Abstract
Controlled expression of cloned φX174 gene E in gram-negative bacteria results in lysis of the bacteria by the formation of a transmembrane tunnel structure built through the cell envelope complex. Production of bacterial ghosts is routinely monitored by classical microbiological procedures. These include determination of the turbidity of the culture and the total number of cells and the number of reproductive cells present during the time course of growth and lysis. Although conceptually simple, these methods are labor intensive and time consuming, providing a complete set of results after the determination of viable cell counts. To avoid culturing methods for bacterial growth, an alternative flow cytometric procedure is presented for the quantification of ghosts and polarized, as well as depolarized, nonlysed cells within a culture. For this method, which is based on the discriminatory power of the membrane potential-sensitive dye bis-(1,3-dibutylbarbituric acid) trimethine oxonol, a staining protocol was developed and optimized for the maximum discrepancy in fluorescence between bacterial ghosts and viable cells. The total quantitative analysis procedure takes less than 2 min. The results derived from classical or cytometric analyses correlate with respect to the total cell numbers and the viability of the culture.
The formation of bacterial ghosts of Escherichia coli is a well-characterized process (19, 24). Initiated by the expression of cloned phage φX174 lysis gene E, a transmembrane tunnel structure is formed in consequence of the oligomerization of protein E. Driven by the high osmotic pressure inside the cell, the cytoplasmic content is expelled into the surrounding medium, thus giving rise to empty bacterial cell envelopes. Except for the lysis hole, the morphology of the bacteria, including all cell surface structures and the cell membranes, is not affected by the lysis event (23). As this procedure is applicable to a diverse spectrum of gram-negative bacteria, the ghosts are under investigation as genetically inactivated candidate vaccines (4, 9) and as carriers of foreign antigens (3, 10).
The procedures for monitoring and characterizing the lysis process are based mainly on classical methods. The lysis of the first bacterial cells is associated with a decrease in the turbidity of the culture, which can be detected by measuring the optical density. As a consequence, the viability of the lysing culture decreases drastically, to reach a minimum at the endpoint of lysis. For E. coli, a correlation between the decrease in optical density (OD) and the reduction of viable cell counts was determined, although it was found not to be accurate. To this day, classical microbiological procedures, like plating or the use of counting chambers, are used to determine the amount of residual viable (reproductive) bacteria and the total number of cells (including ghosts) in time course experiments. As these parameters are critical for the characterization of the growing or lysing culture, a method is desired for the online determination during the lysis process. The rapid and accurate assessment of these parameters would help to optimize the production procedure individually for each strain, resulting in greatly shortened fermentation periods and in ghost fractions containing the minimum number of residual viable cells.
Flow cytometric analysis of bacterial cultures is characterized by the rapid collection of multiple parameters of cells and could therefore be applicable for generating significant quantitative data at various time points during ghost production. The availability of a diverse spectrum of “vital stains” facilitated the cytometric discrimination of certain subpopulations within a bacterial culture in correlation with the physiological status of the cells (1, 14, 15, 18). The cell membrane represents a target structure for the anionic fluorescent dye bis-(1,3-dibutylbarbituric acid) trimethine oxonol [DiBAC4(3)]. The uptake of this membrane potential-sensitive dye is restricted to depolarized cells or cells with disrupted membranes (2, 12). The fluorescent dye accumulates inside the cell by binding to lipid-containing intracellular components, e.g., the bacterial membranes (5). Intact cells with polarized membranes exclude the dye and therefore remain nonfluorescent.
Recently, flow cytometric sorting was applied to separate remaining nonlysed bacteria from E. coli ghost preparations (8). In the present work, a cytometric approach is presented to evaluate the applicability of DiBAC4(3) to support the cytometric discrimination between ghosts and viable and nonlysed but inactivated cells. The results of flow cytometric online quantification of the detected populations are compared to the data obtained by classical microbiological techniques.
MATERIALS AND METHODS
Production of bacterial ghosts.
Lysis plasmid pML1 (20) and E. coli strain NM522 (Stratagene, Amsterdam, The Netherlands) were described previously. The cells were transformed with pML1 and routinely grown in LB broth (17), containing 2% agar for solid medium, supplemented with kanamycin (50 μg/ml) at an incubation temperature of 28°C. To induce the thermosensitive expression system of lysis gene E, the culture was shifted to 42°C after it reached mid-log phase. The growth and lysis of the bacteria were monitored by measuring the OD at 600 nm (OD600). Periodically, samples were taken and (i) analyzed by standard microbiological procedures to determine the total number of cells or the number of reproductive cells or (ii) characterized by cytometric analysis subsequent to the staining procedure described below. In online monitoring experiments, the lysis process was allowed to proceed for 120 min under induced conditions. For the production of ghosts, which were used in calibration experiments, the ghost preparation was harvested by centrifugation (15 min, 10,000 × g, 4°C) after the minimum OD600 was reached. The cells were resuspended in 0.9% NaCl solution and stored at 4°C.
Classical microbiological procedures for cell enumeration.
A counting chamber (Neubauer; chamber depth, 0.02 mm; minisquare area, 0.0025 mm2; Brand GmbH & Co. KG, Wertheim, Germany) was used to quantify the total number of cells by direct microscopy. The average cell count per minisquare multiplied by the reciprocal of the corresponding volume in milliliters equaled the concentration of cells in the diluent (7). To determine the number of reproductive bacteria, the standard procedure of plating serial dilutions on nonselective agar plates was used. The plates were incubated for 15 h at 28°C to permit permissive conditions with regard to the expression system of gene E.
Staining protocol.
The membrane potential-sensitive dye DiBAC4(3) (Molecular Probes, Leiden, The Netherlands) was dissolved in dimethyl sulfoxide to prepare a 1 mM stock solution, which was stored at −20°C. For samples taken at various time points during ghost production or analyzed subsequently to further chemical or heat treatments, 10 μl of bacterial culture was added to 979 μl of FACS-FLOW solution (Becton Dickinson, Heidelberg, Germany) and supplemented with 10 μl of a solution containing fluorescent alignment beads (2.5 μm; extinction, 488 nm; emission range, 515 to 660 nm; Molecular Probes) at a concentration of 1.3 × 108 beads/ml. The mixture was completed by the addition of 1 μl of the DiBAC4(3) stock solution to give a dye concentration of 1 μM. Subsequently, the samples were analyzed by flow cytometry with no further incubation step.
In calibration experiments, 100 μl of a ghost preparation (stored in 0.9% NaCl solution at 4°C), log-phase growing cells of E. coli NM522(pML1), or a mixture of both was stained with DiBAC4(3) to give the desired dye concentration in a final volume of 1 ml of FACS-FLOW. The preparations were either analyzed immediately or incubated at room temperature for 1 to 20 min prior to flow cytometric analysis.
Flow cytometric analysis.
All experiments were performed with a FACScalibur flow cytometer (four-color system; Becton Dickinson) equipped with an air-cooled laser providing 15 mW at 488 nm and the standard filter setup. For acquisition and analysis of data, the CellQuest software package (version 3.3; Becton Dickinson) was used. Forward scatter (FSC), right-angle light scatter (side scatter; SSC), and fluorescence were collected as pulse height signals (four decades of a logarithmic scale). The green fluorescence of DiBAC4(3) was collected in the FL-1 channel (530 ± 15 nm), whereas the fluorescence of the alignment beads was collected in the FL-2 channel (585 ± 21 nm). Detector voltages were set to “E02”/gain 1 (FSC), 582 V/gain 1 (SSC), 600 V/gain 1 (FL-1), and 550 V/gain 1 (FL-2). SSC served as the primary detection parameter (threshold, 230). Cell populations were gated on the basis of FL-1 versus SSC, thereby excluding the background signal and debris. Because the FACScalibur flow cytometer does not quantify the analyzed volume, alignment beads were used as an external standard in quantitative assays. A 10-μl bead solution volume with a known concentration (1.3 × 108/ml) was added to a 990-μl cell suspension (in FACS-FLOW) prepared for flow cytometry. As the cytometric analysis was stopped after 1.3 × 104 alignment beads were counted, the corresponding volume of analyzed FACS-FLOW solution could be calculated as 10 μl, which equaled 0.1 μl of the original bacterial culture, which was investigated for each sample. With this 100-fold dilution of the original growing culture, the maximum flow rate could be limited to 3,500 bacterial particles per second during quantitative assays.
Chemical and heat treatment of E. coli.
For all of the procedures described in this section, cells of E. coli NM522(pML1) grown at 28°C until mid-log phase were used. To portions of the bacterial culture, either ethanol (final concentration, 20% [vol/vol]), formaldehyde (final concentration, 0.1% [vol/vol]), the antibiotic ampicillin (final concentration, 1 mg/ml), the uncoupler 2,4-dinitrophenol (final concentration, 2 mM; Sigma-Aldrich, Taufkirchen, Germany), or purified pore-forming colicin E1 (final concentration, 55 U/ml; Sigma-Aldrich) was added. For 2,4-dinitrophenol, the cells were washed and stored in a solution of 0.9% NaCl prior to treatment. The bacterial samples were incubated for 1 h at 28°C with slow agitation, except the colicin E1-treated culture, which was incubated for only 30 min. An aliquot of the growing E. coli cells was heat treated at 70°C for 20 min. All of the treatments described in this section—except for dinitrophenol—completely or nearly abolished reproduction, which was confirmed by plating indicating a reduction in the number of reproductive cells of at least 6 orders of magnitude. For the protonophore 2,4-dinitrophenol, a concentration and incubation conditions were chosen that had been demonstrated to result in destruction of the transmembrane proton motive force (21). Treatment of E. coli with the uncoupler was accompanied by only a minor reduction in the rate of reproduction (<10%), as determined via plating. The protocol for preparation of the outer membrane fraction, originally developed for Haemophilus influenzae by Lam and colleagues (13), was applied to E. coli NM522(pML1) in an unmodified manner. Subsequent to heat or chemical treatment, all samples were subjected to DiBAC4(3) staining and flow cytometric analyses.
RESULTS
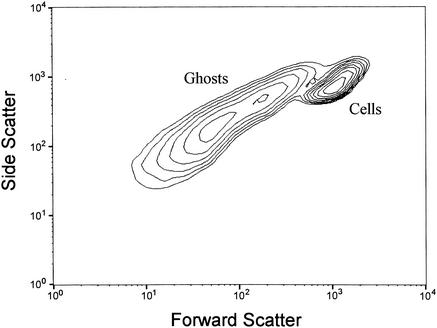
Ghost preparations and log-phase growing cells of E. coli NM522(pML1) were analyzed by flow cytometry either separately (data not shown) or as a mixed sample (Fig. 1). Compared to growing cells, the ghosts clearly exhibited less FSC and reduced SSC. Both populations could be discriminated from debris and background signals (data not shown) and from each other, although a zonal overlap was detected by using scatter analyses exclusively.
FIG. 1.
Flow cytometric analysis of a mixture of log-phase growing cells and bacterial ghosts of E. coli NM522(pML1). The FSC and SSC of 105 bacterial particles were acquired and are presented as a contour plot excluding debris and the background signal.
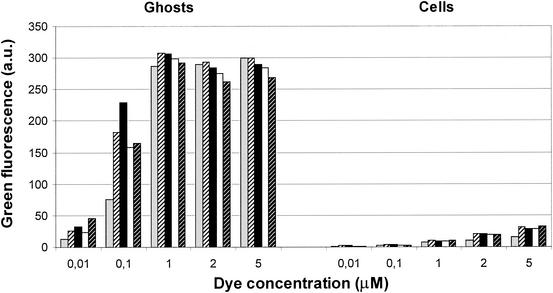
To improve the discriminatory quality of the flow cytometric analysis, a further parameter was introduced by the use of the membrane potential-sensitive dye DiBAC4(3). To establish a staining protocol optimized for samples taken during the time course of lysis, aliquots of ghosts and growing cells were incubated separately with different dye concentrations and incubation periods prior to cytometric analyses (Fig. 2). Maximum fluorescence of ghosts could be determined with dye concentrations ranging from 1 to 5 μM, with only minor variations in intensity caused by the prolonged incubation time of up to 20 min. Dye concentrations of less than 1 μM resulted in a decrease in fluorescence for the ghosts. With increasing dye concentrations, elevated levels of fluorescence were detected for the growing cells; however, the signals derived from the ghosts were always 10- to 30-fold higher. For optimal discrimination between ghosts and polarized cells, as well as to limit the time delay between sampling and analysis, a dye concentration of 1 μM in combination with immediate cytometric analysis was chosen for all of the further experiments described in this work.
FIG. 2.
Cytometric analysis of DiBAC4(3)-derived green fluorescence of log-phase growing cells and bacterial ghosts of E. coli NM522(pML1). Ghosts or cells were incubated separately with DiBAC4(3) at concentrations ranging from 0.01 to 5 μM. Cytometric analysis was performed immediately (gray bars) after dye addition or after an incubation period of 1 (bars with thin black stripes), 5 (black bars), 10 (white bars), or 20 (bars with thin white stripes) min. For each individual analysis, the fluorescence of 104 ghosts or cells was quantified. The dye concentration is given in micromolar units, whereas the mean values of green fluorescence are presented in arbitrary units (a.u.).
Qualitative cytometric characterization of ghost formation.
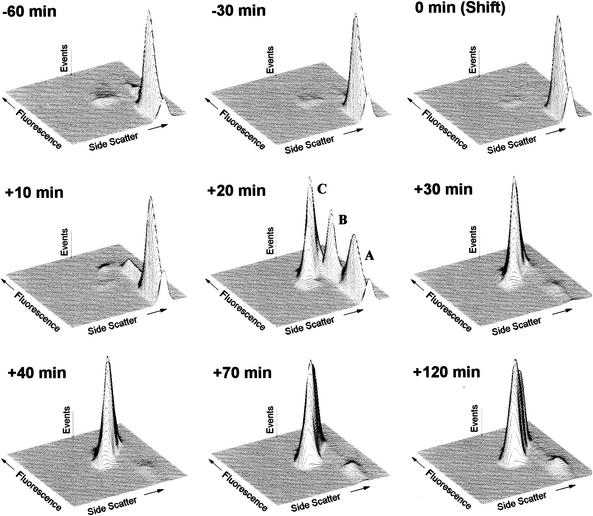
At the beginning of the time course experiment, the growing cells could clearly be detected as a distinct nonfluorescent population, representing the majority of cells within the sample (Fig. 3, −60 min). Also, two minor, highly fluorescent populations were found, one with scatter values typical of ghosts, the other one with scatter values typical of nonlysed cells. During the next hour of incubation, the total ratios of the two fluorescent populations decreased. The sample taken 10 min after lysis induction was characterized by the sudden occurrence of two populations consisting of fluorescent and therefore depolarized cells similar to the fluorescent populations observed in the growing culture. Ten minutes later, a decrease in OD600 was observed for the first time as a result of ongoing lysis. At this time point, all three populations could clearly be discriminated cytometrically by presenting two populations with SSCs typical of nonlysed cells, one of them nonfluorescent (Fig. 3, population A) and the other one brightly fluorescent (Fig. 3, population B), with intensities nearly as high as the third population of cells with greatly decreased scatter values typical of ghosts (Fig. 3, population C). All populations other than the one consisting of ghosts almost disappeared 30 min postinduction of lysis. Although the nonlysed cells were still detectable, they represented only a minor fraction of the lysed culture. Until the end of the time course, the results remained qualitatively the same whereas the OD600 continued to decrease for an additional 30 min, reaching a minimum 1 h after lysis induction. The three cell populations detected could be well discriminated from smaller cellular fragments, debris, or background signals at every time point during the time course.
FIG. 3.
Cytometric online monitoring of bacterial ghost production of E. coli NM522(pML1). Bacteria were grown at 28°C to mid-log phase and then shifted to 42°C for lysis induction [0 min (Shift)]. During the time course of 3 h, various samples were collected and analyzed cytometrically after DiBAC4(3) staining. The bacterial particles within 0.1 μl of the original culture were characterized by means of light scatter and green fluorescence. The sampling time points indicated refer to time points postinduction of lysis [0 min (Shift)]. During ghost production, three populations of bacterial particles were detected: nonlysed, polarized cells (A); nonlysed, depolarized cells (B); and bacterial ghosts (C). The flow cytometric analysis data are presented as three-dimensional plots (SSC versus fluorescence, both on a logarithmic scale) excluding debris and the background signal.
Cytometric analysis of E. coli after heat or chemical treatment.
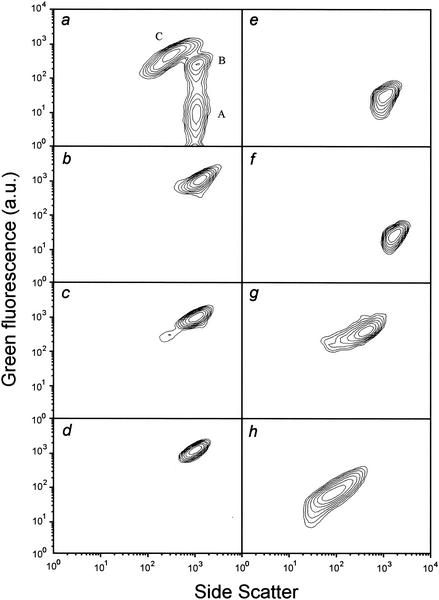
Further characterization of the detected populations with respect to their physiological and morphological properties was desirable. Therefore, growing cells of E. coli NM522(pML1) were subjected to several treatments exerting well-known effects on bacterial cells. After application of the standard protocol of DiBAC4(3) staining, the cytometric properties of the newly generated populations were compared to those of the populations detected during lysis (Fig. 4a). Heat-treated cells (Fig. 4b) or cells treated with ethanol (Fig. 4c), the uncoupler 2,4-dinitrophenol (Fig. 4d), pore-forming colicin E1 (Fig. 4e), or formaldehyde (Fig. 4f) were characterized by light scatters typical of nonlysed cells and exhibited DiBAC4(3)-mediated fluorescence. Compared to nonlysed, fluorescent, and therefore depolarized cells (Fig. 4a, population B), which are detectable mainly shortly after lysis induction, the bacterial fluorescence after heat, ethanol, or 2,4-dinitrophenol treatment was at least equal or greater in intensity. After treatment with formaldehyde or colicin E1, cellular fluorescence was reduced, giving intensities lying between the values of growing cells (background fluorescence; Fig. 4a, population A) and those of nonlysed but depolarized bacteria. The cells treated with the antibiotic ampicillin (Fig. 4g) gave results similar to those of the population classified as ghosts (Fig. 4a, population C) with regard to light scatter and fluorescence. Also, by microscopic examination, the similarity to the bacterial ghosts was confirmed by showing empty cell envelopes devoid of cytoplasmic content. Compared to these populations, the particles derived from outer membrane preparations of E. coli were characterized by reduced fluorescence and SSC (Fig. 4h) and by FSC signals very close to the background signal (data not shown).
FIG. 4.
Flow cytometric characterization of E. coli NM522(pML1) with regard to SSC and DiBAC4(3)-derived fluorescence after chemical and heat treatment and comparison to the bacterial populations (a) detected 20 min after lysis induction: nonlysed, polarized cells (A); nonlysed, depolarized cells (B); and bacterial ghosts (C). (Note: panel a corresponds to Fig. 3, +20 min.) Log-phase growing cells were treated with heat (b), ethanol (c), 2,4-dinitrophenol (d), pore-forming colicin E1 (e), formaldehyde (f), or ampicillin (g). An aliquot of growing cells was subjected to a procedure developed by Lam and colleagues (13) for the preparation of outer membrane fractions of H. influenzae (h). The cytometric analysis data, which are derived from 104 bacterial particles (b to h) or the particles within 0.1 μl of the lysing culture (a), are presented as contour plots (SSC versus green fluorescence in arbitrary units [a.u.], both on a logarithmic scale) excluding debris and the background signal.
Quantitative cytometric analysis and comparison to classical methods.
To determine the variation between samples of the cytometric enumeration of bacteria, aliquots of growing E. coli NM522(pML1) were subjected to numerous runs of cytometric quantification. A total volume of 10 μl of FACS-FLOW cell suspension was analyzed by counting approximately 1.7 × 105 cells per run. The coefficient of variation between individual runs was determined to be 1.4% with regard to the mean number of bacteria counted.
For all three bacterial subpopulations detected during ghost production, regions for cytometric enumeration could be defined, as they were clearly separated from each other and from the background or debris. Depending on the sampling time point, the cytometrically determined absolute cell count ranged between 1 × 104 and 3.2 × 104 per run, which equaled the total number of cells in a 0.1-μl culture volume. During the growth period, the calculated total number of cells increased from 1 × 108 to 2.1 × 108/ml (Fig. 5b) whereas the ratio of polarized cells in these samples increased from an initial 86 to 96%. In parallel, the ratio of cells belonging to one of the DiBAC4(3)-stained (depolarized) populations decreased from 14 to 4%. The results obtained by classical enumeration procedures reflected the same trend (Fig. 5a): increases in the total number of cells and the number of reproductive cells within orders of magnitude similar to those determined via cytometry.
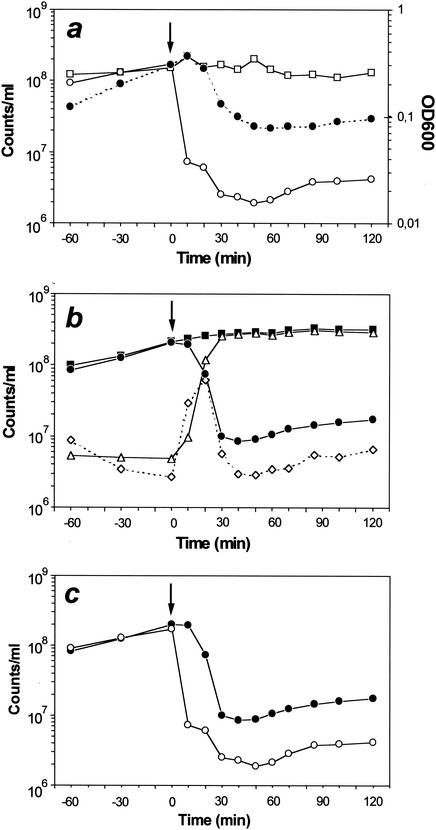
FIG. 5.
Comparison of quantitative analyses of ghost production by classical microbiological procedures (a) and flow cytometry (b). E. coli NM522(pML1) was grown at 28°C to mid-log phase and then shifted to 42°C for lysis induction at the time indicated by arrows. During the 3-h time course, OD600 (closed circles on dotted lines) was monitored and various samples were collected and analyzed to determine the total number of cells (□) and the number of reproductive bacteria (○) by classical procedures. Flow cytometric analysis included determination of the total number of cells (▪); the number of nonlysed and polarized (•) or nonlysed but depolarized cells (⋄); and the number of ghosts (▵). The data used to quantify the viable (reproductive or polarized) bacteria, derived from the classical or flow cytometric analyses, are presented separately (c). Bacterial numbers are shown as counts per milliliter. The sampling time points indicated refer to time points postinduction of lysis (0 min).
Within the first 10 min after lysis induction, the number of reproductive cells dropped from an initial 1.7 × 108/ml (100%) to 7.3 × 106/ml (4%) before reaching a minimum of 1.9 × 106/ml 50 min after lysis induction, accounting for roughly 1% of the initial value (Fig. 5a and c). The number of polarized bacteria decreased slightly from 2 × 108/ml (100%) at the time point of lysis induction to 1.9 × 108/ml (95%) 10 min later (Fig. 5b and c). After an additional 20 min, 1 × 107 (5%) polarized cells were detected per milliliter, before the minimum of 8.5 × 106/ml (4%) was reached 40 min after the heat induction of the lysis process. The viability of the culture then increased continuously again until the end of the time course, which roughly resulted in a duplication of the determined numbers of reproductive, as well as polarized, cells within 1 h. With regard to the population of nonlysed, depolarized cells, an increase of more than 20-fold, ranging from 2.7 × 106/ml, determined at the time point of lysis induction, to a maximum of 6.1 × 107/ml, determined 20 min later, was detected. After reaching a minimum of 2.9 × 106/ml, the number of depolarized cells started to increase again, simultaneously with the population of polarized cells, to reach a twofold elevated number at the end of the experiment. The total number of cells, determined after induction of lysis via the counting chamber, decreased from an initial 1.5 × 108/ml (100%) to 1.3 × 108/ml (87%). In contrast, flow cytometric enumeration of the total cell number reflected an increase from an initial 2.1 × 108/ml (100%) to 3.2 × 108/ml (152%) at the end of the time course. The ratio of bacterial ghosts within the culture started to increase shortly after lysis induction with 9.6 × 106/ml (4%), 1.2 × 108/ml (46%), and 2.6 × 108/ml (94%), determined cytometrically at 10, 20, and 30 min, respectively, after lysis induction and decreased slightly to 92% of the total cell number until the end of the experiment, indicating the rising numbers of nonlysed cells.
DISCUSSION
A flow cytometric procedure for online monitoring of ghost production was developed that supports not only the quantification of polarized cells but also subdivision of the population of depolarized cells into ghosts and nonlysed cells. The DiBAC4(3) staining protocol is devoid of any washing or incubation step, requires a minimum of dye consumption, and enables immediate cytometric analysis. The total procedure required to gain one set of quantitative results can be completed in less than 2 min.
When E. coli was subjected to certain chemical or physical treatments, the cells treated with heat, ethanol, or 2,4-dinitrophenol best reflected the cytometric properties of the depolarized but nonlysed cells detectable during lysis. Heat (11) and ethanol (16) were shown to have a membrane-disrupting effect on gram-negative bacteria, whereas treatment with dinitrophenol led to cell depolarization by uncoupling of oxidative phosphorylation (6). As the population of depolarized cells was mainly detectable within a short span of time after the induction of gene E expression and almost completely disappeared when ghosts made up the majority of cellular particles, we suggest that this population represents an intermediate stage within the initial lysis phase. It is assumed that protein E integrates into the cytoplasmic membrane, which causes the collapse of the transmembrane potential (22) as the first step in the protein E-mediated lysis process. At the time point of minimum culture turbidity, the ratio of nonlysed, depolarized cells was roughly 1%, which correlated well with the results of a recent cytometric study (8) in which nonlysed, nonreproductive bacteria were shown to represent 1% of the total cell number within an E. coli(pML1) ghost preparation.
One of the most prominent differences between the cytometric and classical analyses becomes apparent with the dramatic decline of 96% in the number of reproductive bacteria determined via plating 10 min after lysis induction. Although the cytometric approach reflected the onset of lysis at the same time point, only a minor reduction of 5% in the number of polarized cells could be observed. In the experiments performed, the majority of polarized cells were detectable for longer time periods compared to the reproductive cells (Fig. 5c). As the cytometric classification of the bacteria reflects the actual physiological status of the cells immediately after sample preparation, this approach is more accurate than plating, where single cells have to undergo numerous cell divisions to become visible as a colony. The absolute numbers of polarized or reproductive bacteria ranged within the same order of magnitude, although they were fourfold lower for the latter one when reaching minimum values, whereas samples taken prior to lysis induction gave almost identical numbers. It has been reported that starving or stressed bacteria are restricted in their reproductive ability to form colonies on agar plates even if they maintain a polarized membrane (12, 14). We propose that the majority of the polarized cells within the ghost preparation suffer from restrictions of cellular functions other than depolarization and are therefore no longer able to form colonies on plates. The quantitative discrepancies between classical and cytometric analyses question the accuracy of classical microbiological methods in the monitoring of the ghost production.
Acknowledgments
We are indebted to Jennelle M. Kyd for critical reading of the manuscript.
REFERENCES
- 1.Bunthof, J. C., S. van der Braak, P. Breeuwer, F. M. Rombouts, and T. Abee. 1999. Rapid fluorescence assessment of the viability of stressed Lactococcus lactis. Appl. Environ. Microbiol. 65:3681-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comas, J., and J. Vives-Rego. 1997. Assessment of the effects of gramicidin, formaldehyde, and surfactants on Escherichia coli by flow cytometry using nucleic acid and membrane potential dyes. Cytometry 29:58-64. [DOI] [PubMed] [Google Scholar]
- 3.Eko, F. O., A. Witte, V. Huter, B. Kuen, S. Fürst-Ladani, A. Haslberger, A. Katinger, A. Hensel, M. P. Szostak, S. Resch, H. Mader, P. Raza, E. Brand, J. Marchart, W. Jechlinger, W. Haidinger, and W. Lubitz. 1999. New strategies for combination vaccines based on the extended recombinant bacterial ghost system. Vaccine 17:1643-1649. [DOI] [PubMed] [Google Scholar]
- 4.Eko, F. O., U. B. Mayr, S.R. Attridge, and W. Lubitz. 2000. Characterization and immunogenicity of Vibrio cholerae ghosts expressing toxin-coregulated pili. J. Biotechnol. 83:115-123. [DOI] [PubMed] [Google Scholar]
- 5.Epps, D. E., M. L. Wolfe, and V. Groppi. 1994. Characterization of the steady-state and dynamic fluorescent properties of the potential-sensitive dye bis-(1,3-dibutyl barbituric acid) trimethine oxonol (DiBAC4(3)) in model systems and cells. Chem. Phys. Lipids 69:137-150. [DOI] [PubMed] [Google Scholar]
- 6.Gage, D. J., and F.C. Neidhardt. 1993. Adaptation of Escherichia coli to the uncoupler of oxidative phosphorylation 2,4-dinitrophenol. J. Bacteriol. 175:7105-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilstrap, M., J. G. Kleyn, and E. W. Nester. 1983. Experiments in microbiology, 2nd ed. CBS College Publishing, Philadelphia, Pa.
- 8.Haidinger, W., M. P. Szostak, W. Beisker, and W. Lubitz. 2001. Green fluorescent protein (GFP)-dependent separation of bacterial ghosts from intact cells by FACS. Cytometry 44:106-112. [PubMed] [Google Scholar]
- 9.Hensel, A., V. Huter, A. Katinger, P. Raza, C. Strnistschie, U. Roesler, E. Brand, and W. Lubitz. 2000. Intramuscular immunization with genetically inactivated (ghosts) Actinobacillus pleuropneumoniae serotype 9 protects against homologous aerosol challenge and prevents carrier state. Vaccine 18:2945-2955. [DOI] [PubMed] [Google Scholar]
- 10.Huter, V., M. P. Szostak, J. Gampfer, S. Prethaler, G. Wanner, F. Gabor, and W. Lubitz. 1999. Bacterial ghosts as drug carrier and targeting vehicles. J. Control. Release 61:51-63. [DOI] [PubMed] [Google Scholar]
- 11.Jepras, R. I., J. Carter, S. C. Pearson, F. E. Paul, and M. J. Wilkinson. 1995. Development of a robust flow cytometric assay for determining numbers of viable bacteria. Appl. Environ. Microbiol. 61:2696-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jepras, R. I., F. E. Paul, S.C. Pearson, and M. J. Wilkinson. 1997. Rapid assessment of antibiotic effects on Escherichia coli by bis-(1,3-dibutylbarbituric acid) trimethine oxonol and flow cytometry. Antimicrob. Agents Chemother. 41:2001-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam, J. S., D. M. Granoff, J. R. Gilsdorf, and W. Costerton. 1980. Immunogenicity of outer membrane derivatives of Haemophilus influenzae type B. Curr. Microbiol. 3:359-364. [Google Scholar]
- 14.Lopez-Amoros, R., S. Castel, J. Comas-Riu, and J. Vives-Rego. 1997. Assessment of E. coli and Salmonella viability and starvation by confocal laser microscopy and flow cytometry using rhodamine 123, DiBAC4(3), propidium iodide, and CTC. Cytometry 29:298-305. [DOI] [PubMed] [Google Scholar]
- 15.Mason, D. J., R. Lopez-Amoros, R. Allman, J. M. Stark, and D. Lloyd. 1995. The ability of membrane potential dyes and calcafluor white to distinguish between viable and non-viable bacteria. J. Appl. Bacteriol. 78:309-315. [DOI] [PubMed] [Google Scholar]
- 16.Mason, D. J., E.G.M. Power, H. Talsania, I. Phillips, and V. A. Gant. 1995. Antibacterial action of ciprofloxacin. Antimicrob. Agents Chemother. 39:2752-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Nebe-von Caron, G., P. Stevens, and R. A. Badley. 1998. Assessment of bacterial viability status by flow cytometry and single cell sorting. J. Appl. Microbiol. 84:988-998. [DOI] [PubMed] [Google Scholar]
- 19.Schön, P., G. Schrot, G. Wanner, W. Lubitz, and A. Witte. 1995. Two-stage model for integration of lysis protein E of bacteriophage PhiX174 into the cell envelope of Escherichia coli. FEMS Microbiol. Rev. 17:207-212. [DOI] [PubMed] [Google Scholar]
- 20.Szostak, M. P., A. Hensel, F. O. Eko, R. Klein, T. Auer, H. Mader, A. Haslberger, S. Bunka, G. Wanner, and W. Lubitz. 1996. Bacterial ghosts: non-living candidate vaccines. J. Biotechnol. 44:161-170. [DOI] [PubMed] [Google Scholar]
- 21.Witte, A., W. Lubitz, and E. P. Bakker. 1987. Proton-motive-force-dependent step in the pathway to lysis of Escherichia coli induced by bacteriophage φX174 gene E product. J. Bacteriol. 169:1750-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witte, A., and W. Lubitz. 1989. Biochemical characterization of φX174-protein-E-mediated lysis of Escherichia coli. Eur. J. Biochem. 180:393-398. [DOI] [PubMed] [Google Scholar]
- 23.Witte, A., G. Wanner, and W. Lubitz. 1992. Dynamics of PhiX174 protein E-mediated lysis of Escherichia coli. Arch. Microbiol. 157:381-388. [DOI] [PubMed] [Google Scholar]
- 24.Witte, A., G. Schrot, P. Schön, and W. Lubitz. 1997. Proline 21, a residue within the α-helical domain of φX174 lysis protein E, is required for its function in Escherichia coli. Mol. Microbiol. 26:337-346. [DOI] [PubMed] [Google Scholar]