Abstract
The resistance of polylactide to biodegradation and the physical properties of this polymer can be controlled by adjusting the ratio of l-lactic acid to d-lactic acid. Although the largest demand is for the l enantiomer, substantial amounts of both enantiomers are required for bioplastics. We constructed derivatives of Escherichia coli W3110 (prototrophic) as new biocatalysts for the production of d-lactic acid. These strains (SZ40, SZ58, and SZ63) require only mineral salts as nutrients and lack all plasmids and antibiotic resistance genes used during construction. d-Lactic acid production by these new strains approached the theoretical maximum yield of two molecules per glucose molecule. The chemical purity of this d-lactic acid was ∼98% with respect to soluble organic compounds. The optical purity exceeded 99%. Competing pathways were eliminated by chromosomal inactivation of genes encoding fumarate reductase (frdABCD), alcohol/aldehyde dehydrogenase (adhE), and pyruvate formate lyase (pflB). The cell yield and lactate productivity were increased by a further mutation in the acetate kinase gene (ackA). Similar improvements could be achieved by addition of 10 mM acetate or by an initial period of aeration. All three approaches reduced the time required to complete the fermentation of 5% glucose. The use of mineral salts medium, the lack of antibiotic resistance genes or plasmids, the high yield of d-lactate, and the high product purity should reduce costs associated with nutrients, purification, containment, biological oxygen demand, and waste treatment.
Lactic acid, a widely used specialty chemical in the food and pharmaceutical industries, has recently emerged as a potential bulk chemical for the production of renewable, biodegradable plastics from sugars (8, 17, 24). A Cargill Dow LLC manufacturing facility is currently under construction to produce up to 140,000 metric tons of polylactic acid per year, more than doubling the worldwide capacity for this chemical (26). Lactic acid has a chiral center and occurs as d-(−) and l-(+) enantiomers. Enantiomeric purity is important for industrial uses, and the greatest demand is for the l isomer. Deliberate blending of the enantiomers provides an effective method to control both the physical properties of polylactic acid (7, 44) and the rate of biodegradation (3, 19). The need for optically pure isomers favors the production of lactic acid monomers by biological processes rather than chemical processes which yield racemic mixtures (7, 44).
Further expansion of the lactic acid industry as a competitor of petroleum-based plastics will depend in part on the availability of low-cost sugar substrates, such as hexose and pentose sugars derived from lignocellulosic residues (4, 13, 24). Although many microorganisms produce lactic acid, commercially important strains, such as Lactobacillus strains, have been particularly useful due to their high acid tolerance and their ability to be genetically engineered for selective production of d-(−) or l-(+) optical isomers (5, 20, 28, 29). However, lactic acid bacteria also have undesirable traits, such as a requirement for complex nutrients which complicates acid recovery and incomplete or negligible pentose utilization (28, 29). Other promising biocatalysts are being developed for lactic acid production. These include strains of Rhizopus (41), Bacillus (33), Escherichia (11, 21), Saccharomyces (1), and Kluyveromyces (6, 36). Each biocatalyst, however, could benefit from one or more additional improvements, such as a broader substrate range, improved yield and productivity, reduction of nutritional requirements, elimination of plasmids and antibiotic markers, or improved optical purity of the product.
Escherichia coli, the workhorse of the biotechnology industry (13), can readily metabolize hexose and pentose sugars using only mineral salts as nutrients. During sugar fermentation, however, E. coli produces a mixture of organic acids (d-lactate, acetate, succinate, and formate) and ethanol to accommodate reducing equivalents generated during glycolysis (14, 18). Previous studies have demonstrated the feasibility of engineering E. coli for the production of l-lactate (11, 21) and d-lactate (11). The best E. coli strain reported for d-lactate production, strain JP203 (pta::Tn5 phoA′-lacZ ppc::cat supE hsdS ara proA lacY galK rpsL xyl mtl), contains multiple antibiotic resistance genes (kan and tet) and has an auxotrophic requirement for tricarboxylic acid pathway intermediates or amino acids and mutations blocking the utilization of pentoses and other sugars. Complex nutrients were used to evaluate d-lactate production by JP203, due in part to the complex nutritional requirements resulting from inactivation of phosphoenolpyruvate carboxylase (22, 43). During fermentation, high concentrations of d-lactate (63 g/liter; 700 mmol/liter) were produced from approximately 10% glucose with a volumetric productivity of 1 g/liter per h (11 mmol/liter per h). Although considerable glucose remained at the end of fermentation, the yields based on the amount of glucose metabolized ranged from 70 to 80% on a carbon basis (the maximum theoretical yield is 100%).
In this study, we developed new biocatalysts that convert sugars to d-lactic acid (optical purity, >99.8%) at 97 to 99% of the theoretical yield by using only mineral salts as nutrients. All antibiotic resistance markers and plasmids used during construction were eliminated in the production strains.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strains and plasmids used in this study are listed in Table 1. E. coli DH5α, TOP10F′, and S17-1 were used as hosts for plasmid construction. E. coli TC20 (ΔadhE::tet), AH218 (ΔfocA-pflB::kan), and SE1706 (ΔfrdBC zid::Tn10) were used as sources of mutations to construct homolactic strains. During plasmid and strain construction, cultures were grown in Luria-Bertani broth or agar (39). Antibiotics were included as appropriate at the following concentrations: kanamycin, 50 μg/ml; tetracycline, 5 or 10 μg/ml; and ampicillin, 50 μg/ml. Selection on fusaric acid plates was used to remove the Tn10-encoded tet gene (34). Fusaric acid plates contained (per liter) 5 g of tryptone, 5 g of yeast extract, 10 g of NaCl, 50 mg of chlortetracycline, 15 g of agar, 100 ml of NaH2PO4 (725 mM), 5 ml of ZnCl2 (20 mM), and 6 ml of fusaric acid (11.2 mM)). Homolactic acid-producing strains were maintained on M9 medium (32) containing 2% glucose and 1.5% agar. Broth cultures were grown in M9 medium containing either 1% glucose (tube experiments and seed cultures) or 5% glucose (pH-controlled fermentors).
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| W3110 | Wild type | ATCC 27325 |
| TOP10F′ | lacIq (episome) | Invitrogen |
| DH5α | ΔlacZM15 recA | Bethesda Research Laboratory |
| S17-1 | thi pro recA hsdR RP4-2-tet::Mu aphA::TnZ λpir | 40 |
| BW25113 | lacIqrrnB ΔlacZ hsdR ΔaraBAD ΔrhaBAD | 16 |
| AH218 | Δ(focA-pflB)::FRT-kan-FRT | This study |
| SE1706 | ΔfrdBC zid::Tn10 | 34 |
| TC20 | ΔadhE::FRT-tet-FRT | This study |
| SZ31 | W3110, Δ(focA-pflB)::FRT-kan-FRT P1(AH218) × W3110 | This study |
| SZ32 | W3110, Δ(focA-pflB)::FRT | This study |
| SZ34 | W3110, Δ(frdBC zid)::Tn10 P1(SE1706) × W3110 | This study |
| SZ35 | W3110, Δ(focA-pflB)::FRT ΔfrdBC zid::Tn10 P1(SE1706) × SZ32 | This study |
| SZ37 | W3110, ΔfrdBC | This study |
| SZ40 | W3110, Δ(focA-pflB)::FRT ΔfrdBC | This study |
| SZ57 | W3110, Δ(focA-pflB)::FRT ΔfrdBC ΔadhE::FRT-tet-FRT P1(TC20) × SZ40 | This study |
| SZ58 | W3110, Δ(focA-pflB)::FRT ΔfrdBC ΔadhE::FRT | This study |
| SZ61 | W3110, ackA::FRT-tet-FRT | This study |
| SZ62 | W3110, Δ(focA-pflB)::FRT ΔfrdBC ΔadhE::FRT ackA::FRT-tet-FRT P1(SZ61) × SZ58 | This study |
| SZ63 | W3110, ΔfocA-pflB::FRT ΔfrdBC ΔadhE::FRT ackA::FRT | This study |
| Plasmids | ||
| pCR2.1-TOPO | bla kan, TOPO TA cloning vector | Invitrogen |
| pFT-A | bla flp temperature-conditional replicon | 37 |
| pKD4 | bla FRT-kan-FRT | 16 |
| pKD46 | bla γ β exo (red recombinase), temperature-conditional replicon | 16 |
| pLOI2065 | bla FRT-tet-FRT | This study |
| pLOI2224 | kan, integration vector with conditional R6K replicon | 31 |
| pLOI2302 | pUC19 containing AscI linkers inserted into blunt NdeI and SapI sites | 46 |
| pLOI2372 | bla ackA | This study |
| pLOI2373 | bla ackA::FRT-tet-FRT | This study |
| pLOI2375 | bla ackA::FRT-tet-FRT conditional R6K replicon | This study |
| pLOI2803 | bla kan ΔadhE::FRT-tet-FRT | This study |
Genetic methods.
Standard methods were used for plasmid construction, transduction with phage P1, PCR, and sequencing (32, 39). Chromosomal DNA from W3110 served as a template to amplify the adhE and ackA genes with primers (ORFmers) obtained from Sigma-Genosys (The Woodlands, Tex.). PCR products were initially cloned into pCR2.1-TOPO by using TOP10F′ as the host.
A removable tetracycline cassette (FRT-tet-FRT) was constructed (pLOI2065) and was analogous to the kanamycin cassette (FRT-kan-FRT) in pKD4 (16). In both cassettes, flanking FRT sites are oriented in the same direction to allow efficient in vivo excision by FLP recombinanase (37). Plasmid pLOI2065 contains two EcoRI sites and two SmaI sites for isolation of the FRT-tet-FRT cassette.
Knockout mutations and chromosomal deletions were constructed by using procedures developed by Posfai et al. (37), Datsenko and Wanner (16), and Martinez-Morales et al. (31). Resistance markers with flanking FRT sites were used to facilitate deletion. Chromosomal integrations and deletions were verified by using appropriate antibiotic markers, PCR analysis, and analysis of fermentation products. Plasmid constructs were verified by sequencing relevant regions.
Deletion of pflB.
A ΔfocA-pflB::FRT mutation was constructed by using the method of Datsenko and Wanner (16). Hybrid primers were designed which are complementary to E. coli chromosomal genes and to the antibiotic cassette (FRT-kan-FRT) in pKD4. The sense primer (TTACTCCGTATTTGCATAAAAACCATGCGAGTTACGGGCCTATAAGTGTAGGCTGGAGCTGCTTC) consisted of an initial 45 bp (boldface type) corresponding to the region from position −130 to position −85 of focA, followed by 20 bp (underlined) corresponding to the primer 1 region of pKD4. The antisense primer (ATAGATTGAGTGAAGGTACGAGTAATAACGTCCTGCTGCTGTTCTCATATGAATATCCTCCTTAG) consisted of an initial 45 bp (boldface type) of the C-terminal end of pflB, followed by 20 bp (underlined) corresponding to the primer 2 region of pKD4. The FRT-kan-FRT cassette was amplified by PCR by using these primers and pKD4 as the template. After purification, amplified DNA was electroporated into E. coli BW25113(pKD46). The resulting kanamycin-resistant recombinant, AH218, contained FRT-kan-FRT in the deleted region of pflB (46 bp remaining). A phage P1 lysate prepared from AH218 (ΔpflB::FRT-kan-FRT) was used to transfer this mutation into W3110 to produce strain SZ31 (ΔpflB::FRT-kan-FRT). After verification of this mutation by analysis of PCR products, fermentation products, and antibiotic markers, the kan gene was removed from the chromosome with FLP recombinase by using a temperature-conditional helper plasmid (pFT-A). After removal of the helper plasmid, the resulting kanomycin-sensitive strain (ΔfocA-pflB::FRT) was designated SZ32.
Deletion of adhE.
To construct an adhE mutant, the coding region (2.68 kbp) was amplified by PCR and cloned into pCR2.1-TOPO. The central region of adhE (1.06 kbp) was deleted by using HincII (two sites) and was replaced with a 1.7-kbp SmaI fragment from pLOI2065 containing the FRT-tet-FRT cassette to produce pLOI2803. This plasmid was linearized by digestion with PvuI and ScaI and served as a template to amplify (with adhE primers) the 3.17-kbp region containing ΔadhE::FRT-tet-FRT. Amplified DNA was purified and introduced into W3110 by electroporation. Recombinants from double-crossover events were identified by using antibiotic markers and were confirmed by analysis of PCR and fermentation products. One clone was selected and designated TC20. Transduction with phage P1 was used to move this mutation from TC20 into SZ40, resulting in SZ57. The tet gene was deleted from SZ57 with FLP recombinase by using pFT-A (flp). After elimination of the helper plasmid by growth at 42°C, one resulting clone (ΔfocA-pflB::FRT ΔfrdBC ΔadhE::FRT) was designated SZ58.
Construction of ackA mutation.
The ackA coding region was amplified, initially cloned into pCR2.1-TOPO, and subcloned (1.2-kbp EcoRI fragment) into the corresponding site of pLOI2302 to produce pLOI2372. A SmaI fragment from pLOI2065 containing FRT-tet-FRT (1.7 kbp) was inserted into the unique, dephosphorylated EcoRV site of pLOI2372 to produce pLOI2373. The 2.8-kbp AscI fragment containing ackA::FRT-tet-FRT was isolated from pLOI2373 and cloned into the AscI site of pLOI2224 (conditional R6K replicon) to complete the integration vector, pLOI2375. Plasmid pLOI2375 was introduced into W3110 by electroporation. Recombinants from double-crossover events were identified by antibiotic markers and were confirmed by analysis of PCR and fermentation products. One strain (ackA::FRT-tet-FRT) was selected and designated SZ61. The ackA mutation was then transferred from SZ61 into SZ58 by P1 transduction to produce SZ62 (ΔfocA-pflB::FRT Δfrd ΔadhE::FRT ackA::FRT-tet-FRT). The tet gene was excised with FLP recombinase by using pFT-A. After removal of pFT-A by growth at 42°C, the resulting strain (ΔfocA-pflB::FRT Δfrd ΔadhE::FRT ackA::FRT) was designated SZ63.
Fermentation.
Cultures in 18-ml screw-cap tubes were used for the initial characterization of gene mutations concerned with fermentation. Single colonies from fresh plates were suspended in 1 ml of M9 medium and used to provide inocula (∼50 μl) for tubes filled to the brim with M9 medium (1% glucose). Fermentation products were analyzed after incubation for 48 h at 37°C.
Seed cultures were prepared for larger (8-liter) fermentations by inoculating colonies from fresh M9 plates into 2-liter flasks containing 600 ml of M9 medium with 1% glucose. After incubation for 20 h (37°C, 200 rpm), a portion of each culture was harvested by centrifugation and used to inoculate a New Brunswick Bioflow 3000 fermentor (33.8 mg [dry weight] of cells per liter) containing 8 liters of M9 medium with 5% glucose. Mixing was provided by a single, midlevel, upflow marine impeller (37°C, 200 rpm). KOH (11.7 M) was automatically added to maintain the pH at 7.0. Samples were removed for analysis of organic acids, ethanol, and cell mass. Fermentations were terminated when base addition was no longer required to maintain the pH.
Analyses.
Organic acid and residual glucose contents were determined by using a Hewlett-Packard high-performance liquid chromatograph (HPLC) (HP 1090 series II) equipped with UV (210-nm) and refractive index detectors. Products were separated by using a Bio-Rad HPX 87H column (injection volume, 10 μl) with 4 mM H2SO4 as the mobile phase (0.5 ml/min, 45°C). The identities of organic acids in fermentation broth were confirmed by nuclear magnetic resonance (45). The ethanol content was measured by gas chromatography (30). Optical isomers of d-(−)- and l-(+)-lactic acids were analyzed by using a Chiralpak MA+ column (Chiral Technologies, Exton, Pa.) as described by Omole et al. (35). Cell mass was estimated by measuring the optical density at 550 nm (1 liter of cells at an optical density at 550 nm of 3 was approximately equal to 1 g [dry weight] of cells) with a Bausch & Lomb Spectronic 70 spectrophotometer with round culture tubes (10 by 75 mm) as cuvettes.
Nucleotide sequence accession number.
The nucleotide sequence of pLOI2065 has been deposited in the GenBank database under accession no. AF521666.
RESULTS
Construction of a homolactate fermentation pathway in E. coli W3110.
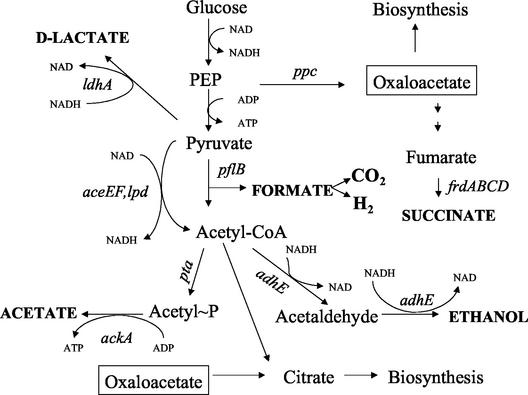
Fermentation of sugars through native pathways in E. coli produces a mixture of organic acids, ethanol, CO2, and H2 (Fig. 1 and 2A). The distribution of carbon among these products is largely determined by the relative in vivo activities of d-lactate dehydrogenase (ldhA gene), pyruvate formatelyase (pfl gene), and phosphoenolpyruvate carboxylase (ppc gene). Acetate and ethanol are typically produced in approximately equimolar amounts from acetyl coenzyme A (acetyl-CoA) to provide redox balance (14, 18). To construct a strain for d-lactate production, we used removable antibiotic resistance genes to sequentially inactivate chromosomal genes encoding alternative pathways.
FIG. 1.
Fermentation of glucose by E. coli. Primary fermentation products are indicated by boldface type. Genes encoding important enzymes are indicated by italics. For clarity, oxaloacetate (enclosed in a box) is shown twice, although it is presumed to exist as a single metabolic pool. PEP, phosphoenolpyruvate.
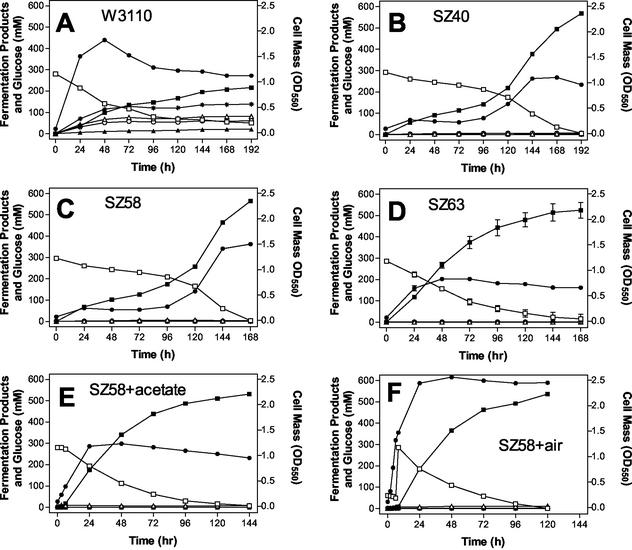
FIG. 2.
Fermentation of 5% glucose by W3110 and derivatives of this strain. (A) W3110 (wild type); (B) SZ40; (C) SZ58; (D) SZ63; (E) SZ58 supplemented with 10 mM acetate; (F) SZ58 with 8 h of (initial) aeration. Symbols: •, cell mass; □, glucose concentration; ▪, lactate concentration; ▴, succinate concentration; ○, ethanol concentration; ▵, acetate; ∗, formate concentration. OD550, optical density at 550 nm.
A deletion was constructed in the pflB gene of W3110 to produce SZ32 (ΔfocA-pflB::FRT). This single mutation eliminated the production of formate, ethanol, and acetate in tube cultures containing 1% glucose (Table 2). Several different mutations can be used to block succinate production (Fig. 1). During fermentation, the tricarboxylic acid pathway serves primarily as a source of carbon skeletons for biosynthesis. Inactivation of the ppc gene was previously utilized for construction of d-lactate-producing biocatalysts (11). However, inactivation of this gene has been shown to create an auxotrophic requirement for amino acids or dicarboxylic acids during growth in mineral salts medium with glucose (22, 43). Previous experience with E. coli B strains (25, 30) engineered for ethanol production showed that a deletion in the frdABCD operon (34, 42) can be used as an alternative method to block succinate production. P1 transduction was used to transfer this mutation (frdBC zid::Tn10) from SE1706 into SZ32, and the resulting strain was designated SZ35 (ΔfocA-pflB::FRT ΔfrdBC zid::Tn10). The tet gene was removed from SZ35 by fusaric acid selection to produce SZ40 (ΔfocA-pflB::FRT ΔfrdBC). This strain remained prototrophic and lacked all antibiotic resistance genes used during construction. For comparison, a W3110 derivative containing the ΔfrdBC mutation alone (SZ37) was also constructed. Homolactate production by SZ40 was confirmed in tube cultures containing M9 medium with 1% glucose (Table 2); only lactate was produced as a fermentation product.
TABLE 2.
Fermentation products from glucosea
| Strain | Relevant mutations | Concn of fermentation products (mM)
|
||||
|---|---|---|---|---|---|---|
| Succinate | Lactate | Formate | Acetate | Ethanol | ||
| W3110 | Wild type | 9.36 ± 1.00b | 30.69 ± 2.49 | 15.26 ± 0.34 | 18.42 ± 0.32 | 14.84 ± 1.77 |
| SZ32 | ΔpflB::FRT | 13.23 ± 1.30 | 49.23 ± 1.26 | <0.5 | <0.5 | <1.0 |
| SZ37 | ΔfrdBC | <0.5 | 37.18 ± 9.05 | 12.95 ± 3.38 | 14.80 ± 1.33 | 13.42 ± 2.08 |
| SZ40 | ΔpflB::FRT ΔfrdBC | <0.5 | 59.07 ± 0.69 | <0.5 | <0.5 | <1.0 |
Fermentation tests were conducted in 18-ml screw-cap culture tubes containing M9 medium supplemented with 1% glucose. Cultures were incubated for 48 h at 37°C.
Mean ± standard deviation.
d-Lactate production by W3110 (wild type) and SZ40 (ΔfocA-pflB::FRT ΔfrdBC).
Lactate production by W3110 and lactate production by SZ40 were compared by using pH-controlled fermentors and M9 medium containing 5% glucose as the sole carbon source (Fig. 2A and B, respectively). Glucose was not fully metabolized by W3110 in 192 h (Table 3). Even after 240 h of incubation, approximately 1% (50 mM) glucose remained. In contrast, SZ40 completed fermentation of 5% glucose within 192 h (Table 3). The lactate yields for SZ40 were more than twice those for W3110 and approached the theoretical maximum, two lactate molecules per glucose molecule (Table 3). Small but measurable amounts of ethanol and acetate were also produced by SZ40 during fermentation of 5% glucose (Table 3). Ethanol (7 mM) was the second most abundant fermentation product and represented less than 1% of the glucose carbon. Although the cell yield for SZ40 (363 mg/liter) was only 60% of the cell yield for the parent (601 mg/liter), the maximum volumetric and maximum specific productivities were higher for SZ40 (2.8- and 6.2-fold, respectively).
TABLE 3.
Summary of fermentation resultsa
| Strain (mutations and conditions) | Cell mass (mg/liter) | Amt of glucose utilized (mM) | Lactate concn (mM) | Yield (% of total; % utilized)b | Maximum volumetric productivityc
|
Maximum specific productivityd
|
Concn of coproducts (mM)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rate (mmol/liter per h) | Time (h) | Rate (mmol/g [dry wt] of cells per h) | Time (h) | Succi- nate | Formate | Acetate | Ethanol | |||||
| W3110 | 601 | 230 | 216 | 39; 47 | 2.33 | 48 | 4.46 | 24 | 21.5 | 138.2 | 81.2 | 60.5 |
| SZ40 (pflB frd) | 363 | 286 | 567 | 97; 99 | 6.41 | 192 | 27.71 | 120 | <0.5 | <0.5 | 4.4 | 7.0 |
| SZ58 (pflB frd adhE) | 495 | 292 | 564 | 95; 97 | 7.31 | 144 | 35.59 | 120 | <0.5 | <0.5 | 3.0 | <1.0 |
| SZ58 (pflB frd adhE; aeration) | 785 ± 79 | 273 ± 11 | 505 ± 31 | 92; 93 ± 2 | 10.80 ± 1.6 | 24 | 15.58 ± 4.78 | 24 | <0.5 | <0.5 | 11.5 ± 0.6 | <1.0 |
| SZ58 (pflB frd adhE; 10 mM sodium acetate) | 406 | 273 | 531 | 95; 97 | 9.11 | 72 | 44.63 | 48 | <0.5 | <0.5 | 7.13 | <1.0 |
| SZ63 (pflB frd adhE ackA) | 284 ± 7 | 276 ± 15 | 539 ± 39 | 96; 98 ± 2 | 5.98 ± 0.70 | 24 | 27.28 ± 5.54 | 24 | <0.5 | <0.5 | 1.7 ± 0.3 | <1.0 |
The values are averages of two or more fermentations. Standard deviations were calculated when three or more replicates were used.
Percentage of the maximum theoretical yield for lactic acid (2 mol of lactate per mol of glucose). The values are percentages of the total sugar added to the fermentation and percentages of the sugar which had been metabolized at the end of fermentation. Only the W3110 culture contained a significant amount of unmetabolized sugar after 192 h (50 mM).
Maximum volumetric productivity for lactate.
Maximum specific productivity for lactate.
The kinetics of growth and lactate production were dramatically altered by the mutations in SZ40 (Fig. 2A and B). W3110 began to grow immediately upon inoculation and reached the maximal cell density within 48 h. With SZ40, an initial doubling of cell mass during the first 24 h was followed by a 2-day lag before growth resumed. The possibility that the resumption of growth after this lag resulted from secondary mutations in SZ40 was investigated in two ways. Cells from the later stage of SZ40 fermentation were serially transferred as an inoculum for a new fermentation. Also, clones were isolated from this culture and used to prepare seed cultures for new fermentations. With both approaches, the growth lag and unusual kinetics persisted, indicating that a mutant population of SZ40 was not responsible for the resumption of growth (data not shown).
Chiral purity of d-lactate.
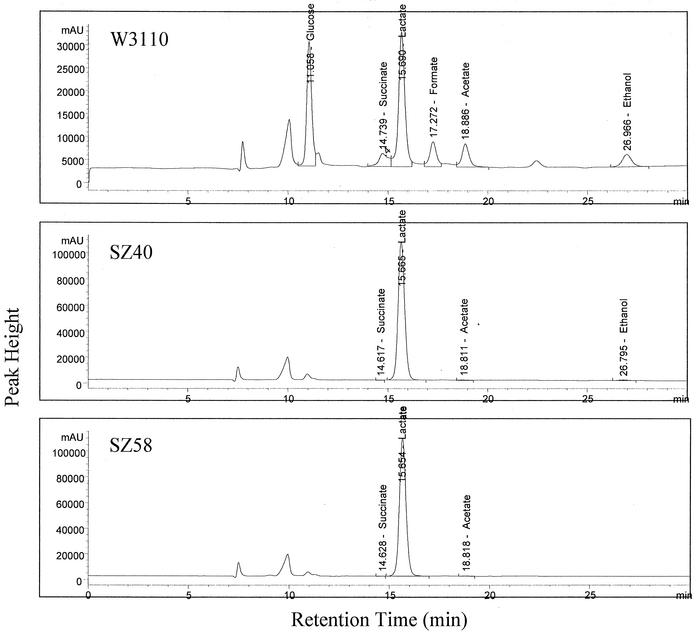
The purity of the d-lactate produced by SZ40 was determined by using the HPLC and a chiral column to separate lactate enantiomers (35). Based on these analyses, the SZ40 product was determined to contain less than 0.2% l-lactate.
Benefits of eliminating ethanol as a minor fermentation product.
Although the amount of ethanol produced by SZ40 was quite small (Table 3; Fig. 3), reducing the level of this compound may significantly increase the lactate yield in large-scale fermentations. P1 transduction and FLP recombinase were used to construct an adhE deletion in SZ40. As expected, deletion of the adhE gene (SZ58) eliminated ethanol production (Fig. 3; Table 3). Two additional improvements were also noted when the mutant was compared to the parent, SZ40 (Fig. 2B and C): (i) there was a 36% increase in the maximum cell density (495 mg/liter, compared to 363 mg/liter for SZ40); and (ii) there was a small increase in lactate productivity. With SZ58, the time required to complete fermentation of 5% glucose was approximately 24 h less than the time required with SZ40. Strain SZ58 exhibited higher maximum volumetric and maximum specific productivities (114 and 128%, respectively) than SZ40. With both strains, the lactate yields remained near the theoretical maximum (two lactate molecules per glucose molecule). The differences in the reported values (99 versus 97%) are within the error of these measurements (Table 3).
FIG. 3.
HPLC profiles (refractive index monitor) from broth samples of W3110, SZ40, and SZ58 cultures at the end of fermentation. The two initial peaks that are not labeled are the inorganic components in M9 medium. The remaining peaks are identified by compound and retention time.
The unusual growth pattern and lag observed for SZ40 persisted in SZ58 (Fig. 2B and C). Despite higher maxima for volumetric and specific productivities, the low level of biocatalyst during the first 96 h extended the time required to complete glucose fermentation. Two possible reasons for this lag were investigated. Sodium bicarbonate (1 g/liter) was added as a supplement to eliminate a potential deficiency of CO2 (2); trace elements (30) were also added. Neither supplement was beneficial for growth or lactate production. The kinetics of fermentation and the lag in growth remained unchanged (data not shown).
Benefits of eliminating acetate as a minor fermentation product.
Both SZ40 and SZ58 accumulated approximately 6 mM acetate in the broth during the initial 96 h of incubation, part of which was subsequently consumed during cell growth (Fig. 4). To reduce acetate production, an ackA mutant of SZ58 was constructed by using P1 transduction followed by treatment with FLP recombinase to excise the tet gene used for selection. Acetate production by the resulting strain (SZ63) was reduced by more than 80% during the initial 120 h of incubation but was not eliminated (Fig. 4).
FIG. 4.
Acetate production and utilization during fermentation. Symbols: ○, SZ58; ▵, SZ58 supplemented with 10 mM acetate; ∗, SZ40; ▪, SZ63.
Addition of the ackA mutation resulted in a dramatic and unexpected improvement in lactate productivity (Fig. 2C and D). The growth lags observed for SZ40 and SZ58 were absent in SZ63. Although the cell yield was lower for SZ63 than for SZ40 and SZ58 (and W3110), the rapid initial growth of SZ63 provided a higher level of biocatalyst early in the fermentation process, which in turn increased the rate of glucose conversion to lactate. The lactate levels for SZ63 after 72 and 96 h were more than twice those for SZ40 and SZ58 (Fig. 5). Like the lactate yields with SZ40 and SZ58, the lactate yield at the end of SZ63 fermentations approached the theoretical maximum yield for glucose.
FIG. 5.
Comparison of lactate production after 72 h (open bars) and 96 h (solid bars). The bacterial strains and additives (if present) are indicated at the bottom. ace, acetate.
One immediate effect of an ackA mutation on metabolism is to increase the pools of acetyl-P and acetyl-CoA (9, 12, 27), providing a possible mode of action. Since both enzymes used for acetate production (phosphotransacetylase and acetate kinase) are known to be reversible (10, 23, 38), this hypothesis was tested by supplementing SZ58 and SZ63 fermentations with sodium acetate (10 mM). Addition of acetate to SZ58 eliminated the lag in growth (Fig. 2C and E) and stimulated lactate production (Fig. 5), but addition of acetate had no effect on SZ63 (data not shown). Addition of acetate to SZ58 increased the maximum volumetric productivity (9.1 mmol/liter per h) and the maximum specific productivity (44.6 mmol/g [dry weight] of cells per h). All growth occurred during the initial 48 h of incubation. During this period, the concentration of acetate declined in the broth (from 10 to 6.6 mM) and then remained essentially constant (Fig. 4). Presumably, the acetate was incorporated into SZ58 cell mass.
Process-based improvements in lactate production.
Within the limitations of our measurements, the engineered biocatalysts catalyzed a one-for-one conversion of glucose carbon to lactic acid carbon, leaving little room for further improvement in yield or product purity (Table 3; Fig. 3). Conversion rates, however, are limited by the level of biocatalyst due to relatively slow growth and low cell yield (Fig. 2). Previous studies have shown (11) that an initial period of aeration in complex media can be used to boost the growth of d-lactate-producing strains of E. coli containing mutations in phosphoenolpyruvate carboxylase (ppc) and phosphotransacetylase (pta) genes and shorten the time required for fermentation. An initial period of aeration (8 h) was investigated with SZ63 and SZ58 in M9 medium containing 5% glucose (1% added initially and the balance added after 8 h). Oxygen levels were maintained at about 20% of air saturation by automatically mixing oxygen and nitrogen while a constant flow of 1 liter/min was maintained (400 rpm).
Initial aeration of an SZ58 culture eliminated the lag in growth and resulted in a 10-fold increase in cell yield within the first 24 h (Fig. 2C and F). This early increase in biocatalyst level accelerated glucose conversion to lactate (Fig. 5) and reduced the time required to complete fermentation of 5% glucose. Maximum volumetric productivity was essentially unchanged by aerobic growth despite the higher cell mass. Accordingly, the maximum specific productivity of cells from the initially aerated cultures was less than one-half that of cells grown anaerobically (after the lag or with acetate). The lactate yield, however, was reduced by initial aeration (Table 3). Unlike SZ40 and SZ58, strain SZ63 did not exhibit an initial growth lag. Growth and lactate production by SZ63 were not improved by an initial period of aeration (data not shown). While no attempt at further optimization was made, it is clear that changes in process conditions have the potential to reduce the time required to complete fermentations without the addition of complex nutrients.
DISCUSSION
The genetic engineering of prototrophic strains of E. coli (lacking plasmids and antibiotic resistance genes) as biocatalysts for the production of chemically pure d-lactic acid (∼98% pure with respect to organic compounds) and optically pure d-lactic acid (>99% enantiomeric purity) has several advantages. The use of mineral salts medium should reduce costs associated with ingredients, product purification, biological oxygen demand reduction, and waste treatment. Native lactic acid bacteria typically require complex nutrients for growth (24) and seldom achieve the enantiomeric purity or product selectivity obtained with SZ40 and its derivatives. The choice of the genes used to inactivate competing fermentation pathways is critical to maintain the yield and to minimize the nutritional requirements. Use of a ppc mutation to reduce succinate production in a previous study (22, 43) severely limited biosynthesis of the aspartate family of amino acids and other cellular constituents, creating an auxotrophic requirement which could be met only by amino acids or intermediates of the tricarboxylic acid pathway (23, 43). In our biocatalysts (SZ40, SZ58, and SZ63), major biosynthetic pathways were left intact by deleting genes encoding two subunits of the fermentative fumarate reductase (ΔfrdBC). The requirements for succinate can readily be met by alternative pathways, such as the glyoxylate cycle.
The cell yields for SZ40, SZ58, and SZ63 remained less than 1 g/liter during anaerobic growth in M9 medium, equivalent to less than 2% of the sugar metabolized. Although metabolic activity can be high (44.6 mmol of lactate/g [dry weight] of cells per h; approximately 1.35 μmol/mg of cell protein per min), the low concentrations of biocatalysts in these fermentations restrict the volumetric rate of lactate production. The low cell yield (low concentration of biocatalyst) was further exacerbated by the delayed growth of SZ40 and SZ58 (Fig. 2B and C). This growth lag was attributed to a problem in carbon partitioning, insufficient acetyl-CoA, or acetyl-P. Although small amounts of acetate were present in the broth of SZ40 and SZ58 at the end of fermentation (Table 3), which is consistent with an excess of acetyl-CoA, a more detailed analysis of HPLC data during fermentation indicated that there was a possible relationship between the growth of SZ40 and SZ58 and assimilation of acetate from the broth (Fig. 4). Prior to the resumption of growth, acetate accumulated slowly to a maximum concentration of about 6 mM, approximately 4 mM of which was used during the late burst of growth (Fig. 2B and C; Fig. 4). Adding 10 mM acetate eliminated the growth lag in SZ58, and one-third of the acetate was used during the initial growth of this strain. The decrease in the amount of acetate during growth (201 mg/liter) was approximately equal to one-half of the dry cell weight of SZ58 (405.9 mg/liter) in acetate-supplemented fermentations. Thus, the accumulation of acetate in the broth cultures of SZ40 and SZ58 appeared to be responsible for the resumption in growth after 96 h. It is interesting that even higher levels of acetate (20 mM) were produced in the broth of SZ58 cultures during initial aeration concurrent with rapid growth (Fig. 2F). Inactivation of ackA (SZ63) eliminated the growth lag by blocking the drain of acetyl-CoA into acetate. This mutation reduced the net acetate accumulation in the broth by up to 80% (Fig. 4). The accumulation of acetate during the growth lag (SZ40 and SZ58), the beneficial effects of supplementing SZ58 cultures with acetate, and the ackA mutation in SZ63 together provide evidence that the availability of acetyl-CoA (or acetyl-P) limits the initial growth of SZ40 and SZ58 during homolactate fermentation. The final cell yield was improved by addition of an adhE mutation (SZ58), declined with further addition of an ackA mutation (SZ63), and was not improved by addition of 10 mM acetate. The cell yield was doubled (SZ58), however, by an initial period of aeration, demonstrating the nutritional adequacy of M9 medium with glucose. Factors other than nutrient limitation must limit cell yield (Table 3).
The stimulation of growth by aeration observed with SZ58 was eliminated by introduction of an ackA mutation (SZ63), a mutation which was beneficial for anaerobic growth. Numerous studies have previously shown that mutations in the acetate pathway (pta, ackA) reduce the growth of E. coli under aerobic conditions (12, 15). Although the basis remains unknown, problems associated with the turnover of acetyl-CoA (or acetyl-P) and other metabolic imbalances have been implicated (9, 12, 27). Addition of a recombinant pathway for polyhydroxybutyrate biosynthesis from acetyl-CoA relieved the detrimental effect of ackA during aerobic growth (12).
The lag in growth and the resultant decrease in lactate production observed with SZ40 and SZ58 are undesirable traits that increase the time required to complete fermentation. Elimination of the lag by initial aeration suggests that further process optimization may increase lactate productivity while preserving high yields. The high d-lactate yields and chiral purity obtained with SZ40, SZ58, and SZ63 are equal to or better than the d-lactate yields and chiral purity previously reported for other biocatalysts (11, 17, 24).
Acknowledgments
We thank Brent Wood (B.C. International, Dedham, Mass.) for analysis of the optical purity of lactate.
This research was supported by the Florida Agricultural Experiment Station, by grants 01-35504-10669 and 00-52104-9704 from the U.S. Department of Agriculture, and by grant FG02-96ER20222 from the U.S. Department of Energy.
Footnotes
Florida Agricultural Experiment Journal Series no. R-08894.
REFERENCES
- 1.Adachi, E., M. Torigoe, M. Sugiyama, J. Nikawa, and K. Shimizu. 1998. Modification of metabolic pathways of Saccharomyces cerevisiae by the expression of lactate dehydrogenase and deletion of pyruvate decarboxylase genes for the lactic acid fermentation at low pH value. J. Ferment. Bioeng. 86:284-289. [Google Scholar]
- 2.Ajl, S. J., and H. H. Werkman. 1948. Replacement of CO2 in heterotrophic metabolism. Arch. Biochem. 19:483-492. [PubMed] [Google Scholar]
- 3.Andriano, K. P., T. Pohjonen, and P. Tormala. 1994. Processing and characterization of absorbable polylactide polymers for use in surgical implants. J. Appl. Biomater. 5:133-140. [DOI] [PubMed] [Google Scholar]
- 4.Arntzen, C. E., and B. E. Dale (co-chairs). 1999. Biobased industrial products. Priorities for research and commercialization. National Academy Press, Washington, D.C. [PubMed]
- 5.Benthin, S., and J. Villadsen. 1995. Production of optically pure d-lactate by Lactobacillus bulgaricus and purification by crystallization and liquid-liquid extraction. Appl. Microbiol. Biotechnol. 426:826-829. [Google Scholar]
- 6.Bianchi, M. M., L. Brambilla, F. Protani, C. Liu, J. Lievense, and D. Porro. 2001. Efficient homolactic fermentation by Kluyveromyces lactis strains defective in pyruvate utilization and transformed with heterologous LDH gene. Appl. Environ. Microbiol. 67:5621-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blomqvist, J. 2001. RIS Metropolis Monte Carlo studies of poly(l-lactic), poly(l, d-lactic) and polyglycolic acids. Polymer 42:3515-3521. [Google Scholar]
- 8.Boswell, C. 2001. Bioplastics aren't the stretch they once seemed. Chem. Market Rep. 260:15-18. [Google Scholar]
- 9.Bouche, S., E. Klauck, D. Fischer, M. Lucassen, K. Jung, and R. Hengge-Aronis. 1998. Regulation of RssB-dependent proteolysis in Escherichia coli: a role for acetyl phosphate in a response regulator-controlled process. Mol. Microbiol. 27:787-795. [DOI] [PubMed] [Google Scholar]
- 10.Brown, T. D. K., M. C. Jones-Mortimer, and H. L. Kornberg. 1977. The enzymatic interconversion of acetate and acetyl∼CoA in Escherichia coli. J. Gen. Microbiol. 102:327-336. [DOI] [PubMed] [Google Scholar]
- 11.Chang, D. E., S. Shin, J. Rhee, and J. Pan. 1999. Homofermentative production of d- or l-lactate in metabolically engineered Escherichia coli RR1. Appl. Environ. Microbiol. 65:1384-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, D. E., S. Shin, J. S. Rhee, and J. G. Pan. 1999. Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl coenzyme A flux for growth and survival. J. Bacteriol. 181:6656-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chotani, G., T. Dodge, A. Hsu, M. Kumar, R. LaDuca, D. Trimbur, W. Weyler, and K. Sanford. 2000. The commercial production of chemicals using pathway engineering. Biochim. Biophys. Acta 1543:434-455. [DOI] [PubMed] [Google Scholar]
- 14.Clark, D. P. 1989. The fermentation pathways of Escherichia coli. FEMS Microbiol. Rev. 63:223-234. [DOI] [PubMed] [Google Scholar]
- 15.Contiero, J., C. Beatty, S. Kumar, C. L. DeSanti, W. R. Strohl, and A. Wolfe. 2000. Effects of mutations in acetate metabolism on high-cell-density growth of Escherichia coli. J. Ind. Microbiol. Biotechnol. 24:421-430. [Google Scholar]
- 16.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datta, R., S. P. Tsai, P. Bonsignore, S. H. Moon, and J. R. Frank. 1995. Technological and economic potential of poly(lactic acid) and lactic acid derivatives. FEMS Microbiol. Rev. 16:221-231. [Google Scholar]
- 18.de Graef, M. R., S. Alexeeva, J. L. Snoep, and M. J. Teixiera de Mattos. 1999. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J. Bacteriol. 181:2351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Jong, S. J., B. van Eerdenbrugh, C. F. van Nostrum, J. J. Kettenes-van den Bosch, and W. E. Hennink. 2001. Physically crosslinked dextran hydrogels by stereocomplex formation of lactic acid oligomers: degradation and protein release behavior. J. Control Release 71:261-275. [DOI] [PubMed] [Google Scholar]
- 20.Demirci, A., and A. L. Pometto. 1992. Enhanced production of d(−)-lactic acid by mutants of Lactobacillus delbrueckii ATCC 9649. J. Ind. Microbiol. Biotechnol. 11:23-28. [Google Scholar]
- 21.Dien, B. S., N. N. Nichols, and R. J. Bothast. 2001. Recombinant Escherichia coli engineered for the production of l-lactic acid from hexose and pentose sugars. J. Ind. Microbiol. Biotechnol. 27:259-264. [DOI] [PubMed] [Google Scholar]
- 22.Gokarn, R. R., M. A. Eiteman, and E. Altman. 2000. Metabolic analysis of Escherichia coli in the presence and absence of the carboxylating enzymes phosphoenolpyruvate carboxylase and pyruvate carboxylase. Appl. Environ. Microbiol. 66:1844-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins, T. E., and M. J. Johnson. 1970. Pathways of anaerobic acetate utilization in Escherichia coli and Aerobacter cloacae. J. Bacteriol. 101:885-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofvendahl, K., and B. Hahn-Hagerdal. 2000. Factors affecting the fermentative lactic acid production from renewable resources. Enzyme Microb. Technol. 26:87-107. [DOI] [PubMed] [Google Scholar]
- 25.Ingram, L. O., H. C. Aldrich, A. C. C. Borges, T. B. Causey, A. Martinez, F. Morales, A. Saleh, S. A. Underwood, L. P. Yomano, S. W. York, J. Zaldivar, and S. Zhou. 1999. Enteric bacterial catalysts for fuel ethanol production. Biotechnol. Prog. 15:855-866. [DOI] [PubMed] [Google Scholar]
- 26.Jarvis, L. 2001. Lactic acid outlook up as polylactide nears market. Chem. Market Rep. 259:5, 14.
- 27.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteomes of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyla-Nikkila, K., M. Hujanen, M. Leisola, and A. Palva. 2000. Metabolic engineering of Lactobacillus helveticus CNRZ32 for production of pure l-(+)-lactic acid. Appl. Environ. Microbiol. 66:3835-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lapierre, L., J. E. Germond, A. Ott, M. Delley, and B. Mollet. 1999. d-Lactate dehydrogenase gene (ldhD) inactivation and resulting metabolic effects in the Lactobacillus johnsonii strains La1 and N312. Appl. Environ. Microbiol. 65:4002-4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez, A., S. W. York, L. P. Yomano, V. L. Pineda, F. C. Davis, J. C. Shelton, and L. O. Ingram. 1999. Biosynthetic burden and plasmid burden limit expression of chromosomally integrated heterologous genes (pdc, adhB) in Escherichia coli. Biotechnol. Prog. 15:891-897. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Morales, F., A. G. Borges, A. Martinez, K. T. Shanmugam, and L. O. Ingram. 1999. Chromosomal integration of heterologous DNA in Escherichia coli with precise removal of markers and replicons during construction. J. Bacteriol. 181:7143-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 33.Ohara, H., H. Okuyama, S. Sawa, Y. Fujii, and K. Hiyama. 2001. Development of industrial production of high molecular weight poly-l-lactate from renewable resources. Nippon Kagaku Kaishi 6:323-331. [Google Scholar]
- 34.Ohta, K., D. S. Beall, J. P. Mejia, K. T. Shanmugam, and L. O. Ingram. 1991. Genetic improvement of Escherichia coli for ethanol production of chromosomal integration of Zymamonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl. Environ. Microbiol. 57:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Omole, O. O., D. R. Brocks, G. Nappert, J. M. Naylor, and G. A. Zello. 1999. High-performance liquid chromatographic assay of (±)-lactic acid and its enantiomers in calf serum. J. Chromatogr. B 727:23-29. [DOI] [PubMed] [Google Scholar]
- 36.Porro, D., M. B. Michele, B. Luca, M. Rossella, B. Davide, C. Vittorio, L. Jefferson, C. L. Liu, M. R. Bianca, F. Laura, and A. Lilia. 1999. Replacement of a metabolic pathway for large-scale production of lactic acid from engineered yeasts. Appl. Environ. Microbiol. 65:4211-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Posfai, G., M. D. Koob, H. A. Kirkpatrick, amd F. C. Blattner. 1997. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rose, I. A., M. Grunberg-Manago, S. R. Korey, and S. Ochoa. 1954. Enzymatic phosphorylation of acetate. J. Biol. Chem. 211:737-756. [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 40.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 41.Skory, C. D. 2000. Isolation and expression of lactate dehydrogenase genes from Rhizopus oryzae. Appl. Environ. Microbiol. 66:2343-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tao, H., R. Gonzales, A. Martinez, M. Rodriguez, L. O. Ingram, J. F. Preston, and K. T. Shanmugam. 2001. Use of expression arrays to investigate the basis for increased glycolytic flux (xylose) in ethanologenic Escherichia coli KO11. J. Bacteriol. 183:2979-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Theodore, T. S., and E. Englesberg. 1964. Mutant of Salmonella typhimurium deficient in the carbon dioxide-fixing enzyme phosphoenolpyruvic carboxylase. J. Bacteriol. 88:946-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuji, F. 2002. Autocatalytic hydrolysis of amorphous-made polylactides: effects of l-lactide content, tacticity, and enantiomeric polymer blending. Polymer 43:1789-1796. [Google Scholar]
- 45.Underwood, S. A., M. L. Buszko, K. T. Shanmugam, and L. O. Ingram. 2002. Flux through citrate synthase limits the growth of ethanologenic Escherichia coli KO11 during xylose fermentation. Appl. Environ. Microbiol. 68:1071-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, S., and L. O. Ingram. 1999. Engineering endoglucanase-secreting strains of ethanologenic Klebsiella oxytoca P2. J. Ind. Microbiol. Biotechnol. 22:600-607. [DOI] [PubMed] [Google Scholar]