Abstract
The emission of methane (1.3 mmol of CH4 m−2 day−1), precursors of methanogenesis, and the methanogenic microorganisms of acidic bog peat (pH 4.4) from a moderately reduced forest site were investigated by in situ measurements, microcosm incubations, and cultivation methods, respectively. Bog peat produced CH4 (0.4 to 1.7 μmol g [dry wt] of soil−1 day−1) under anoxic conditions. At in situ pH, supplemental H2-CO2, ethanol, and 1-propanol all increased CH4 production rates while formate, acetate, propionate, and butyrate inhibited the production of CH4; methanol had no effect. H2-dependent acetogenesis occurred in H2-CO2-supplemented bog peat only after extended incubation periods. Nonsupplemented bog peat initially produced small amounts of H2 that were subsequently consumed. The accumulation of H2 was stimulated by ethanol and 1-propanol or by inhibiting methanogenesis with bromoethanesulfonate, and the consumption of ethanol was inhibited by large amounts of H2; these results collectively indicated that ethanol- or 1-propanol-utilizing bacteria were trophically associated with H2-utilizing methanogens. A total of 109 anaerobes and 107 hydrogenotrophic methanogens per g (dry weight) of bog peat were enumerated by cultivation techniques. A stable methanogenic enrichment was obtained with an acidic, H2-CO2-supplemented, fatty acid-enriched defined medium. CH4 production rates by the enrichment were similar at pH 4.5 and 6.5, and acetate inhibited methanogenesis at pH 4.5 but not at pH 6.5. A total of 27 different archaeal 16S rRNA gene sequences indicative of Methanobacteriaceae, Methanomicrobiales, and Methanosarcinaceae were retrieved from the highest CH4-positive serial dilutions of bog peat and methanogenic enrichments. A total of 10 bacterial 16S rRNA gene sequences were also retrieved from the same dilutions and enrichments and were indicative of bacteria that might be responsible for the production of H2 that could be used by hydrogenotrophic methanogens. These results indicated that in this acidic bog peat, (i) H2 is an important substrate for acid-tolerant methanogens, (ii) interspecies hydrogen transfer is involved in the degradation of organic carbon, (iii) the accumulation of protonated volatile fatty acids inhibits methanogenesis, and (iv) methanogenesis might be due to the activities of methanogens that are phylogenetic members of the Methanobacteriaceae, Methanomicrobiales, and Methanosarcinaceae.
Peat bogs are wetland ecosystems in which biomass production exceeds biodegradative activity, are often acidic, and emit approximately 3 to 7% of the global annual emission of the greenhouse gas methane (CH4) (2, 15). Peat bogs cover less than 3% of the Earth's terrestrial surface (8) but harbor approximately 30% of the global reserves of soil carbon and soil nitrogen, and the underlying peat is often anoxic (18). In anoxic environments with negligible concentrations of inorganic electron acceptors (e.g., sulfate or nitrate), methanogenesis is often the main terminal microbiological process during the biodegradation of organic matter. H2-CO2 and acetate are considered the main substrates for methanogenesis (37, 58, 62); although normally less important than H2-CO2 and acetate, formate and methanol can also be significant substrates for methanogens (4).
It is estimated that about two-thirds of the methane produced in nature originates from acetoclastic methanogenesis (14). However, H2-CO2 appears to be a significant precursor of the methane that is emitted from acidic peats (29, 62). In northern peatlands, although acetoclastic methanogenesis occurs in the upper vegetated zones of the peat, methanogenesis shifts toward the reduction of CO2 in deeper zones (7, 44, 54). Acetate and one-carbon compounds (e.g., formate) can even accumulate in acidic peat (22), indicating that these compounds are not the primary source of methane that is emitted from such habitats. Supplemental acetate can be inhibitory to methanogenesis in acidic peat samples, whereas glucose and H2 can be stimulatory (62).
Little is known about acid-tolerant methanogens (4). Only one acid-tolerant H2-utilizing methanogen, identified as a member of the Methanobacteriaceae, has been reported in the literature (63). The methanogenic microbiota in acidic peat are unresolved. Gene sequences indicative of members of Methanobacteriaceae, Methanococcaceae, Methanosarcinaceae, and Methanomicrobiales have been detected in a blanket bog peat (20, 36); however, the methanogens present in the zone of highest methanogenic activity in this bog peat were not identified (36).
An investigation designed to resolve both the methanogenic community and the flow of carbon and reductant in bog peat has thus far not been reported. The main objectives of this study were to (i) assess precursors of methane during the flow of carbon and reductant in an anoxic acidic bog peat and (ii) enumerate and identify the cultured microorganisms associated with methanogenesis.
MATERIALS AND METHODS
Description of the site and collection of peat.
The acidic bog is located approximately 700 m above sea level (50°08′40" N, 11°51′55" E) in the Fichtelgebirge, a forest with medium-sized mountains in east-central Germany. The soil is Cambic Podzol on granite bedrock with peat accumulation (33), and Picea abies and Sphagnum spp. dominate the vegetation of the site. During the investigation period (1998 and 1999), the bog lacked vegetation and was covered with water (5 to 30 cm depending on the amount of precipitation). The annual mean air temperature at the site approximates 6.5°C, with temperature extremes of approximately 30 and −20°C; annual precipitation averages 1,100 mm per year (33). During the sampling periods, the peat temperature at a depth of 5 to 10 cm was 9.4 ± 2.2°C (n = 10) and the air temperature was 12.6 ± 4.0°C (n = 10) (these data were obtained at approximately 11 a.m. on 10 different days). Peat was collected in sterile airtight vessels and stored on ice until processed (within 8 h); it had a dry weight of 12% ± 2% (dry weights were determined on three different dates, and each determination was done in triplicate).
Assessment of in situ gas emissions.
Gases were collected in round Plexiglas (Degussa, Darmstadt, Germany) chambers (1.9 liters, 227 cm2) (34) that were placed on the water surface above the peat. Gas emissions were analyzed with samples from two chambers at each sampling date. Gas samples (n = 14) were withdrawn at gas outlets with syringes, injected into gastight evacuated bottles, and analyzed for CH4 in the laboratory.
Anoxic microcosms.
Microcosms with large headspaces were 500-ml screw-cap, butyl rubber-stoppered infusion flasks (Merck ABS, Dietikon, Switzerland) containing 15 g (dry wt) of peat per flask under 100% argon. Microcosms with small headspaces were 125-ml infusion flasks containing 12 g (dry wt) of peat per flask. Substrates were added from sterile anoxic stock solutions (adjusted to pH 4.5) or from bottles containing sterile gases. Microcosms were incubated horizontally in the dark at 20°C, and each microcosm experiment was performed in triplicate. Substrate/product stoichiometries were corrected with values obtained in control microcosms lacking supplemental substrates. The pH of peat during incubations was usually between 4.3 and 4.5, with extremes of 4.0 and 4.8.
Cultivation media.
Anoxic media were prepared using modified Hungate techniques (24). Anoxic media were boiled, subsequently cooled under an argon gas phase, dispensed into culture tubes or serum bottles, and autoclaved. Unless otherwise indicated, gas phases were 100% argon. The sterile anoxic mineral solution used for the most-probable-number (MPN) dilution series (see below) contained (in milligrams per liter) (NH4)2SO4, 25; CaCl2 · 2H2O, 10; MgCl2 · 6H2O, 10; NaCl, 200; NH4Cl, 200; and KH2PO4, 200. The pH of the mineral solution was adjusted to 4.5 with HCl. Anoxic tryptic soy broth containing glucose (TSB; Difco Laboratories, Detroit, Mich.) was used at 2.75 g liter−1 (yielding an initial glucose concentration of 1.5 mM); the pH was adjusted to 4.5 (unless otherwise indicated). Undefined medium (UM) was anoxic mineral solution (above) supplemented with (per liter) yeast extract, 0.5 g; tryptone, 0.5 g; vitamin solution (3), 10 ml; and trace element solution (3), 10 ml. The pH of UM was adjusted to 4.5 or 6.0. UM supplemented with antibiotics (UMab) was UM with (per liter) penicillin, 0.3 g; streptomycin, 0.2 g; and kanamycin, 0.2 g. Selective methanogen medium (SMM) contained (per liter) mineral solution, 10 ml; vitamin solution, 10 ml; trace element solution, 10 ml; acetate-free fatty acid solution (61), 1 ml; and cell-free enrichment culture fluid (see below), 100 ml. The pH of SMM was adjusted to 4.5 or (in one case) 6.5. For preparation of cell-free enrichment culture fluid, methanogenic UM enrichment cultures were centrifuged at 10,000 × g for 20 min; the supernatant fluid was freed of particulate matter by microfiltration (pore size 0.2 μm). The filtered supernatant fluid constituted the cell-free enrichment culture fluid and was stored at 4°C under 100% argon. The gas phase of SMM was H2-CO2 (8:2) or argon-H2-CO2 (78:18:4).
Enumeration of cultured microorganisms.
Cultured microorganisms were estimated by the MPN technique (10). Peat was processed inside a Mecaplex (Grenchen, Switzerland) O2-free chamber (the gas phase was 100% N2). For extraction of microorganisms, 50 g (wet wt) of peat and 200 ml of anoxic mineral solution (see above) were placed in 500-ml infusion flasks; the flasks were then sealed with a rubber stopper and placed on an end-over-end shaker (60 cycles per min) for 10 h at 2°C. Tenfold serial dilutions of peat were prepared in sterile anoxic mineral solution by using anoxic techniques (13). Inoculated tubes of anoxic TSB that showed both a consumption of glucose and an increase in optical density at 660 nm (≥0.05) were scored positive for general fermentative anaerobes. Methanogen numbers were estimated in various media with an argon-H2-CO2 (78:18:4) gas phase. MPN tubes for which the stoichiometries of H2 consumed with respect to CH4 produced were approximately 4:1 were scored positive for methanogens. TSB tubes were scored positive for methanogens if at least 0.1 μmol of CH4 was produced per tube (methane was not detected in uninoculated controls). MPN analyses were performed in triplicate. Culture tubes were incubated horizontally at 20°C in the dark.
Enrichment of methanogens under acidic conditions.
A total of 10 g (wet wt) of peat was added to 120-ml serum bottles that contained 50 ml of UM and had an argon-H2-CO2 (78:18:4) gas phase. Enrichments were incubated at 20°C in the dark. After the fourth transfer into the same medium, enrichment cultures that yielded stoichiometries of H2 consumed with respect to CH4 produced of approximately 4:1 were used to inoculate SMM. Experiments with methanogenic enrichments were performed in triplicate with SMM.
Analytical methods.
Gases (CH4, CO2, and H2) were analyzed by gas chromatography, and organic acids and alcohols were quantified by high performance liquid chromatography (28). Unless otherwise indicated, no distinction is made between organic acids and their salt forms. Redox potentials of peat (n = 9) were measured with a Schott PT-6280 platinum redox electrode (Schott Glasgeräte, Hofheim, Germany) with Ag/AgCl as a reference system and a WTW pH 90 digital pH meter (Wissenschaftlich Technische Werkstätten, Weilheim, Germany). Redox potentials were calculated by using the following equation: 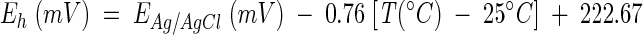 (55). The pH of peat (n = 10) was measured with a U457-S7/110 combination pH electrode (Ingold, Steinbach, Germany). Oven-dried (60°C) peat was homogenized with a ball mill (MM2; Retsch, Haan, Germany) and analyzed for total carbon content (n = 3) with an element analyzer (CHN-O rapid; Foss-Heraeus, Hanau, Germany). The total inorganic carbon content of peat (n = 1) was determined after heating peat at 500°C for 12 h. The aqueous phase of bog peat was analyzed for dissolved organic carbon (n = 1) with a liquiTOC (Foss-Heraeus). The dry weight of peat (n = 6) was determined by weighing peat before and after drying at 60°C for 48 h. Ammonium (n = 4) and nitrate plus nitrite (n = 4) were determined by flow injection analysis (QuickChemAE; Lachat Instruments, Milwaukee, Wis.). Sulfate (n = 1) was measured by ion chromatography (DX-100 with an AS4 A column [Dionex, Sunnyvale, Calif.]). Acetate (n = 3) was quantified by high-performance liquid chromatography (28).
(55). The pH of peat (n = 10) was measured with a U457-S7/110 combination pH electrode (Ingold, Steinbach, Germany). Oven-dried (60°C) peat was homogenized with a ball mill (MM2; Retsch, Haan, Germany) and analyzed for total carbon content (n = 3) with an element analyzer (CHN-O rapid; Foss-Heraeus, Hanau, Germany). The total inorganic carbon content of peat (n = 1) was determined after heating peat at 500°C for 12 h. The aqueous phase of bog peat was analyzed for dissolved organic carbon (n = 1) with a liquiTOC (Foss-Heraeus). The dry weight of peat (n = 6) was determined by weighing peat before and after drying at 60°C for 48 h. Ammonium (n = 4) and nitrate plus nitrite (n = 4) were determined by flow injection analysis (QuickChemAE; Lachat Instruments, Milwaukee, Wis.). Sulfate (n = 1) was measured by ion chromatography (DX-100 with an AS4 A column [Dionex, Sunnyvale, Calif.]). Acetate (n = 3) was quantified by high-performance liquid chromatography (28).
FISH and identification of methanogens.
Fluorescently labeled oligonucleotide probes (Table 1) were obtained from Interactiva (Ulm, Germany). Cells were fixed in 4% formaldehyde solution, immobilized on gelatin-coated slides, and dehydrated with ethanol at increasing concentrations (50, 80, and 98%). Fluorescent in situ hybridization (FISH) and quantification of hybridized cells in comparison to total cell counts (as determined by staining with 4′,6-diamidino-2-phenylindole [DAPI]) were done by published protocols (52); formamide and NaCl concentrations in the buffers are listed in Table 1. Methanogens were also microscopically identified by the fluorescence of coenzyme F420 (11). The following reference organisms were used for FISH: E. coli (DSM 423), Methanosarcina sp. strain WH1 (DSM 4659), Methanobacterium bryantii M.o.H.G. (DSM 862), Methanococcus voltae PS (DSM 1537), and Methanogenium cariaci JR1 (DSM 1497). The reference organisms were grown as suggested by the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany [http://www.dsmz.de]).
TABLE 1.
Sequences of oligonucleotides used as PCR primers and for FISH with corresponding concentrations of formamide and NaCl
| Probe or primer | Sequence (5′-3′) | Target organisms | % Formamide in hybridization buffer | Concn of NaCl in washing buffer (mM) | Reference |
|---|---|---|---|---|---|
| EUB338 | GCTGCCTCCCGTAGGAGT | Bacteria | 0 | 900 | 1 |
| ARC915 | GTGCTCCCCCGCCAATTCCT | Archaea | 20 | 225 | 53 |
| MS821 | CGCCATGCCTGACACCTAGCGAGC | Methanosarcina | 30 | 112 | 45 |
| MSMX860 | GGCTCGCTTCACGGCTTCCCT | Methanosarcinales | 30 | 112 | 45 |
| NONa | ACTCCTACGGGAGGCAGC | NAb | NA | NA | 32 |
| Arch21Fa | TCCGGTTGATCCYGSCRGc | Archaea | NA | NA | 9 |
| ARC344Fa-Clamp | CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG-ACGGGGYGCAGCAGc | Archaea | NA | NA | 45 |
| GM4R | TACCTTGTTACGACTT | All organisms | NA | NA | 40 |
| 341F-Clamp | CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG-CCTACGGGAGGCAGCAG | Bacteria | NA | NA | 39 |
| 907R | CCGTCAATTCMTTTGAGTTT | Bacteria | NA | NA | 39 |
Used as negative control.
NA, not applicable.
Sequence modified from original publication.
Phylogenetic analyses.
DNA was extracted from methanogenic enrichments and highest CH4-positive MPN dilutions by using the FastDNA SPIN Kit (Bio 101, Carlsbad, Calif.) as specified by the manufacturer. PCR was performed in a T-Gradient cycler (Biometra, Göttingen, Germany) by using 33 cycles of 94°C for 0.5 min, 55°C for 0.5 min, and 72°C for 1.5 min. The final PCR extension step was at 72°C for 5 min. To construct an archaeal 16S rRNA gene library, almost full-length archaeal 16S rRNA gene fragments were amplified using primers Arch21Fa and GM4R (Table 1). PCR products were ligated into pGEM-T (Promega, Mannheim, Germany) and transformed into E. coli JM109 (Promega) as specified by the manufacturer. For screening of the gene library by denaturing gradient gel electrophoresis (DGGE), 570-bp 16S rRNA gene fragments were directly amplified from 1 μl of resuspended clones using the primer pair ARC344Fa-Clamp plus ARC915 (Table 1). The Wizard Plus Minipreps DNA purification system (Promega) was used for the preparation of plasmids, and DNA was sequenced commercially at MWG Biotech (Ebersberg, Germany). DGGE analysis was also directly performed on extracted DNA from methanogenic enrichments or the highest CH4-positive MPN dilutions (10−6 to 10−7). Fragments (470 to 570 bp) of 16S rRNA genes were amplified by PCR using the primer pairs ARC344Fa-Clamp plus ARC915 for Archaea, ARC344Fa-Clamp and MSMX860 for Methanosarcinales, ARC344Fa-Clamp and MS821 for Methanosarcina, and 341F-Clamp/907R for Bacteria (Table 1).
DGGE was performed by following a published protocol (39); the temperature was 60°C, the denaturant (urea and formamide) gradient was 20 to 80%, the electrophoresis time was 18 h, and the voltage was 100 V. Gels were stained with SYBR Gold (Molecular Probes, Eugene, Oreg.). Selected DGGE bands were excised, and the DNA was extracted with water, reamplified, and sequenced commercially at MWG Biotech.
Sequences were aligned and phylogenetically analyzed using the program package ARB (Department of Microbiology, Technical University Munich, Germany [http://www.arb-home.de]). For calculation of phylogenetic trees, neighbor-joining, parsimony, and maximum-likelihood algorithms were applied to 16S rRNA gene sequences that were longer than 1.4 kb; a consensus tree was drawn using consistent branchings from all three methods, while inconsistent branches were drawn as multifurcations. Partial sequences were added to the consensus tree by maximum-parsimony analysis without changing the tree topology.
Nucleotide sequence accession numbers.
All 16S rRNA gene sequences have been deposited in the EMBL nucleotide sequences database. Accession numbers AJ459876 to AJ459902 have been assigned to the following archaeal sequences: AMC 3, AEC 11, AEC 10, AEC 9, AEC 8, AMC 6, AEC 3, AEC 6, AEC 2, AMC 7, AEC 4, AMC 4, AMC 5, AEC 5, AED 4, AED 9, AED 7, AED 8, AED 5, AED 6, AED 10, AED 3, AMC 2, AMC 1, AEC 1, AED 1, and AED 2. Accession numbers for bacterial sequences are given in the dendrogram.
RESULTS
Chemical characteristics of acidic bog peat.
The peat had a pH of 4.4 ± 0.2 and a mean total carbon content of 319 ± 30 g of C kg (dry wt) of soil−1. The organic and inorganic carbon contents in the peat were 329 and 1.2 g of C kg (dry wt) of soil−1, respectively. In the aqueous phase, the ammonium concentration was below 0.1 mM, the nitrate plus nitrite and sulfate concentrations were always below 0.01 mM, and the acetate concentration never exceeded 0.2 mM. The in situ redox potential of the bog was 550 to 650 mV in the water layer above the peat and 10 to 98 mV at a depth of 4 to 7 cm below the water/peat interface. The steepest redox gradient was observed at the water/peat interface (data not shown). The redox potential did not change significantly below a depth of 7 cm; the lowest redox potential measured was −50 mV.
In situ emission of methane at the bog.
CH4 was emitted at a rate of 1.3 mmol of CH4 m−2 day−1 in September 1998. This rate was within the range of CH4 emission rates (0.02 to 15 mmol of CH4 m−2 day−1) that were obtained at the same site in 1995 (August through October) and 1996 (June through November) (C. Kuhner and H. L. Drake, unpublished data).
Methanogenesis and the production of H2 by peat.
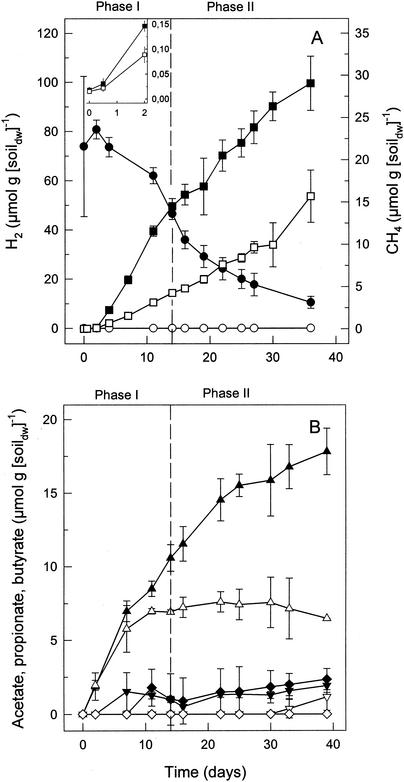
In anoxic microcosms at in situ pH (4.3 to 4.5), CH4 was produced at rates that ranged from 0.36 to 1.71 μmol of CH4 g (dry weight) of soil−1 day−1 (four sampling dates). Supplemental H2-CO2 stimulated the linear production of CH4 (Fig. 1A). CH4 was initially produced in stoichiometries that averaged 3.1 mol of H2 consumed per mol of CH4 formed (Fig. 1A, phase I), and approximately 1 mol of CH4 was formed per mol of CO2 consumed in H2-supplemented microcosms (data not shown). These stoichiometries were indicative of hydrogenotrophic methanogenesis 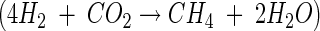 . In addition to CH4, acetate and small amounts of butyrate and propionate were formed in H2-supplemented microcosms (Fig. 1B). Acetate production in H2-supplemented microcosms exceeded that in nonsupplemented microcosms, indicating that significant amounts of H2-derived reductant were recovered in acetate (Fig. 1B, phase II).
. In addition to CH4, acetate and small amounts of butyrate and propionate were formed in H2-supplemented microcosms (Fig. 1B). Acetate production in H2-supplemented microcosms exceeded that in nonsupplemented microcosms, indicating that significant amounts of H2-derived reductant were recovered in acetate (Fig. 1B, phase II).
FIG. 1.
Effects of supplemental H2 on the production of CH4 (A) and volatile fatty acids (B) in bog peat microcosms. Solid symbols indicate microcosms amended with H2; open symbols indicate controls (unamended); •, ○, H2; ▪, □, CH4; ▴, ▵, acetate; ▾, ▿, propionate; ⧫, ◊, butyrate. A detailed view of the production of CH4 during the first 2 days is shown in the inset.
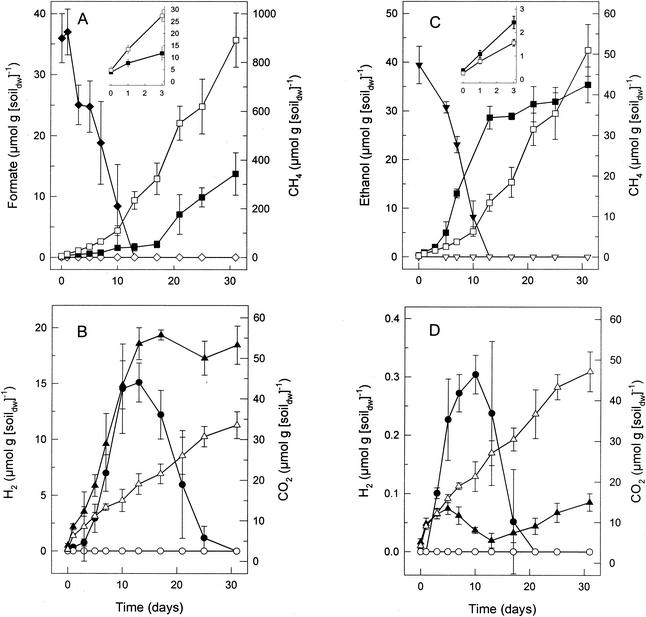
Supplemental formate was readily consumed, stimulated the production of H2, CO2, and acetate, and initially inhibited methanogenesis in the peat (Fig. 2A and B and data not shown). During the initial phase of activity when formate was utilized, the ratio of formate consumed to CO2, H2, and acetate produced approximated 8:6:4:1 (data not shown). In contrast, supplemental acetate and methanol persisted in anoxic microcosms, and acetate inhibited methanogenesis (data not shown). Although supplemental formate was not readily utilized for methanogenesis, the consumption of H2 and the reengagement of methanogenesis occurred after formate was consumed to depreciable levels (Fig. 2A and B).
FIG. 2.
Effects of supplemental formate (A and B) or ethanol (C and D) on the production of CH4 (A and C), H2 (B and D), and CO2 (B and D) in bog peat microcosms. Solid symbols indicate amended microcosms; open symbols indicate controls (unamended); ⧫, ◊, formate; ▪, □, CH4; ▾, ▿, ethanol; •, ○, H2; ▴, ▵, CO2. A detailed view of the production of CH4 during the first 3 days is shown in the insets (A and C).
Ethanol and 1-propanol were readily consumed and stimulated the production of CH4 (Fig. 2C and data not shown). The consumption of ethanol or 1-propanol was concomitant with the stoichiometric production of acetate and propionate, respectively (data not shown), indicating that these substrates were oxidized. Ethanol- and 1-propanol-supplemented peat initially produced trace amounts of H2 that were subsequently consumed. CH4 production was concomitant with the apparent consumption of CO2 (Fig. 2C and D and data not shown). When H2 (70 μmol g [dry wt] of soil−1) and ethanol (40 μmol g [dry wt] of soil−1) were added simultaneously to peat microcosms, the ethanol was not consumed until the H2 was depleted (data not shown), suggesting that methanogens did not use ethanol as a substrate.
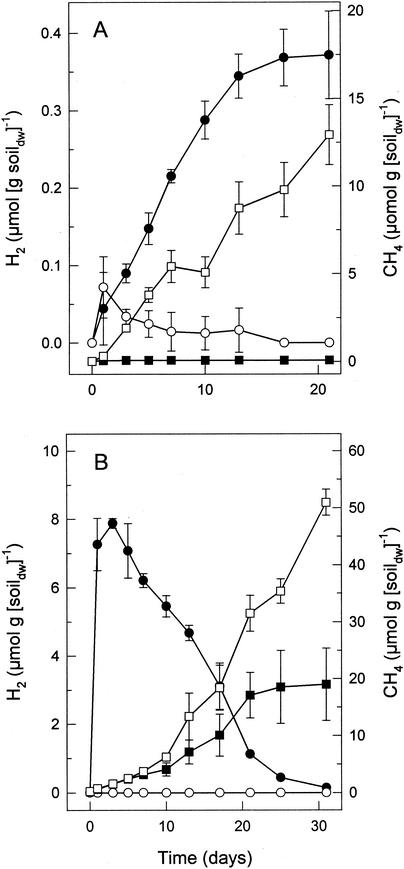
The results described above indicated that H2 was a significant precursor of CH4. Therefore, the capability of the peat-associated microbes to produce H2 from endogenous substrates was evaluated. In microcosms with small headspaces (see Materials and Methods), unsupplemented peat initially produced trace amounts of H2 that were subsequently consumed (Fig. 3A). The amount of H2 increased significantly when bromoethanesulfonate (20 mM), an inhibitor of methanogenesis (19), was added to the microcosms (Fig. 3A).
FIG. 3.
Effects of 20 mM bromoethanesulfonate (A) or easily fermentable carbon (TSB containing glucose) (B) on the production of H2 and CH4 in bog peat microcosms. Solid symbols indicate amended microcosms; open symbols indicate controls (unamended); •, ○, H2; ▪, □, CH4.
When peat was supplemented with TSB, large amounts of H2 transiently accumulated and the production of methane decreased significantly (Fig. 3B); ethanol (0.5 mM) was initially produced during the first 5 days and was consumed by day 15 (data not shown). The glucose (1.5 mM) present in TSB was depleted after 24 h. Acetate (9 mM), propionate (3 mM), butyrate (3 mM), isovalerate (1 mM), and isobutyrate (1 mM) were produced within 15 days and were not consumed within the first 31 days of incubation. However, analysis of microcosms after 80 days of incubation indicated that about half of the 9 mM acetate that initially accumulated during the first 31 days of incubation was consumed between days 31 and 80. In contrast, the acetate that accumulated in TSB-supplemented microcosms during the initial incubation period of 31 days was not subsequently consumed when such microcosms contained 20 mM bromoethanesulfonate (data not shown). The consumption of acetate that was produced in microcosms containing TSB and H2-CO2 occurred only after H2 was consumed to trace levels, and acetate was totally consumed within 80 days (data not shown). Methanogenesis was inhibited by 80 to 96% in microcosms amended with acetate, propionate, or butyrate (5 mM each) (Table 2). These supplemental fatty acids were not consumed during 31 days of incubation.
TABLE 2.
Effects of supplemental volatile fatty acids on the production of CH4 in acidic bog peat (pH 4.5)
| Expt | Volatile fatty acid (5 mM) | pKa | Mean amt of CH4 produced (nmol g [dry wt] of soil−1 day−1) ± SDa |
|---|---|---|---|
| Ab | None | NAc | 268 ± 19 |
| Formate | 3.75 | 28 ± 4 | |
| Acetate | 4.76 | 8 ± 4 | |
| Bd | None | NA | 541 ± 88 |
| Propionate | 4.88 | 96 ± 11 | |
| Butyrate | 4.82 | 112 ± 28 |
Rates were calculated from the initial phase of activity (7 to 11 days of incubation; a minimum of five data points were used to calculate rates) in anoxic microcosms (0.95 ≤ r2 ≤ 0.98).
Bog peat sample was collected on 5 May 1999.
NA, not applicable.
Bog peat sample was collected on 25 June 1999.
Enumeration of cultured methanogens from peat and product analyses of MPN series.
An MPN value of 3 × 107 methanogens g (dry wt) of soil−1 was obtained for hydrogenotrophic methanogens, a value that was approximately 1% of the MPN value for general fermentative anaerobes and 30 times greater than the MPN value for unspecified methanogens obtained in TSB (Table 3). MPN values for unspecified methanogens in TSB at pH 6.0 and pH 6.8 were similar to those obtained at pH 4.5 (data not shown). Antibiotics (streptomycin, kanamycin, and penicillin) greatly decreased the MPN values obtained for hydrogenotrophic methanogens (Table 3).
TABLE 3.
MPN of cultured anaerobes and methanogens in acidic bog peat
| Metabolic type | Medium | Substrate | MPN of organisms g (dry wt) of soil−1a |
|---|---|---|---|
| General fermentative anaerobes | TSB (pH 4.5) | Complex | 1 × 109 (2 × 108-5 × 109) |
| Methanogens | TSB (pH 4.5) | Complex | 1 × 106 (3 × 105-6 × 106) |
| H2-CO2-utilizing methanogens | UM (pH 4.5) | H2-CO2 | 3 × 107 (6 × 106-1 × 108) |
| H2-CO2-utilizing methanogens | UMab (pH 4.5) | H2-CO2 | 3 × 102 (7 × 101-2 × 103) |
Values in parentheses represent confidence intervals with 95% probability.
Acetate and propionate were the only soluble fermentation products detected in all of the growth-positive MPN dilutions (10−2 to 10−9) when TSB (containing glucose) was used as the growth medium (data not shown). Butyrate and isovalerate were detected in the 10−2 to 10−6 MPN dilutions. Ethanol was detected only in the 10−5 to 10−6 MPN dilutions of bog peat, indicating that ethanol producers outnumbered ethanol degraders in the peat. CH4 was detected in TSB-MPN tubes up to dilutions of 10−6 (Table 3). However, the largest amounts of CH4 were detected in the 10−2 TSB-MPN dilutions, in which the concentrations of acetate (and ethanol) were lower than those obtained in higher TSB-MPN dilutions (data not shown). Collectively, the MPN analyses indicated that organisms that might link the consumption of acetate or ethanol to methanogenesis (via acetoclastic methanogens, syntrophic acetate oxidzers, syntrophic ethanol oxidiers, etc.) at pH 4.5 occurred in numbers that were substantially smaller than the number of hydrogenotrophic methanogens that were viable at this pH.
Enrichment of acid-tolerant methanogens from peat.
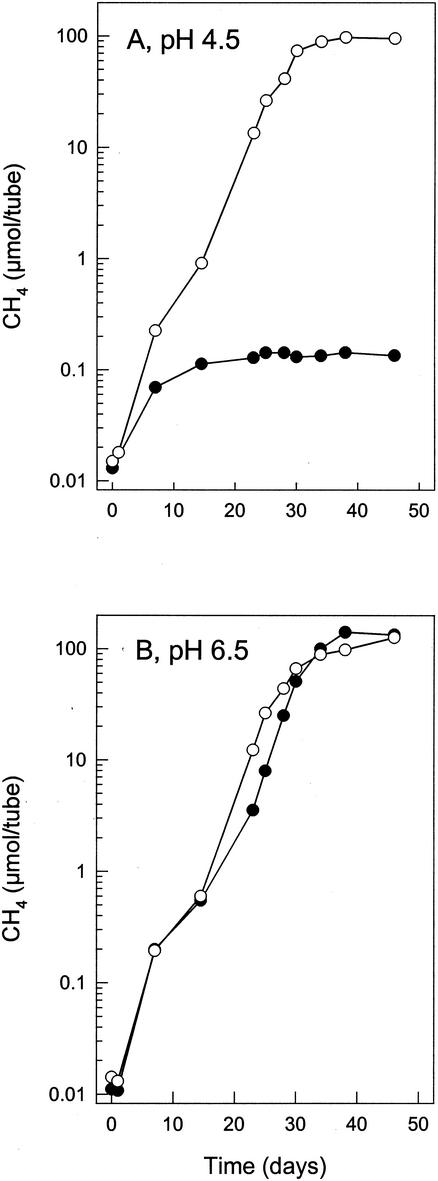
Attempts to obtain methanogenic enrichments were unsuccessful with UM (pH 4.5 and 6.0) supplemented with either 5 mM acetate or 5 mM formate. Successful methanogenic enrichments were obtained with H2-CO2-supplemented UM (pH 4.5) and subsequently maintained in either H2-CO2-supplemented SMM or H2-CO2-supplemented UM (pH 4.5). The ending pH approximated 5.5. After prolonged enrichment, the relative number of Archaea and Bacteria detected by FISH was 70 and 7%, respectively. Approximately 4 mol of H2 was consumed per mol of CH4 produced in methanogenic enrichments. Methanogens identified by coenzyme F420 fluorescence or FISH were mainly small, straight rods, some of which were aggregated in short chains of up to four cells. Long, curved rods were also observed. Neither acetate nor ethanol was consumed by methanogenic enrichments. Methanogenesis by the enriched methanogens was similar at pH 4.5 and 6.5; however, methanogenesis was significantly inhibited by 5 mM acetate at pH 4.5 but not at pH 6.5 (Fig. 4). The production of CH4 from H2-CO2 by the methanogenic enrichments was completely inhibited by the antibiotics streptomycin, kanamycin, and penicillin (data not shown).
FIG. 4.
Effect of 5 mM acetate on the production of CH4 from H2-CO2 by the methanogenic enrichment culture at pH 4.5 (<pKaacetic acid) (A) and pH 6.5 (≫pKaacetic acid) (B). Values are the means of triplicates. Symbols: •, amended with acetate; ○, controls (unamended).
Phylogenetic analyses of cultured methanogens.
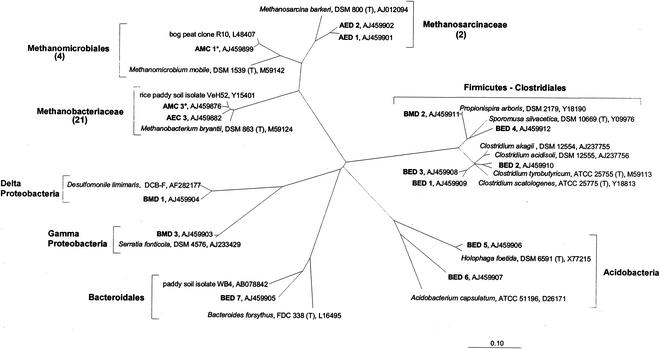
A total of 27 different archaeal 16S rRNA gene sequences were obtained from methanogenic enrichments and methanogenic MPN dilutions. 16S rRNA gene sequences included 10 sequences obtained from excised DGGE bands and 17 clone sequences. Most of these 27 sequences were closely related to known phylogenetic groups of methanogens. A total of 21 sequences (AMC 3 to AMC 7, AEC 2 to AEC 6, AEC 8 to AEC 11, and AED 4 to AED 10) clustered with Methanobacteriaceae, 4 (AMC 1, AMC 2, AEC 1, and AED 3) clustered with Methanomicrobiales, and 2 (AMC 1 and AMC 2) clustered with Methanosarcinaceae. For each group, one or two representative sequences are displayed in Fig. 5. Five clone sequences of the Methanobacteriaceae cluster were from the highest CH4-positive MPN dilutions of peat in UM (AMC 3 to AMC 7) and were closely related to the VeH52 isolate from rice paddy soil (97 to 99% sequence similarity). Two clone sequences from the highest CH4-positive peat dilution (10−6) in TSB (AMC 1 and AMC 2) clustered with sequences of Methanomicrobiales and were closely related to a clone sequence retrieved from a Welsh bog peat (97% sequence similarity). Clone sequences from methanogenic enrichments were affiliated with the same methanogenic groups to those for the clone sequences that were obtained from the highest CH4-positive MPN dilutions. Likewise, all partial 16S rRNA gene sequences that were obtained from DGGE bands were affiliated with the same methanogenic groups as the clone sequences, i.e., Methanobacteriaceae and Methanomicrobiales, except for two sequences (AED 1 and AED 2); these sequences were retrieved after (semi)specific PCR amplifications with primer pair ARC344Fa-clamp and MS821 and fell into the Methanosarcinaceae cluster. AED 1 was found to be very similar to an archaeal 16S rRNA gene sequence (98% sequence similarity) of an uncultured rice field soil archaeon; in contrast, AED 2 was only distantly related to this sequence.
FIG. 5.
Phylogenetic positions of bacterial and representative archaeal sequences obtained in this study (bold) as inferred from comparative analysis of 16S rRNA gene sequence data. Values in parentheses are the number of sequences obtained for a given group. The codes used for the sequences obtained in this study are as follows: A, archaeal; B, bacterial; M, from highest CH4-positive MPN dilution of bog peat; E, from methanogenic enrichment culture; C, sequences (1,400 bp) from clone library; D, sequences (approximately 400 bp) from excised bands of DGGE analyses. Sequences that are almost complete (i.e., approximately 1,400 bp) are marked with an asterisk. The bar represents a 0.1 estimated change per nucleotide.
Seven bacterial 16S rRNA gene sequences were obtained from methanogen enrichments in UM; they were phylogenetically related to Bacteroidales (BED 7), the Acidobacterium phylum (BED 5 and BED 6), and various strains and clone sequences of the Clostridiales (BED 1 to BED 4) (Fig. 5). The three bacterial 16S rRNA gene sequences obtained from the highest CH4-positive MPN dilution (BMD 1 to BMD 3) were most similar to Serratia fonticola (BMD 3; 99% sequence similarity), Propionispira arboris (BMD 2; 96% sequence similarity), and an uncultured delta proteobacterium (BMD 1; 98% sequence similarity) (Fig. 5).
DISCUSSION
Hydrogenotrophic methanogenesis and the interspecies transfer of H2.
In contrast to terrestrial environments and pH-neutral wetlands, in which CH4 is produced predominantly by acetoclastic methanogens (6, 14, 30, 59), H2 was the main substrate for methanogenesis in the acidic bog peat investigated in this study. Indeed, acetate inhibited methanogenesis, and attempts to obtain acetoclastic, methanogenic enrichments at both pH 4.5 and 6.0 were unsuccessful. These results are consistent with earlier reports (29, 62) and with recent isotope data (7, 44) obtained from acidic bog peats and northern peatlands. Acetate is a main intermediate during the anaerobic decomposition of organic matter (38) and can accumulate to high levels in the absence of acetoclastic methanogenesis in peatlands (22, 51, 58). In the present study, in situ accumulation of acetate was not evident, presumably because under in situ conditions, acetate was oxidized to CO2 via aerobic respiration or other oxidative microbial processes (e.g., via the dissimilation of iron or nitrate) or oxidized syntrophically to CO2 by the concerted activity of acetate-oxidizing anaerobes and hydrogenotrophic methanogens. The latter possibility is indirectly supported by the results from TSB-supplemented anoxic microcosms (Fig. 3B). In microcosms, about 50% of the acetate produced from glucose was consumed slowly after an extensive incubation period. Bromoethanesulfonate inhibited both the consumption of acetate and the production of methane, indicating that acetate degradation was indeed linked to methanogenesis. In contrast, supplemental H2 stimulated the consumption of acetate after H2 was consumed, indicating that the initial stimulation of hydrogenotrophic methanogens enhanced the usually slow syntrophic degradation of acetate. The syntrophic degradation of acetate under anoxic conditions can occur in methanogenic bioreactors (50) and lake sediments (42).
Syntrophic interactions of certain anaerobic microorganisms are based on the transfer of H2 to a hydrogenotrophic partner (37, 48). The acidic bog peat evaluated in the present study produced small, transient amounts of H2 from endogeneous sources (Fig. 3A), and the amount of this transiently produced H2 increased significantly in TSB-supplemented bog peat (Fig. 3B). The production of H2 became more evident when methanogenesis was inhibited or when the peat was amended with primary alcohols. Since H2 was transiently produced concomitant with the consumption of alcohols and the production of CH4, H2 appears to be an important substrate for the production of CH4 in alcohol-supplemented microcosms (Fig. 2B and C). This conclusion was corroborated by the observation that ethanol was not consumed in the presence of high partial pressures of H2. The MPN values for hydrogenotrophic methanogens were approximately 30-fold greater than the MPN values for the unspecified methanogens that were obtained in TSB lacking supplemental H2 (Table 3). The absence of CH4 in 10−7 TSB-MPN dilutions indicated that no suitable precursor for methanogenesis (i.e., H2) was produced beyond TSB-MPN dilutions of 10−6. These collective observations indicate that H2 is an intermediate in the overall decomposition of organic matter in this acidic bog and that hydrogenotrophic methanogens outnumber H2-producing microorganisms in the bog peat.
Primary alcohols are produced by fermentation and are intermediates in the anoxic decomposition of organic matter to CH4 and CO2 (38). Alcohols were quickly consumed in anoxic peat microcosms, demonstrating that the anaerobic microbiota of the peat had a substantial capacity to consume alcohols. Anaerobic bacteria that degrade primary alcohols in syntrophic association with H2-scavenging anaerobes have been isolated from pH-neutral environments and have pH optima that approximate pH 7 (47, 48, 60, 65). The syntrophic methanogenic consortia of the bog peat characterized in the present study is moderately acid tolerant and is probably composed of microorganisms that have not been described yet. The moderately acid-tolerant hydrogenotrophic methanogens appear to be syntrophically associated with H2-producing anaerobes and essential to the normal flow of both carbon and reductant in this acidic bog peat.
Effects of volatile fatty acids and pH.
Methanogens can tolerate up to 100 mM volatile fatty acids at neutral pH; however, they are very sensitive to volatile fatty acids at mildly acidic pH (56, 57). It was therefore not surprising that acetate and other volatile fatty acids inhibited methanogenesis in bog peat at pH 4.5 but not at pH 6.5 (Table 2; Fig. 4). The methanogenic capacities of tundra wetland soils and mildly acidic peat are likewise inhibited by volatile fatty acids under mildly acidic conditions (26, 29, 62). At an extracellular pHout of <4.7 (pKaacetic acid = 4.7), undissociated acetic acid is abundant and can permeate the cellular membrane. Inside the cell, at an assumed near-neutral intracellular pHin ≫ pKaacetic acid, the acid dissociates and leads to a decoupling of the membranous proton motive force. The resulting decrease in the intracellular pH in poorly buffered cells and the decoupling of the proton motive force can be lethal to the cell (31, 46). Thus, the decrease in methanogenesis in TSB-supplemented microcosms was probably due to the inhibitory effects of the fatty acids that were produced from TSB (which contained glucose). Microorganisms capable of producing volatile fatty acids and H2 include clostridial (23) and Propionispira (49) species; both types of bacteria were detected in bog peat (Fig. 5). Novel, acid-tolerant clostridial strains have been isolated from similar bog peats (27, 35).
The inhibition of methanogenesis by formate was relieved after formate had been consumed. The ratio of formate consumed to CO2, H2, and acetate produced approximated 8:6:4:1 (reaction 1), a stoichiometry that is consistent with the combined activities of formate dehydrogenase and hydrogenase (reaction 2) and formate- or H2-dependent acetogenesis (reactions 3 and 4 respectively) (12):
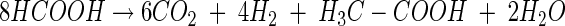 |
(1) |
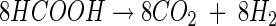 |
(2) |
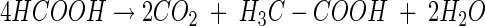 |
(3) |
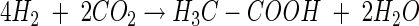 |
Although these results suggest that acetogens occur in this acidic habitat, it should be noted that most known acetogens have relatively neutral pH optima (12).
Most known methanogens grow optimally at near-neutral pH (4, 65). However, an unclassified Methanobacterium isolate (63) and a strain of Methanosarcina barkeri (5) can tolerate pH 4. The in situ emission of CH4 from the bog at pH 4.3 to 4.5 and the relative abundance of culturable methanogens in the acidic peat (approximately 1% of the general fermentative anaerobes at a cultivation pH of 4.5 [Table 3]) demonstrated that the methanogens of this bog are functional under these acidic conditions. Indeed, an increase in pH from 4.5 to 6.5 did not result in higher rates of CH4 production by methanogenic enrichments (Fig. 4). In contrast, methanogens obtained from other peats or acidic bog sediments are significantly stimulated by neutral pH and account for only 0.1% of the cultured anaerobes (17, 64).
Effect of antibiotics.
The production of CH4 by cultured methanogens was inhibited by streptomycin, kanamycin, and the β-lactam antibiotic penicillin (Table 3 and data not shown), indicating that the cultured methanogens were sensitive to at least one of these antibiotics. Some methanogens are sensitive to the antibiotics used. Methanotrix soehngenii and the moderately acid-tolerant Methanobacterium espanolae are sensitive to β-lactam antibiotics and kanamycin, respectively (25, 43). Additional antibiotics that inhibit protein synthesis also inhibit certain methanogens (21). Thus, antibiotics do not always selectively inhibit only bacteria and cannot be used to definitively differentiate between bacterial and methanogenic activities.
Phylogenetic identification of methanogens.
Much of the experimental evidence outlined above indicated that H2-CO2 was the main precursor for methanogenesis in the peat, and the main goal of the cultivation experiments was to identify in situ relevant hydrogenotrophic methanogens. Most of the archaeal sequences obtained from MPN dilutions and enrichments were phylogenetically related to Methanobacteriaceae and Methanomicrobiales; most known species of these two groups are capable of hydrogenotrophc methanogenesis (4, 16). Methanobacteriaceae- and Methanomicrobiales-related methanogens were also detected in an acidic Welsh peat and a Minnesota peatland (41, 63), and three of the archaeal bog peat sequences (AMC 1, AMC 2, and AEC 1) obtained in the present study were closely affiliated with a Methanomicrobiales-related sequence that was derived from one of these peats (20). Based on the MPN analyses and 16S rRNA gene sequences that were retrieved from the highest CH4-positive MPN dilutions of bog peat, cultivated cell numbers of Methanobacterium- and Methanomicrobiales-related species approximated 3 × 107 and 1 × 106 organism g (dry wt) of soil−1, respectively, indicating that these organisms contribute to the in situ production of CH4 in this bog peat.
In contrast, Methanosarcinales-related species appear to occur only in small numbers in this acidic peat since the sequences that were retrieved from the CH4-positive MPN dilutions were not affiliated with the Methanosarcinales cluster and since (semi) specific PCR amplifications were necessary to obtain Methanosarcinales-related sequences (AED 1 and AED 2) from the methanogenic bog peat enrichments. Methanosarcinales-related species might play a role in other peatlands (20, 36). Regional differences in the structure and function of the methanogenic communities in acidic peats have not been investigated.
In conclusion, the results of the present study indicate that in situ relevant acid-tolerant methanogens of this acidic peat belong to the Methanomicrobiales and Methanobacteriaceae, are hydrogenotrophic, and are inhibited by small amounts of acetate or other volatile fatty acids at in situ pH. Hydrogenotrophic methanogens may be symbiotically associated with hydrogen-producing anaerobes and keep H2 levels low during the normal flow of carbon and reductant in this bog peat.
Acknowledgments
We thank Anja Grießhammer, Petra Dietrich, Bettina Popp, and Kerstin Moser for technical assistance.
Support for this study was provided by the German Ministry of Education, Research and Technology (BEO 51-0339476C).
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aselmann, I., and P. J. Crutzen. 1989. Global distribution of natural freshwater wetlands and rice paddies, their net primary productivity, seasonality and possible methane emission. J. Atmos. Chem. 8:307-359. [Google Scholar]
- 3.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boone, R. B., W. B. Whitman, and P. Rouviere. 1993. Diversity and taxonomy of methanogens, p. 35-80. In J. G. Ferry (ed.), Methanogenesis. Chapman & Hall, New York, N.Y.
- 5.Bryant, M. P., and D. R. Boone. 1987. Emended description of strain MST (DSM 800T), the type strain of Methanosarcina barkeri. Int. J. Syst. Bacteriol. 37:169-170. [Google Scholar]
- 6.Capone, D. G., and R. P. Kiene. 1988. Comparison of microbial dynamics in marine and freshwater sediments: contrasts in anaerobic carbon metabolism. Limnol. Oceanogr. 4:725-749. [Google Scholar]
- 7.Chasar, L. S., J. P. Chanton, P. H. Glaser, D. I. Siegel, and J. S. Rivers. 2000. Radiocarbon and stable carbon isotopic evidence for transport and transformation of dissolved organic carbon, dissolved inorganic carbon, and CH4 in a northern Minnesota peatland. Global Biogeochem. Cycles 14:1095-1108. [Google Scholar]
- 8.Clymo, R. S. 1987. The ecology of peatlands. Sci. Pro. 71:593-614. [Google Scholar]
- 9.De Long, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Man, J. C. 1975. The probability of most probable numbers. Eur. J. Appl. Microbiol. 1:67-78. [Google Scholar]
- 11.Doddema, H. J., and G. D. Vogels. 1978. Improved identification of methanogenic bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 36:752-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drake, H. L. 1994. Acetogenesis, acetogenic bacteria, and the acetyl-CoA “Wood/Ljungdahl” pathway: past and current perspectives, p. 3-60. In H. L. Drake (ed.), Acetogenesis. Chapman & Hall, New York, N.Y.
- 13.Drake, H. L., N. G. Aumen, C. Kuhner, C. Wagner, A. Grießhammer, and M. Schmittroth. 1996. Anaerobic microflora of Everglades sediment: effects of nutrients on population profiles and activities. Appl. Environ. Microbiol. 62:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferry, J. G. 1992. Methane from acetate. J. Bacteriol. 174:5489-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fung, I., J. John, J. Lerner, E. Matthews, M. Prather, L. P. Steele, and P. J. Fraser. 1991. Three-dimensional model synthesis of the global methane cycle. J. Geophys. Res. 96:13033-13065. [Google Scholar]
- 16.Garcia, J. L., B. K. C. Patel, and B. Ollivier. 2000. Taxonomic, phylogenetic, and ecological diversity of methanogenic Archaea. Anaerobe 6:205-226. [DOI] [PubMed] [Google Scholar]
- 17.Goodwin, S., and J. G. Zeikus. 1987. Ecophysiological adaptions of anaerobic bacteria to low pH: analysis of anaerobic digestion in acidic bog sediments. Appl. Environ. Microbiol. 53:57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorham, E. 1991. Northern peatlands: role in the carbon cycle and probable responses to climate warming. Ecol. Appl. 1:182-195. [DOI] [PubMed] [Google Scholar]
- 19.Gunsalus, R. P., J. A. Romesser, and R. S. Wolfe. 1978. Preparation of coenzyme M analogues and their activity in the methyl coenzyme M reductase system of Methanobacterium thermoautotrophicum. Biochem. J. 17: 2374-2377. [DOI] [PubMed] [Google Scholar]
- 20.Hales, B. A., C. Edwards, D. A. Ritchie, G. Hall, R. W. Pickup, and J. R. Saunders. 1996. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilpert, R., J. Winter, W. Hammes, and O. Kandler. 1981. The sensitivity of archaebacteria to antibiotics. Zentbl. Bakteriol. Mikrobiol. Hyg. 1 Abt Orig. C 2:21-31. [Google Scholar]
- 22.Hines, M. E., K. N. Duddleston, and R. P. Kiene. 2001. Carbon flow to acetate and C-1 compounds in northern wetlands. Geophys. Res. Lett. 28:4251-4254. [Google Scholar]
- 23.Hippe, H., J. R. Andreesen, and G. Gottschalk. 1992. The genus Clostridium—nonmedical, p. 1800-1866. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer Verlag, New York, N.Y.
- 24.Hungate, R. E. 1969. A roll tube method for cultivation of strict anaerobes. Methods Microbiol. 3B:117-132. [Google Scholar]
- 25.Huser, A. A., K. Wuhrmann, and A. J. B. Zehnder. 1982. Methanotrix soehngenii gen. nov. sp. nov., a new acetotrophic, non-hydrogen-oxidizing methane bacterium. Arch. Microbiol. 132:1-9. [DOI] [PubMed] [Google Scholar]
- 26.Kotsyurbenko, O. R., A. N. Nozhevnikova, T. I. Soloviova, and G. A. Zavarzin. 1996. Methanogenesis at low temperatures by microflora of tundra wetland soil. Antonie Leeuwenhoek 69:75-86. [DOI] [PubMed]
- 27.Kuhner, C. H., C. Matthies, G. Acker, M. Schmittroth, A. Gößner, and H. L. Drake. 2000. Clostridium akagii sp. nov. and Clostridium acidisoli sp. nov.: acid-tolerant, N2-fixing clostridia isolated from acidic forest soil and litter. Int. J. Syst. Evol. Microbiol. 50:873-881. [DOI] [PubMed] [Google Scholar]
- 28.Küsel, K., and H. L. Drake. 1995. Effects of environmental parameters on the formation and turnover of acetate by forest soils. Appl. Environ. Microbiol. 61:3667-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landsdown, J. M., P. D. Quay, and S. L. King. 1992. CH4 production via CO2 reduction in a temperate bog: a source of 13C-depleted CH4. Geochim. Cosmochim. Acta 56:3493-3503. [Google Scholar]
- 30.Lovely, D. R., and M. J. Klug. 1986. Model for the distribution of sulfate reduction and methanogenesis in freshwater sediments. Geochim. Cosmochim. Acta 50:11-18. [Google Scholar]
- 31.Luli, G. W., and W. R. Strohl. 1990. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Environ. Microbiol. 56:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 33.Matschonat, G., and E. Matzner. 1996. Soil chemical properties affecting NH4+ sorption in forest soils. Z. Pflanzenernaehr. Bodenkd. 159:505-511. [Google Scholar]
- 34.Matthies, C., A. Grießhammer, M. Schmittroth, and H. L. Drake. 1999. Evidence for involvement of gut-associated denitrifying bacteria in emission of nitrous oxide (N2O) by earthworms obtained from garden and forest soils. Appl. Environ. Microbiol 65:3599-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthies, C., C. H. Kuhner, G. Acker, and H. L. Drake. 2001. Clostridium uliginosum sp. nov., a novel acid-tolerant, anaerobic bacterium with connecting filaments. Int. J. Syst. Evol. Microbiol. 51:1119-1125. [DOI] [PubMed] [Google Scholar]
- 36.McDonald, I. R., M. Upton, G. Hall, R. W. Pickup, C. Edwards, J. R. Saunders, D. A. Ritchie, and J. C. Murrell. 1999. Molecular ecological analysis of methanogens and methanotrophs in blanket peat bog. Microb. Ecol. 38:225-233. [DOI] [PubMed] [Google Scholar]
- 37.McInerney, M. J., and M. P. Bryant. 1980. Syntrophic associations of hydrogen utilizing methanogenic bacteria and hydrogen producing alcohol and fatty-acid degrading bacteria in anaerobic degradation of organic matter, p. 117-126. In G. Gottschalk, N. Pfennig, and H. Werner (ed.), Anaerobes and anaerobic infections. Gustav Fischer Verlag, Munich, Germany.
- 38.McInerny, M. J., and M. P. Bryant. 1981. Basic principles of bioconversions in anaerobic digestion and methanogenesis, p. 277-296. In S. S. Sofer and O. R. Zaborsky (ed.), Biomass conversion processes for energy and fuels. Plenum, New York, N.Y.
- 39.Muyzer, G., T. Brinkhoff, U. Nübel, C. M. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, 3rd ed., vol. 3.4.4. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 40.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 41.Nercessian, D., A. Upton, D. Lloyd, and C. Edwards. 1999. Phylogenetic analysis of peat bog methanogen populations. FEMS Microbiol. Lett. 173:425-429. [DOI] [PubMed] [Google Scholar]
- 42.Nüsslein, B., K. J. Chin, W. Eckert, and R. Conrad. 2001. Evidence for anaerobic syntrophic acetate oxidation during methane production in the profundal sediment of subtropical Lake Kinneret (Israel). Environ. Microbiol. 3:460-470. [DOI] [PubMed] [Google Scholar]
- 43.Patel, G. B., G. D. Sprott, and J. E. Fein. 1990. Isolation and characterization of Methanobacterium espanolae sp. nov., a mesophilic, moderately acidiphilic methanogen. Int. J. Syst. Bacteriol. 40:12-18. [Google Scholar]
- 44.Popp, T. J., J. P. Chanton, G. J. Whiting, and N. Grant. 1999. Methane stable isotope distribution at a Carex dominated fen in north central Alberta. Global Biogeochem. Cycles 13:1063-1077. [Google Scholar]
- 45.Raskin, L., J. M. Stromley, B. E. Rittman, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russell, J. B. 1991. Intracellular pH of acid-tolerant ruminal bacteria. Appl. Environ. Microbiol. 57:3383-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schink, B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 64:262-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schink, B. 1992. Syntrophism among prokaryotes, p. 276-299. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer Verlag, New York, N.Y.
- 49.Schink, B., T. E. Thompson, and J. G. Zeikus. 1982. Characterization of Propionispira arboris gen. nov. sp. nov., a nitrogen-fixing anaerobe common to wetwoods of living trees. J. Gen. Microbiol. 128:2771-2779. [Google Scholar]
- 50.Schnürer, A., G. Zellner, and B. H. Svensson. 1999. Mesophilic syntrophic acetate oxidation during methane formation in biogas reactors. FEMS Microbiol. Ecol. 29:249-261. [Google Scholar]
- 51.Shannon, R. D., and J. R. White. 1996. The effects of spatial and temporal variations in acetate and sulfate on methane cycling in two Michigan peats. Limnol. Oceanogr. 41:435-443. [Google Scholar]
- 52.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K.-H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes in bacterial systematics, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 54.Svensson, B. H. 1984. Different temperature optima for methane formation when enrichments from acid peat are supplemented with acetate or hydrogen. Appl. Environ. Microbiol. 48:389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urquhart, C., and A. J. P. Gore. 1973. The redox characteristics of four peat profiles. Soil Biol. Bichem. 5:659-672. [Google Scholar]
- 56.Van den Berg, L., G. B. Patel, D. S. Clarke, and C. P. Lentz. 1976. Factors affecting rate of methane formation from acetic acid by enriched methanogenic cultures. J. Can. Microbiol. 22:1312-1319. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Y. S., W. S. Odle, W. E. Eleazer, and M. A. Barlaz. 1997. Methane potential of food waste and anaerobic toxicity of leachate produced during food waste decomposition. Waste Manage. Res. 15:149-167. [Google Scholar]
- 58.Westermann, P. 1993. Wetland and swamp microbiology, p. 215-238. In T. E. Ford (ed.), Aquatic microbiology: an ecological approach. Blackwell Scientific Publications, Oxford, United Kingdom.
- 59.Whiticar, M. J., E. Faber, and M. Schoell. 1986. Biogenic methane formation in marine and freshwater environments: CO2 reduction vs. acetate fermentation - isotope evidence. Geochim. Cosmochim. Acta 50:693-607. [Google Scholar]
- 60.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer Verlag, New York, N.Y.
- 61.Widdel, F., G. W. Kohring, and F. Mayer. 1983. Studies on dissimilatory sulfate- reducing bacteria that decompose fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov. sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 134:286-294. [Google Scholar]
- 62.Williams, R. T., and R. L. Crawford. 1984. Methane production in Minnesota peatlands. Appl. Environ. Microbiol. 47:1266-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams, R. T., and R. L. Crawford. 1985. Methanogenic bacteria, including an acid-tolerant strain, from peatlands. Appl. Environ. Microbiol. 50:1542-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams, R. T., and R. L. Crawford. 1983. Microbial diversity of Minnesota peatlands. Microb. Ecol. 9:201-214. [DOI] [PubMed] [Google Scholar]
- 65.Zinder, S. H. 1993. Physiological ecology of methanogens, p. 128-206. In J. G. Ferry (ed.), Methanogenesis. Chapman & Hall, New York, N.Y.