophage resistance, and exopolysaccharide production (7, 23). While a number of tools have been designed to generate chromosomal mutations in lactic acid bacteria (13, 19), less consideration has been given to the potentially enormous benefits of engineering plasmid-carried genes in a food-grade manner. Efficient plasmid engineering would allow the removal or replacement of undesirable genes, permit the functional analysis of a particular plasmid gene(s), or allow the introduction of point mutations to determine the importance of individual nucleotides and amino acids.
In addition to the limitations associated with the engineering of food-grade plasmids, the lack of a readily selectable marker to allow their transfer is another aspect that often restricts their use. Due to the restrictions associated with using antibiotic markers in food applications, three alternative types of selection have been used. The first of these, involving the use of dominant markers such as bacteriocin production and immunity (1, 24) or heavy metal resistance (12, 14), is relatively straightforward but depends upon the presence of these selectable markers on the plasmid of interest. The second alternative involves complementation, which for example might depend on lactose (15, 20), thymidine (5), or pyrimidine (22) auxotrophies, a strategy requiring the initial mutation of chromosomally encoded traits and the subsequent complementation of this phenotype by plasmid-carried genes. The third and most recent development has involved the utilization of two plasmid systems. The use of such a system by Emond et al. (4) involved the separation of the antibiotic marker (on a RepB− plasmid) from the RepB+ vector into which the genes of interest were cloned. Both plasmids are involved in the gene transfer step, but the marker plasmid can be subsequently removed by curing (4). A coelectroporation method has also been used for the isolation of cryptic plasmids. This method reduces the number of colonies that need to be screened on the premise that optimization of the ratio between the marker and cryptic plasmid results in an average cotransformation frequency of 20%. Once again the marker plasmid is subsequently lost by removing the selective pressure (2).
Here we describe a method that combines SOEing (splicing by overlap extension) PCR (10) and a system originally designed to generate unlabeled gene replacements in bacterial chromosomes (13) to create gene knockouts and/or replacements on native plasmids in a food-grade manner. This technique takes advantage of the temperature-sensitive RepA+ plasmid pVE6007 (16) and the RepA− vector pORI280 (13) and relies crucially on the temporary integration of pORI280 into the target plasmid (facilitated by using a pORI280 derivative containing an insert bearing homology to the target plasmid). This cointegrate is stable in the presence of an antibiotic marker, but its resolution can be readily detected by screening for the loss of β-galactosidase activity or the erythromycin (ERY) resistance (Eryr) phenotype associated with pORI280 when the selective pressure is removed.
To illustrate the utility of this strategy, we have chosen the 60.2-kb conjugative plasmid pMRC01 (3, 21), which carries the genes responsible for production of and immunity to the lantibiotic lacticin 3147 (Fig. 1A) (17, 18). To date, functional analysis of the bacteriocin gene cluster has been performed on a subclone, pOM02, containing 10 genes arranged in two divergent operons. This analysis has the limitation of assuming that no other pMRC01-carried genes are involved in bacteriocin production or immunity. In order to address this limitation, we have initiated a systematic functional analysis of the native 60.2-kb plasmid. We also wished to create an expression host that possesses all the lacticin biosynthetic and immunity machinery but lacks the structural genes, so that we can express alternate structural genes against this background; thus, our initial experiments focused on deriving a version of pMRC01 in which both structural genes, ltnA1 and ltnA2, have been precisely deleted.
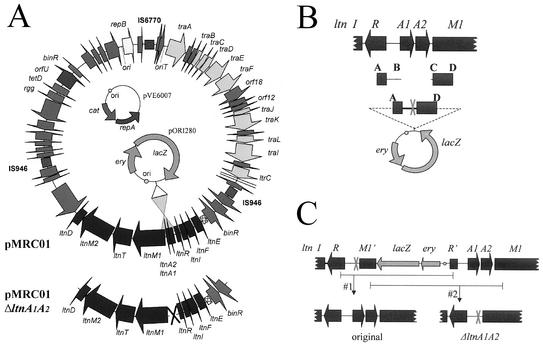
FIG. 1.
(A) Physical map of pMRC01, pORI280, and pVE6007, showing the cointegration strategy. (B) The primer pairs ltnA1A2SOEA/B and ltnA1A2SOEC/D, on either side of the ltnA1A2 genes, generated two PCR products of equal size. A subsequent PCR with these products and the SOEA/D primers generated a spliced DNA fragment, lacking ltnA1A2, which was cloned into the Eryr lac+ RepA− vector pORI280. (C) Depiction of the integration of pORI280ΔltnA1A2A-D into pMRC01. Resolution can result in either the reversion of pMRC01 to its original form (#1) or the creation of pMRC01ltnA1A2 (#2).
To generate a SOEing product to facilitate an ltnA1A2 in-frame deletion, primers ltnA1A2SOEA, -B, -C, and -D were designed (Fig. 1 has a graphic overview and Table 1 has a list of primers). Vent polymerase (New England Biolabs) was used to amplify fragments for cloning to minimize mutation due to nucleotide misincorporation, and Taq polymerase (BioTaq) was used in all other cases. Initially two fragments (SOEAB and SOECD), each 291 bp in length, on either side of the ltnA1A2 genes were amplified from pMRC01. The resulting products were gel extracted by using the Qiaex gel extraction kit (Qiagen), mixed in a 1:1 ratio, and reamplified by PCR, with the ltnA1A2SOEA and -D primers, resulting in the production of a spliced A-D product. The splicing reaction was facilitated by the presence of an 18-bp overhang on the SOEC primer that is the reverse complement of the SOEB primer. This overhang on the CD product hybridizes with the corresponding region on the AB product, allowing splicing to occur. This strategy was also used to generate an SOEing product splicing regions on either side of the ltnA1 gene for the purpose of deleting that gene in an in-frame manner. This required primers ltnA1A2SOEA, ltnA1A2SOEB, ltnA1SOEC, and ltnA1SOED.
TABLE 1.
Primers used in this study
| Name | Sequence (5′ to 3′) |
|---|---|
| ltnA1A2SOEA | AACTGCAGTTATATATTTGCGGCa |
| ltnA1A2SOEB | GATATGAACCTCCTTATT |
| ltnA1A2SOEC | AATAAGGAGGTTCATATCTTAGGAGTAAAAAATGAAb |
| ltnA1A2SOED | ACGAATTCTCTTACAGAGTTc |
| ltnA1SOEC | AATAAGGAGGTTCATATCGTTAATAACAAATTTTTAATTAAT |
| ltnA1SOED | TGTCGAATTCATTTCTGGAAAAACc |
| pORI280 For | CTCGTTCATTATAACCCTC |
| pORI280 Rev | CGCTTCCTTTCCCCCCAT |
Boldface represents changes made to incorporate a PstI site.
Underlined sequence represents an overhang that is the reverse complement of the corresponding SOEB primer.
Boldface represents changes made to incorporate an EcoRI site.
The final A-D PCR products were digested with PstI and EcoRI, sites which had been incorporated into the A and D primers, respectively, and then cloned into pORI280 (a RepA− ori+ β-galactosidase-positive Eryr integration vector) (Fig. 1B). These were transformed into Escherichia coli EC101 (RepA+) (11). Blue colonies on Luria-Bertani agar plates containing ERY (150 μg/ml) and 5-bromo-4-chloro-3-indolyl-β-d-galactosidase (X-Gal; 40 μg/ml) were selected, and the primer pair pORI280F/R, situated on either side of the multiple cloning site, was used to confirm the correct insert. pORI280(A-D) was electroporated into competent L. lactis MG1363 cells (6, 9) containing both pMRC01 and the temperature-sensitive Cmr RepA+ helper plasmid pVE6007.
Integration of pORI280 into pMRC01 by single crossover (Fig. 1C) was selected for by either of two methods. The first method, used to generate pMRC01ΔltnA1A2, took advantage of the self-transmissible nature of pMRC01 (21). A conjugation was performed with MG1363 containing all three plasmids (pVE6007, pORI280ΔltnA1A2SOEA-D, and pMRC01) as donor and Strr L. lactis MG1614 (6) as recipient. Both strains were grown to mid-log phase (optical density at 600 nm of 0.5 to 1), the cells were harvested (1 ml), and pellets were washed once in GM17. Each pellet was resuspended in 25 μl of GM17, mixed, and spotted on a nonselective GM17 agar plate. Controls were prepared by spotting 25 μl of unmixed donor and recipient in the same manner. After overnight incubation at 30°C, the cells were cut from the plate and resuspended in GM17 broth, diluted, and plated on a selective agar of GM17 containing streptomycin (500 μg/ml), ERY (5 μg/ml), and X-Gal (160 μg/ml). This selects for recipients which have presumably received a cointegrate of both pMRC01 (supplying the conjugation mechanism) and pORI280 (providing both Eryr and β-galactosidase activity). Transconjugants were scored for the absence of the Cmr phenotype associated with pVE6007. Excision and loss of pORI280 were selected after continuous passage in GM17 at 37°C, in the absence of antibiotic selection, and spreading at intervals onto GM17-X-Gal at the same temperature. Individual colonies were transferred to 10 ml of GM17 broth and grown overnight. After 12 consecutive transfers (0.1% inocula), white colonies appeared at a frequency of 16%, indicating a second crossover event leading to the loss of pORI280. While the loss of β-galactosidase activity can be used as a marker in non-lactose-fermenting backgrounds, one can also use replica plating to differentiate between Eryr and Erys colonies in lactose-fermenting backgrounds.
Depending on where the second crossover event occurs, either the lactococcal plasmid will revert to its original composition or the SOEing fragment will be incorporated, resulting in the deletion-replacement of the target gene(s) (Fig. 1C). The likelihood that these events will occur in a 50:50 ratio is enhanced by designing the SOEAB and SOECD products to be of similar size. A simple PCR, with primers SOEA and SOED, can discriminate between reversion and deletion, though sequencing was also employed to ensure the integrity of the constructs (results not shown). In this specific example a well diffusion assay was used to identify events leading to the deletion of the structural genes. To perform this assay, overnight cultures were grown in GM17 broth and centrifuged, and the cell-free supernatant was removed. This supernatant was heat treated at 80°C for 15 min (a regimen which fails to inactivate bacteriocin activity) to eliminate viable cells. Molten agar was cooled to 48°C and seeded with the indicator strain, L. lactis HP (approximately 106 fresh overnight-grown cells/ml). The inoculated medium was dispensed into sterile petri plates, allowed to solidify, and dried. Wells (approximately 4.6 mm in diameter) were then bored in the seeded agar plates. Aliquots (50 μl) of the cell-free supernatant were dispensed into the wells, and the plates were incubated overnight at 30°C. In those instances where the ltnA1A2 genes were deleted, bacteriocin production was eliminated and no zone of inhibition was observed (Fig. 2). In those instances where bacteriocin production was retained, the result was the transfer of the original 60.2-kb plasmid to MG1614 with Eryr and β-galactosidase activity as temporary selective markers. Lacticin 3147 production in MG1614 was equal to that seen for MG1363 (Fig. 2), confirming that no changes had occurred in the transferred plasmid. Thus, the food-grade transfer of pMRC01 was achieved without requiring a native selectable marker.
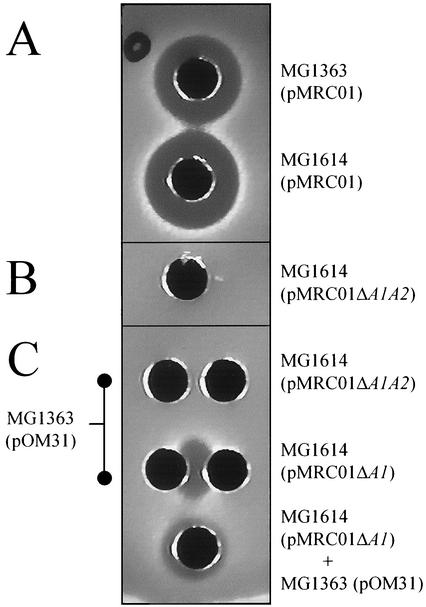
FIG. 2.
Well diffusion assay, with the L. lactis indicator strain HP, to test the inhibitory activity of 50-μl volumes of cell-free supernatant from a number of strains. (A) The zone size produced by MG1614(pMRC01) supernatant is identical to that of MG1363(pMRC01), demonstrating that plasmid transfer did not impact lacticin 3157 production. (B) Deletion of ltnA1A2 eliminates inhibitory activity. (C) Supernatant from MG1363(pOM31), which produces LtnA1 but not LtnA2, does not produce zones [a phenotype that cannot be complemented by supernatant from MG1614(pMRC01ΔltnA1A2)]. However, supernatant from MG1363(pOM31) and MG1363(pMRC01ΔltnA1) combined by either diffusion from two wells (each containing 50 μl of supernatant) or the mixing of the two supernatants in one well (25 μl of each supernatant) results in activity, demonstrating that LtnA2 is being produced by the latter and thus that the SOEing deletion of ltnA1 is not polar.
The strategy used to mutate ltnA1A2 took advantage of the conjugative nature of pMRC01. To determine whether the same system could be used to engineer nonconjugative plasmids, a second strategy was attempted which took advantage of the temperature-sensitive nature of pVE6007. Once again, a strain with three plasmids was constructed (pMRC01, pORI280-SOEA-D, and pVE6007) and then subjected to temperature-induced curing of pVE6007. After curing, the only Eryr cells should have been those in which pORI280 had integrated into pMRC01 (at the site of homology). Using this strategy, we attempted to create a pMRC01 vector lacking only ltnA1, which would again eliminate bacteriocin production. An MG1363 derivative containing three plasmids (pVE6007, pORI280ΔltnA1SOEA-D, and pMRC01) was serially passaged in GM17-ERY (5 μg/ml) broth prewarmed to 39°C and subsequently streaked onto prewarmed GM17-ERY-X-Gal agar at the same temperature, with scoring for blue colonies (indicating the retention of pORI280ΔltnA1SOEA-D through integration with pMRC01). Blue colonies resulting from these processes were confirmed as Cms to ensure the loss of pVE6007. As before, resolution of the cointegrate and loss of pORI280 were achieved by continuous passaging, plating, and selecting for white colonies. On this occasion 1% of colonies were white after nine consecutive transfers. The SOEA and SOED PCR primers were used to identify mutants lacking ltnA1 (data not shown). Supernatant from these mutants did not produce zones due to the absence of the structural protein LtnA1. An advantage of deleting ltnA1 but retaining ltnA2 was that it was possible to check if the mutation was precise and nonpolar. This is possible because both LtnA1 and LtnA2 are required for activity. If the removal of ltnA1 had a polar effect on the rest of the operon, then active LtnA2 would not be produced (18). Supernatant from a strain containing pOM31 (18), which produces only LtnA1, was used to determine whether active LtnA2 was present. As expected, supernatant from MG1363(pOM31), when combined with that of MG1363(pMRC01ΔltnA1), resulted in antimicrobial activity. This confirms the production of LtnA2 and thus the nonpolarity of the ΔltnA1 deletion (Fig. 2).
This study represents a significant advance in our ability to modify lactococcal plasmids. Due to the nature of the procedure the creation of food-grade deletions is possible, as there is no exogenous DNA left following double-crossover recombination. The system can also be used to replace plasmid genes, or even to introduce new genes, in order to confer an advantage in industrial applications and to generate engineered plasmids in order to conduct functional analysis. It could also be manipulated to permit the introduction of point mutations to determine the importance of individual nucleotides and amino acids by incorporating changes into both the SOEB primer and the overhang of the SOEC primer with the resultant change, rather than a deletion, being incorporated. The procedure also benefits from permitting the introduction of successive alterations sequentially to the same plasmid. Genetic manipulation of an intact plasmid, without the need for subcloning, overcomes a number of potential problems. Cloning can be difficult if the area of interest is particularly large, if all the relevant genes are not all located within one region, or if one is limited by the restriction sites on the cloning vector. In addition, despite the use of polymerase with proofreading abilities, the risk of incorporating errors increases with the size of the PCR product generated. When cloning into pORI280 is being done, the PCR fragment that needs to be amplified is relatively small (500 to 600 bp), thus minimizing the possibility of nucleotide misincorporation and facilitating the sequencing of inserts to ensure the integrity of the constructs again. The system described in this paper also overcomes the need to identify, for the purpose of subcloning, all of the genes which play a role in the phenotype of interest, since presumably all of these will accompany the plasmid when it is transferred. It should be noted when modifying high-copy-number plasmids that it may be necessary to design SOE primers such that the SOE-AB product is smaller than the SOE-CD equivalent, to minimize the possibility of mutated and wild-type revertant plasmids existing in the same cell. Following integration PCR could be used to identify the less-likely event of crossover occurring within the AB region only. There is then a strong likelihood that the second crossover event will occur within the CD region. Subsequent extensive passaging should result in the isolation of a cell containing only the mutated plasmid. Alternatively, in situations where a high-copy-number native plasmid is self-transmissible, it may be possible to separate the mutant and wild-type plasmids through conjugation.
In addition to plasmid engineering, this strategy can also be adapted to provide a reversible selection for natural plasmids that are potentially of industrial importance but lack a selectable trait. pORI280 can be integrated for the purposes of screening for plasmid transfer, after which revertants can be readily identified as β-galactosidase negative or Erys variants. This method also represents an improvement over coelectroporation, as its use will ensure that following plasmid transfer 100% of antibiotic-resistant colonies will have the native plasmid of interest. While knowledge of the sequence of at least part of the plasmid to be transferred is useful, the cloning of randomly digested plasmid fragments would also allow integration and transfer. This ability to temporarily mark plasmids should also prove useful when one is attempting to stack plasmids with similar phenotypes in a single strain, for example, combining plasmids encoding different phage resistance mechanisms or plasmids encoding different bacteriocins. Additionally, the strategy should not be restricted to naturally conjugative plasmids. A relatively simple modification can be suggested in which an origin of transfer (e.g., oriT from pMRC01 [8]) is cloned onto pORI280, rendering the plasmid, and any cointegrate, mobilizable by pMRC01. Thus, we can envisage moving nonconjugative plasmids between strains in a food-grade manner. Lastly, cointegrate plasmids (pORI280::native replicon) could be introduced to commercial cultures via electroporation, though this be may limited by difficulties in transforming large cointegrate plasmids or by the fact that many industrial strains remain recalcitrant to electrotransformation. In addition, electrotransformation may not be regarded as a food-grade procedure in some countries.
In this communication we have described a system that can precisely engineer native plasmids while providing the additional advantages of permitting multiple alterations and maintaining the food-grade status of the plasmid as well as facilitating the transfer of such native plasmids between hosts. We envisage that the system described herein will prove to be a useful resource for plasmid biologists working with lactococci and other gram-positive organisms.
Acknowledgments
This project was funded by the Irish Government under the National Development Plan 2000-2006.
We thank Jan Kok, University of Groningen, for providing plasmids pVE6007 and pORI280 and the strain EC101.
REFERENCES
- 1.Allison, G. E., and T. R. Klaenhammer. 1996. Functional analysis of the gene encoding immunity to lactacin F, lafI, and its use as a Lactobacillus-specific, food-grade genetic marker. Appl. Environ. Microbiol. 62:4450-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corneau, N., C. Dube, G. LaPointe, and E. Emond. 2001. A coelectroporation method for the isolation of cryptic plasmids from Lactococcus lactis. Lett. Appl. Microbiol. 33:7-11. [DOI] [PubMed] [Google Scholar]
- 3.Dougherty, B. A., C. Hill, J. F. Weidman, D. R. Richardson, J. C. Venter, and R. P. Ross. 1998. Sequence and analysis of the 60kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol. Microbiol. 29:1029-1038. [DOI] [PubMed] [Google Scholar]
- 4.Emond, E., R. Lavallee, G. Drolet, S. Moineau, and G. Lapointe. 2001. Molecular characterization of a theta replication plasmid and its use for development of a two-component food-grade cloning system for Lactococcus lactis. Appl. Environ. Microbiol. 67:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu, X., and J. G. Xu. 2000. Development of a chromosome-plasmid balanced lethal system for Lactobacillus acidophilus with thyA gene as a selective marker. Microbiol. Immunol. 44:551-556. [DOI] [PubMed] [Google Scholar]
- 6.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasson, M. J., and G. F. Fitzgerald. 1994. Gene transfer systems and transposition, p. 1-51. In M. J. Gasson and W. M. de Vos (ed.), Genetics and biotechnology of lactic acid bacteria. Blackie Academic and Professional, London, United Kingdom.
- 8.Hickey, R. M., D. P. Twomey, R. P. Ross, and C. Hill. 2001. Exploitation of plasmid pMRC01 to direct transfer of mobilizable plasmids into commercial lactococcal starter strains. Appl. Environ. Microbiol. 67:2853-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holo, H., and I. F. Nes. 1989. High-frequency transformation by electroporation of Lactococcus lactis subsp. cremoris strains grown with glycine in osmotically stable media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-534. [PubMed] [Google Scholar]
- 11.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. Appl. Environ. Microbiol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leelawatcharamas, V., J. G. Chia, P. Charoenchai, N. Kunajakr, C.-Q. Liu, and N. W. Dunn. 1997. Plasmid-encoded copper resistance in Lactococcus lactis. Biotechnol. Lett. 19:639-643. [Google Scholar]
- 13.Leenhouts, K., G. Buist, A. Bolhuis, A. ten Berge, J. Kiel, I. Mierau, M. Dabrowska, G. Venema, and J. Kok. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253:217-224. [DOI] [PubMed] [Google Scholar]
- 14.Liu, C.-Q., V. Leelawatcharamas, M. L. Harvey, and N. W. Dunn. 1996. Cloning vectors for lactococci based on plasmid encoding resistance to cadmium. Curr. Microbiol. 33:35-39. [DOI] [PubMed] [Google Scholar]
- 15.MacCormick, C. A., H. G. Griffin, and M. J. Gasson. 1995. Construction of a food-grade host/vector system for Lactococcus lactis based on the lactose operon. FEMS Microbiol. Lett. 127:105-109. [DOI] [PubMed] [Google Scholar]
- 16.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McAuliffe, O., C. Hill, and R. P. Ross. 2000. Identification and overexpression of ltnI, a novel gene which confers immunity to the two-component lantibiotic lacticin 3147. Microbiology 146:129-138. [DOI] [PubMed] [Google Scholar]
- 18.McAuliffe, O., C. Hill, and R. P. Ross. 2000. Each peptide of the two-component lantibiotic lacticin 3147 requires a separate modification enzyme for activity. Microbiology 146:2147-2154. [DOI] [PubMed] [Google Scholar]
- 19.Mills, D. A. 2001. Mutagenesis in the post genomic era: tools for generating insertional mutations in the lactic acid bacteria. Curr. Opin. Biotechnol. 12:503-509. [DOI] [PubMed] [Google Scholar]
- 20.Platteeuw, C., I. Van Alen-Boerrigter, S. van Schalkwijk, and W. M. de Vos. 1996. Food-grade cloning and expression system for Lactococcus lactis. Appl. Environ. Microbiol. 62:1008-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan, M. P., M. C. Rea, C. Hill, and R. P. Ross. 1996. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl. Environ. Microbiol. 62:612-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorensen, K. I., R. Larsen, A. Kibenich, M. P. Junge, and E. Johansen. 2000. A food-grade cloning system for industrial strains of Lactococcus lactis. Appl. Environ. Microbiol. 66:1253-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steele, J. L., and L. L. McKay. 1989. Conjugative transfer of genetic material in lactococci; a review. J. Dairy Sci. 72:3388-3397. [Google Scholar]
- 24.von Wright, A., S. Wessels, S. Tynkknen, and M. Saarela. 1990. Isolation of a replicon region of a large lactococcal plasmid and use in cloning of a nisin resistance determinant. Appl. Environ. Microbiol. 56:2029-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]