Abstract
The fate of transplastomic (chloroplast genome contains the transgene) tobacco plant DNA in planta was studied when the plant leaves were subjected to decay conditions simulating those encountered naturally, including grinding, incubation with cellulase or enzymes produced by Erwinia chrysanthemi, and attack by the plant pathogen Ralstonia solanacearum. Direct visualization of DNA on agarose gels, gene extraction yield (the number of amplifiable aadA sequences in extracted plant DNA), and the frequency that recipient bacteria can be transformed by plant DNA were used to evaluate the quality and quantity of plant DNA and the transgene. These measurements were used to monitor the physical and biological degradation of DNA inside decaying plant tissues. Our results indicate that while most of the DNA will be degraded inside plant cells, sufficient DNA persists to be released into the soil.
The development of transgenic plants in which the chloroplast genome contains the transgenes has reduced the probability of transgene transfer through pollen dispersal. Another advantage of transplastomic plants is the hyperexpression of the cloned genes (8). On the other hand, this emerging and promising plant technology increases the number of copies of any transgenes to thousands more than nuclear genetic engineering does and thus increases the frequency of gene transfer to competent bacterial cells that colonize plant tissue (15).
Most studies attempting to detect gene transfer from transgenic plants to environmental bacteria have been done with soil environments where soil bacteria might be transformed by plant DNA. Two arguments justify studies focusing on soil environments: (i) the number of prokaryotes in soil is very high, up to 109 g of soil−1, and thus, the presence of competent cells could be significant for possible bacterial transformation; (ii) transformation-mediated gene transfer could be expected to occur in soil due to the significant amount of soil extracellular DNA (11) that results from its passive release from dead microbial, vegetable, and animal cells or its active release by living cells (5, 14).
Persistence of DNA in soils is probably due to adsorption onto soil components such as sand particles, clay minerals, or humic compounds, thus protecting DNA against nuclease degradation (1, 7, 16, 18, 21). Numerous experiments have been conducted with pure DNA adsorbed onto soil components in order to study the influence of various biotic and abiotic parameters, including the size and conformation of DNA (10, 13, 16, 22-24). Other studies have used ground plant material (29) or whole leaves (28) buried in soils for various periods under field and laboratory conditions before plant DNA was quantitatively analyzed. When experiments were conducted with soil microcosms, PCR tests indicated that most of the plant DNA was rapidly lost, although a proportion remained detectable in the soil (28, 29). Although others have found detectable transgenic plant sequences in field soil, where the presence of plant tissues was no longer detectable (12, 20), the decaying plant process certainly affects DNA stability and persistence over time (27). During plant senescence, active nucleases can degrade DNA. During plant decay in soil, the activity of vegetable nucleases continues because these enzymes tend to remain active for some time after cell death (17). Moreover, during plant decay, plant DNA can be also degraded by nucleases of microbial cells that degrade plant residues. In addition to nucleases, other microbial enzymes (cellulases, proteases, ligninases, etc.) involved in the degradation of plant polymers are active during plant decay, and their activity might facilitate the access of microbial nucleases to plant DNA. No information is available on the effects of nucleases and other enzymes on transgenic DNA persistence. Another limitation to most of the studies described above is that they have not considered the possible biological activity (transforming ability) of the PCR-detected DNA.
The objective of this paper was to study the fate of DNA in transgenic plant cells throughout the decay process of plant material under conditions simulating those encountered in nature. For this purpose, we used transplastomic (chloroplast genome contains transgene) tobacco plant leaves containing the aadA gene. Construction of the transplastomic tobacco plant Nicotiana tabacum cv. PBD6 was described previously (15). This plant was generated by particle bombardment as described by Staub and Maliga (26) and contained the aadA gene conferring resistance to spectinomycin and streptomycin integrated in the plastid genome. The leaves were incubated with extracellular enzymes from the plant pathogen Erwinia chrysanthemi strain A576, which was cultured overnight in 250 ml of M63 medium at 28°C until reaching an optical density at 600 nm (OD600) of 2.10 according to the method described by Moulard et al. (19). The culture was centrifuged at 5,000 × g for 20 min, and the supernatant, containing a cocktail of pectinolytic enzymes, was stored for several hours before use at 4°C in the presence of some drops of chloroform. The leaves were also incubated with a cellulase-containing solution from Trichoderma viride (Merck-Eurolab), which was prepared at concentrations of 0.08 g ml−1 in 0.1 M Na-acetate buffer (pH 4) according to the manufacturer's recommendations. The DNase activity of each enzymatic solution was determined with plasmid pLEP01 or purified genomic tobacco DNA as a substrate according to the method of Moulard et al. (19). At various stages, the plant DNA was extracted by cutting green leaves into equivalent pieces of about 0.5 g. These pieces were incubated in two separate Erlenmayer flasks in the presence of 100 ml of cellulase or pectinolytic solution. At least three plant pieces were recovered at each time point after 0, 1, 24, and 72 h of incubation at 23°C. The plant pieces were ground in liquid nitrogen, weighed, and stored in Eppendorf tubes at −20°C until undergoing DNA extraction with the DNeasy plant DNA miniextraction kit (Qiagen). This kit was also used to extract and purify DNA from plant leaves that were used as controls, which included incubation of plant pieces in each buffer (M63 medium and 0.1 M Na-acetate buffer [pH 4]) without the enzymes. Finally, green leaf pieces were directly ground in liquid nitrogen, and the resulting dry material was incubated at room temperature and DNA extracted to assess DNA degradation over time.
Finally, tobacco leaves removed from the plant were also inoculated with pathogenic Ralstonia solanacearum strain K60. The inoculum consisted of R. solanaceraum cells (OD600 of 1.52) grown overnight in rich BG medium (4) supplemented with 12 μg of gentamicin ml−1 (Sigma). This inoculum was centrifuged, rinsed with sterile water, centrifuged again, and resuspended in the appropriate amount of sterile water to provide a concentration of 8.8 × 108 cells ml−1. Three hundred microliters of the bacterial suspension was inoculated into the central and secondary veins of the leaf as described by Bertolla et al. (2), and plant infection was left to proceed at room temperature for more than 1 week. Pieces of the leaf were cut on days 0, 5, and 8 and treated as previously described for storage and DNA extraction.
The concentration of extracted DNA was measured spectrophotometrically (OD260), and aliquots were used to estimate DNA degradation by electrophoresis on agarose gels. The persistent transgene DNA sequences were quantified by real-time PCR. DNA was amplified with the Light Cycler thermocycler (Roche, France) with initial denaturation at 95°C for 480 s before 45 cycles of 95°C for 10 s, 55°C for 8 s, and 72°C for 16 s. Primers p415 (5′-ATTCCGTGGCGTTAT-3′) and p416 (5′-TGACGGGCTGATACT-3′) were complementary to part of the aadA gene amplifying a 382-bp fragment. Plasmid pLEP01, previously used to transform plants, and which contained the aadA marker gene, was serially diluted in ultrapure water from 3.2 × 100 ng to 3.2 × 10−5 ng and used as a PCR template for definition of calibration curves. Data were analyzed with the Light Cycler software (version 5.3). The crossing points were assessed and plotted versus concentrations of standards. All samples were tested in triplicate, and the average value of the three replicates was used for quantification. Quantity and relative loss were calculated in order to evaluate the number of aadA target sequences in the total plant DNA during the degradation processes.
Finally, the loss of the transformation potential of the extracted DNA was measured by using each DNA solution to transform Acinetobacter sp. strain BD413, which harbors the recombinant plasmid pBAB2 (15), in which plastid sequences have been cloned to favor homologous recombination with transplastomic tobacco sequences. Acinetobacter sp. strain BD413 (DSM586) was cultured on Luria-Bertani (LB) medium (10g of Bacto tryptone, 5 g of yeast extract, and 5 g of NaCl in 1 liter of distilled water) containing 20 μg of nalidixic acid ml−1 (Sigma Chemical Co., St. Louis Mo.). An overnight culture of the strain was diluted 25-fold into fresh medium and cultured for an additional 2 h at 28°C. A bacterial suspension volume of 180 μl was added to 20 μl of concentrated plant DNA solutions of 20 and 40 μg ml−1, providing total amounts of 400 and 800 ng, respectively. The resulting mixtures were thoroughly mixed and incubated for 120 min at 28°C. In control experiments, DNA was replaced by sterile water. Transformants were selected on LB medium containing nalidixic acid (20 μg ml−1), ampicillin (50 μg ml−1), and spectinomycin (50 μg ml−1), while recipient cells were grown on the same medium without spectinomycin. Colonies were counted after 2 to 3 days of incubation at 28°C. Three repetitions were carried out for each sample.
The presence of chloroplast and aadA gene sequences in recombinant clones was controlled by PCR with p1531cpl2up (5′-TTTCTATTGTTGTCTTGGAT-3′) and p416 (5′-TGACGGGCTGATACT-3′) as the forward and reverse primers, respectively. The cycle consisted of a touchdown PCR in which denaturing and elongation steps were done at 95 and 72°C and the annealing temperature decreased by 2°C after 2 cycles starting from 60°C to 50°C, followed by 25 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min. The last cycle was done at 72°C with a 7-min extension temperature before storage at 4°C. All treatments and analyses were done in triplicate, allowing calculation of the average and standard deviation parameters.
Plant DNA degradation kinetics in ground plant leaf.
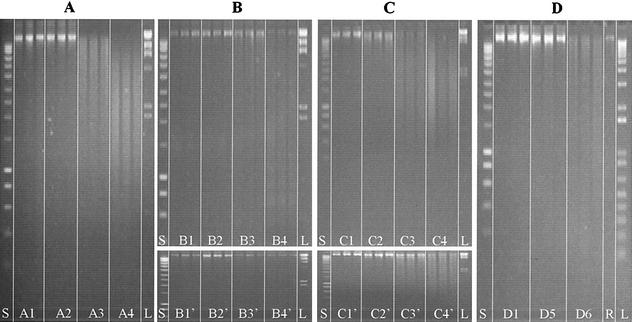
After being ground in liquid nitrogen, the resulting plant material was incubated at room temperature before the DNA was extracted and analyzed. Agarose gel electrophoresis indicates that DNA degradation occurred rapidly, as illustrated by the size distribution, which differed significantly after 24 h from those after 0 and 1 h (Fig. 1A). Degradation was even more significant after 72 h, when the longest DNA fragments had disappeared completely. Total DNA yield, as estimated by spectrophotometry (OD260), was found to remain nearly constant for 24 h (67.3 ng mg−1) before a drop at 72 h (22.2 ng mg−1) (Table 1), corresponding to a 57% genomic DNA loss. The specific aadA sequence was quantified by real-time PCR to be 6.77 × 104 (± 1.53 × 103) sequences ng of total DNA−1 (Table 2), which corresponds to a relative gene loss of 98.2% after 72 h. Thus, both the total DNA and the gene aadA appear to decrease drastically in concentration between 24 and 72 h, indicating that plant nucleases were active. Interestingly, the relative loss of total DNA was about 50% over 72 h, while that of the aadA sequences was >98%, indicating that the shearing process decreased the average size of the DNA fragments, thus preventing amplification of aadA sequences. Transformation tests with Acinetobacter calcoaceticus (pBAB2) were also carried out with 400 or 800 ng of DNA extracted from the various samples (Table 2). Transformation frequencies with the lowest quantity of DNA were reduced by 2 orders of magnitude. DNA extracted directly after grinding or after 1 h of incubation provided transformation frequencies ranging across the same order of magnitude (7.7 × 10−4 ±1.9 × 10−4 and 9.1 × 10−4± 3.8 × 10−4). The DNA from samples incubated for 24 h had a significantly lower transformation frequency (by about 3 orders of magnitude: 0.21 × 10−7 ± 0.15 × 10−7), and the frequency remained at that level when DNA originated from samples incubated for an additional 48 h (t = 72 h), indicating that DNA lost most of its biological potential due to plant nuclease activity (Table 2).
FIG. 1.
Kinetics of in planta DNA degradation as estimated by agarose gel electrophoresis of DNA extracted from plant tissues. Three replicates were electrophoresed for each treatment, including incubation times of 0 h (lanes A1, B1, B1′, C1, C1′, and D1), 1 h (lanes A2, B2, B2′, C2, and C2′), 24 h (lanes A3, B3, B3′, C3, and C3′), 72 h (lanes A4, B4, B4′, C4, and C4′), 5 days (lane D5), or 8 days (lane D6). In panel A, plant leaves were ground in liquid nitrogen and left to incubate at room temperature. In panel B, tobacco leaves were incubated with the supernatant of E. chrysanthemi cultures containing pectate lyase and nuclease enzymes, and in panel B′, the leaves were incubated with the sterile M63 culture medium as a control. In panel C, plant leaves were submitted to a commercial cellulase preparation, and in panel C′, they were submitted to the corresponding dilution buffer as a control. Finally, in panel D, plant leaves that had been cut out of the plant were infected in vitro with 108 cells of R. solanacearum strain K60. Lane S, smart ladder; lane L, marker II; and lane R, genomic DNA from R. solanacearum.
TABLE 1.
Kinetics of DNA degradation inside plant leaves subjected to grinding, incubation with enzymatic solutions, and infection by R. solanacearum as estimated by extracted DNA yield
| Plant leaf treatment group | Extracted DNA yield (ng mg of plant leaf−1)
|
|||
|---|---|---|---|---|
| Grinding | Incubation with:
|
Infection with R. solanacearum | ||
| E. chrysanthemi enzymes | Cellulase enzymes | |||
| 0 h | ||||
| Treated | 51.6 ± 16.2 | 42.8 ± 17.8 | 73.5 ± 39.3 | 50.2 ± 3.3 |
| Control | 39.8 ± 7.7 | 32.3 ± 6.6 | ||
| 1 h | ||||
| Treated | 59.5 ± 13.9 | 42.4 ± 7.8 | 71.3 ± 10.7 | NDa |
| Control | 19.9 ± 5.2 | 39.6 ± 1.2 | ||
| 24 h | ||||
| Treated | 67.3 ± 2.2 | 103.3 ± 15.9 | 62 ± 8 | ND |
| Control | 46.2 ± 9.9 | 24.8 ± 6.1 | ||
| 72 h | ||||
| Treated | 22.2 ± 4 | 26.8 ± 0.9 | 12.3 ± 8.9 | ND |
| Control | 64.9 ± 2.4 | 15.2 ± 1.3 | ||
| 5 days | ||||
| Treated | ND | ND | ND | 236.5 ± 74.8 |
| Control | ||||
| 8 days | ||||
| Treated | ND | ND | ND | 509.6 ± 22.5 |
| Control | ||||
ND, not determined.
TABLE 2.
Kinetics of DNA degradation inside plant leaves subjected to grinding, incubation with enzymatic solutions, and infection by R. solanacearum as estimated by the number of amplifiable aadA gene sequences and transformation frequency of a recipient Acinetobacter sp. bacteriuma
| Plant leaf treatment time | Grinding
|
Incubation with:
|
Infection with R. solanacearum
|
|||||
|---|---|---|---|---|---|---|---|---|
|
E. chrysanthemi enzymes
|
Cellulase enzymes
|
|||||||
| aadA gene no. | Transformation frequency | aadA gene no. | Transformation frequency | aadA gene no. | Transformation frequency | aadA gene no. | Transformation frequency | |
| 0 | 3.74 × 106 (1.21 × 106) | 7.7 × 10−4 (1.9 × 10−4) | 1.73 × 106 (5.66 × 105) | 2.5 × 10−5 (2 × 10−5) | 6.60 × 106 (1.1 × 106) | 6.2 × 10−6 (9.9 × 10−5) | 1.54 × 106 (8.01 × 105) | 1.2 × 10−5 (3 × 10−5) |
| 1 h | 6.11 × 106 (7.92 × 105) | 9.1 × 10−4 (3.8 × 10−4) | 2.85 × 106 (6.09 × 105) | 1.9 × 10−5 (2.8 × 10−5) | 2.96 × 106 (3.7 × 105) | 3.0 × 10−6 (3.8 × 10−5) | NDb | ND |
| 24 h | 8.87 × 105 (1.52 × 105) | 2.1 × 10−6 (1.5 × 10−6) | 2.08 × 106 (1.05 × 105) | 7.9 × 10−7 (3.8 × 10−6) | 1.46 × 106 (1.4 × 105) | 4.9 × 10−7 (1.5 × 10−5) | ND | ND |
| 72 h | 6.77 × 104 (1.53 × 103) | 2.3 × 10−6 (4.6 × 10−6) | 5.00 × 104 (8.17 × 103) | 0.0 × 100 (0) | 5.68 × 105 (2.6 × 103) | 1.1 × 10−8 (1.2 × 10−7) | ND | ND |
| 5 days | ND | ND | ND | ND | ND | ND | 4.61 × 106 (1.57 × 106) | 4.5 × 10−6 (1 × 10−5) |
| 8 days | ND | ND | ND | ND | ND | ND | 4.97 × 106 (7.35 × 105) | 4.4 × 10−8 (5.4 × 10−7) |
Standard deviation is indicated in parentheses.
ND, not determined.
Plant DNA degradation kinetics in pectinase-treated plant leaves.
In nature, enzymes such as pectinases or cellulases are produced by saprophytic or pathogenic organisms and are involved in the decay process of plant materials. To simulate such enzymatic attacks, we tested a cocktail of compounds produced by the pathogen E. chrysanthemi by using the supernatant of the growth medium in which the bacterium was cultured. According to Moulard et al. (19), such a solution contains not only pectinases and pectin methyl esterases, but also nucleases, as confirmed by the ability of this solution to degrade pure plasmid or plant DNA (results not shown). Leaves incubated with the supernatant of E. chrysanthemi cultures remained green for 2 days before turning to a light brown color on the 3rd day. Control samples treated with the M63 culture medium did not undergo this color change. Agarose gel electrophoresis of DNA solutions extracted from the leaf pieces indicated that DNA degradation could be observed only after 72 h. Control samples incubated in the sterile M63 medium provided very similar DNA patterns throughout the incubation time, thus indicating the degradation observed previously was actually due to E. chrysanthemi extracellular solution (Fig. 1B). Spectrophotometric quantification of DNA (Table 1) indicated that there was no loss of DNA in cells treated for 24 h with the pectinase-containing E. chrysanthemi extracellular solution (103.3 ± 15.9 ng mg−1).
Moreover, incubation of plant leaves for 1 h in either the sterile or E. chrysanthemi culture M63 medium increased the efficiency of the DNeasy plant DNA miniextraction kit, leading to greater quantities of DNA extracted after incubation (Table 1). However, the total amount of extracted DNA decreased to more than 37% after incubation for 72 h (26.8 ± 0.9 ng mg−1) in the pectinase-containing solution, confirming electrophoresis results from degradation of DNA occurring at this stage. In addition, the real-time PCR-estimated aadA sequence number, which remained nearly unchanged until 24 h (2.08 × 106 ±1.05 × 105), dropped more than 97% after 3 days (5.00 × 104 ± 8.17 × 103). This decrease was not observed in control leaves incubated in the sterile M63 medium (results not shown).
The transformation frequency when recovered DNA was used to transform Acinetobacter sp. strain BD413 containing plasmid pBAB2 dropped significantly after 24 h (Table 2), corresponding to a transformation frequency loss of 98%, and then continued to decrease by more than 2 orders of magnitude after 72 h.
All of these results are consistent with the concept that plant DNA still present inside plant cells was subjected to degradation.
Plant DNA degradation kinetics in cellulase-treated plant leaves.
The color of tobacco leaves submitted to a commercial cellulase preparation darkened within the first 12 h, and the leaves lost their rigidity, becoming soft and flaccid, whereas samples incubated in sterile Na-acetate buffer remained green and rigid. DNA degradation occurring inside the transgenic tobacco plant cells treated with cellulase was faster than when plants were treated with pectinase-containing E. chrysanthemi extracellular solution. DNA smears were detected after incubation for only 1 h in the cellulase-containing solution (Fig. 1C). Larger DNA fragments present after 1 h were gradually degraded during their incubation. After 72 h, most of the fragments were below 2 kb in size (Fig. 1C). Incubation of leaf pieces in sterile Na-acetate buffer also resulted in DNA degradation within 24 h. However, smear patterns indicated that DNA was much less degraded in these control samples than in those treated with cellulase-containing solution (Fig. 1C and C′). The amounts of recovered and spectrophotometrically quantified DNA indicated that cellulase-treated leaves experienced a gradual loss of DNA inside the plant cells (Table 1). However, the incubation of plant leaf pieces in sterile Na-acetate buffer also resulted in a decrease in significant amounts of DNA (15.2 ng mg−1, corresponding to a loss of 52.8%), although not as much as with the cellulase solution (12.3 ng mg−1, corresponding to a loss of 83.2%). A gradual decrease in the aadA gene sequences was also quantified by real-time PCR amplification (Table 2). After 72 h of incubation in cellulase and Na-acetate buffer, losses of 91.4 and 99.4%, respectively, were observed.
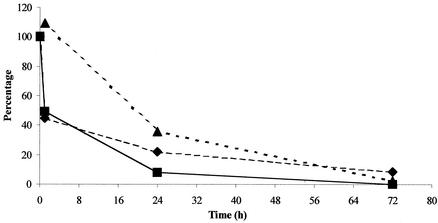
The transformation frequency of Acinetobacter sp. strain BD413, which contained the plasmid pBAB2, with DNA from cellulase-treated leaves decreased to 1.1 × 10−8 after 72 h (Table 2), which corresponds to a 99.8% decrease. In this case, we calculated the kinetics of transformation efficiency, which can be defined as the ratio between transformation frequency and the number of aadA gene sequences. A regular decrease in transformation efficiency during the decay process (Fig. 2) suggests that the number of aadA sequences quantified by real-time PCR is only a relative indicator of the biological potential. These data also confirm the need for additional studies to determine if persistent transgenic sequences detected in field soils (12, 20) maintain their biological potential.
FIG. 2.
Percentage change of transformation frequency (▪), gene number (♦), and transformation efficiency (as transformation frequency/number of aadA genes) (▴) for the experiment with 800 ng of DNA during exposure to the cellulase-containing solution.
Plant DNA degradation kinetics in R. solanacearum-infected plant leaves.
Tobacco leaves, which were cut out of the plant and infected in vitro with 2.64 × 108 cells of R. solanacearum strain K60, exhibited wilting symptoms similar to those observed in greenhouse plants infected by R. solanacearum. Extracted DNA exhibited patterns on agarose gel (Fig. 1D) indicating that the DNA maintained a high level of integrity for at least 5 days following bacterial inoculation (Fig. 1D, lanes D5). However, at t = 8 days, a smear could be detected, the loss of the higher fragments indicating that degradation occurred (Fig. 1D, lanes D6). Spectrophotometric quantification of DNA indicated that the amounts of DNA material extracted from contaminated plants increased regularly and significantly. An increase of more than 1 order of magnitude between control plants and those infected by R. solanacearum for 8 days was observed (Table 1). When the various extracted DNA samples were used as templates for real-time PCR, a small increase in the plant-specific aadA gene sequences was detected (from 1.54 × 106 ± 8.01 × 105 to 4.97 × 106 ± 7.35 × 105)(Table. 2). The transformation frequency, which was 1.2 × 10−5 ± 3 × 10−5 at t = 0, was found to decrease when plant DNA extracted 5 days (4.5 × 10−6 ± 1 × 10−5) and 8 days (4.4 × 10−8 ± 5.4 × 10−7) after bacterial infection was used.
These results indicate an increase in the total DNA yield, while the two other approaches, which target plant DNA specifically (PCR- and transformation-based determination of the number of plant aadA sequences), indicated that plant DNA was degraded. These data could be related to a release of bacterial DNA during plant infection. Moreover, the strong decrease (by 2 orders of magnitude) in transformation frequency could be directly related to the presence of R. solanacearum genomic DNA that diluted the aadA sequences in the transforming DNA solution. However, the possibility that leaf colonization by the bacteria leads to changes in plant cell structure that would improve the efficiency of DNA extraction cannot be excluded.
A potential transfer of aadA sequences from the plant to the bacteria is not a valid hypothesis, because transformation of R. solanacearum in planta (3) is not favored. Moreover, the strain used in this study does not harbor sequences in which homologous recombination with plastid sequences could occur.
DNA smears and DNA loss could be related to the “programmed cell death” (PCD) involved in plants during reproduction, embryogenesis, senescence, and other processes, such as a hypersensitive response to pathogen attacks (30). Genome fragmentation was observed in plant cells submitted to PCD (30) due to endonuclease activation, thus, ensuring the removal of unwanted DNA and the recycling of N and P sources (6).
In conclusion, the kinetics of DNA degradation in plant cells submitted to decay processes could vary, depending on various biotic and abiotic factors. This confirms the interest in studying plant DNA after plant decay, but before the DNA enters the soil. Actually, this “residuesphere,” defined as the interface between decaying plant material and the soil matrix (9, 25), was shown to be a “ hot spot ” for bacterial growth and conjugal gene transfer. Even if most of the DNA is degraded in planta, enough DNA would persist to contribute to the pool of extracellular and potentially biologically active DNA in soil.
Acknowledgments
We thank S. Reverchon for the gift of the E. chrysanthemi strain A576.
This work was done as part of the programs “Thématiques Prioritaires, Domaine Santé” from Région Rhône Alpes, “Impact des biotechnologies dans les agro-ecosystèmes” from CNRS, and TRANSBAC QLK3-CT-2001-02242 from the EU (5th RTD Program). M. T. Ceccherini is a Ph.D. student in a coordinated program involving the University of Florence and the University of Lyon I.
REFERENCES
- 1.Aardema, B. W., M. G. Lorenz, and W. E. Krumbein. 1983. Protection of sediment-adsorbed transforming DNA against enzymatic inactivation. Appl. Environ. Microbiol. 46:417-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertolla, F., A. Frostegard, B. Brito, X. Nesme, and P. Simonet. 1999. During infection of its host, the plant pathogen Ralstonia solanacearum naturally develops a state of competence and exchanges genetic material. Mol. Plant-Microbe Interact. 12:467-472. [Google Scholar]
- 3.Bertolla, F., R. Pepin, E. Passelegue-Robe, E. Paget, A. Simkin, X. Nesme, and P. Simonet. 2000. Plant genome complexity may be a factor limiting in situ the transfer of transgenic plant genes to the phytopathogen Ralstonia solanacearum. Appl. Environ. Microbiol. 66:4161-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher, C. A., P. A. Barberis, A. P. Trigalet, and D. A. Demery. 1985. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5 induced avirulent mutants. J. Gen. Microbiol. 131:2449-2457. [Google Scholar]
- 5.Catlin, B. W. 1956. Extracellular deoxyribonucleic acid of bacteria and deoxyribonuclease inhibitor. Science 124:441-442. [DOI] [PubMed] [Google Scholar]
- 6.Choi, J. J., S. J. Klosterman, and L. A. Hadwiger. 2001. A comparison of the effects of DNA-damaging agents and biotic elicitors on the induction of plant defense genes, nuclear distortion, and cell death. Plant. Physiol. 125:752-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crecchio, C., and G. Stotzky. 1998. Binding of DNA on humic acids: effect on transformation of Bacillus subtilis and resistance to DNase. Soil Biol. Biochem. 30:1060-1067. [Google Scholar]
- 8.Daniell, H., B. Muthukumar, and S. B. Lee. 2001. Marker free transgenic plants: engineering the chloroplast genome without the use of antibiotic selection. Curr. Genet. 39:109-116. [DOI] [PubMed] [Google Scholar]
- 9.Delipthay, J. R., T. Barkay, and S. J. Sorensen. 2001. Enhanced degradation of phenoxyacetic acid in soil by horizontal transfer of the tfdA gene encoding a 2,4-dichlorophenoxyacetic acid dioxygenase. FEMS Microbiol. Ecol. 35:75-84. [DOI] [PubMed] [Google Scholar]
- 10.Demanèche, S., L. Jocteur-Monrozier, H. Quiquampoix, and P. Simonet. 2001. Evaluation of biological and physical protection against nuclease degradation of clay-bound plasmid DNA. Appl. Environ. Microbiol. 67:293-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frostegård, A., S. Courtois, V. Ramisse, S. Clerc, D. Bernillon, F. Le Gall, P. Jeannin, X. Nesme, and P. Simonet. 1999. Quantification of bias related to the extraction of DNA directly from soils. Appl. Environ. Microbiol. 65:5409-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gebhard, F., and K. Smalla. 1999. Monitoring field releases of genetically modified sugar beets for persistence of transgenic plant DNA and horizontal gene transfer. FEMS Microbiol. Ecol. 28:261-272. [Google Scholar]
- 13.Greaves, M. P., and M. J. Wilson. 1969. The adsorption of nucleic acids by montmorillonite. Soil Biol. Biochem. 1:317-323. [Google Scholar]
- 14.Herdman, M., and N. G. Carr. 1971. Recombination in Anacystis nidulans mediated by extracellular DNA/RNA complex. J. Gen. Microbiol. 68:16-20. [Google Scholar]
- 15.Kay, E., T. M. Vogel, F. Bertolla, R. Nalin, and P. Simonet. 2002. In situ transfer of antibiotic resistance genes from transgenic (transplastomic) tobacco plants to bacteria. Appl. Environ. Microbiol. 68:3345-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khanna, M., and G. Stotzky. 1992. Transformation of Bacillus subtilis by DNA bound on montmorillonite and effect of DNase on the transforming ability of bound DNA. Appl. Environ. Microbiol. 58:1930-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ladd, J. N., R. C. Foster, P. Nannipieri, and J. M. Oades. 1996. Soil structure and biological activity. Soil Biochem. 9:23-78.
- 18.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moulard, M., G. Condemine, and J. Robert-Baudouy. 1993. Characterization of the nucM gene coding for a nuclease of the phytopathogenic bacteria Erwinia chrysanthemi. Mol. Microbiol. 8:685-695. [DOI] [PubMed] [Google Scholar]
- 20.Paget, E., M. Lebrun, G. Freyssinet, and P. Simonet. 1998. The fate of recombinant plant DNA in soil. Eur. J. Soil. Biol. 34:81-88. [Google Scholar]
- 21.Paget, E., and P. Simonet. 1994. On the track of natural transformation in soil. FEMS Microbiol. Ecol. 15:109-118. [Google Scholar]
- 22.Pietramellara, G., L. Dal Canto, C. Vettori, E. Gallori, and P. Nannipieri. 1997. Effects of air-drying and wetting cycles on the transforming ability of DNA bound on clay minerals. Soil Biol. Biochem. 29:55-61. [Google Scholar]
- 23.Pietramellara, G., M. Franchi, E. Gallori, and P. Nannipieri. 2001. Effect of molecular characteristics of DNA on its adsorption and binding on homoionic montmorillonite and kaolinite. Biol. Fert. Soils 33:402-409. [Google Scholar]
- 24.Poly, F., C. Chenu, P. Simonet, J. Rouiller, and L. Jocteur Monrozier. 2000. Differences between linear chromosomal and supercoiled plasmid DNA in their mechanisms and extent of adsorption on clay minerals. Langmuir 16:1233-1238. [Google Scholar]
- 25.Sengelov, G., G. A. Kowalchuk, and S. J. Sorensen. 2000. Influence of fungal-bacterial interactions on bacterial conjugation in the residuesphere. FEMS Microbiol. Ecol. 31:39-45. [DOI] [PubMed] [Google Scholar]
- 26.Staub, J. M., and P. Maliga. 1993. Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbA mRNA. EMBO J. 12:601-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas, H., and J. L. Stoddart. 1980. Leaf senescence. Annu. Rev. Plant Physiol. 31:83-111. [Google Scholar]
- 28.Widmer, F., R. J. Seidler, K. K. Donegan, and G. L. Reed. 1997. Quantification of transgenic plant marker gene persistence in the field. Mol. Ecol. 6:1-7. [Google Scholar]
- 29.Widmer, F., R. J. Seidler, and L. S. Watrud. 1996. Sensitive detection of transgenic plant marker gene persistence in soil microcosms. Mol. Ecol. 5:603-613. [Google Scholar]
- 30.Wood, N. T. 2001. Apoptosis: a way of life for plants? Trends Plant Sci. 6:451-452. [DOI] [PubMed] [Google Scholar]