Abstract
Terminal restriction fragment length polymorphism (T-RFLP) analysis is a widely used method for profiling microbial community structure in different habitats by targeting small-subunit (SSU) rRNA and also functional marker genes. It is not known, however, whether relative gene frequencies of individual community members are adequately represented in post-PCR amplicon frequencies as shown by T-RFLP. In this study, precisely defined artificial template mixtures containing genomic DNA of four different methanogens in various ratios were prepared for subsequent T-RFLP analysis. PCR amplicons were generated from defined mixtures targeting not only the SSU rRNA but also the methyl-coenzyme M reductase (mcrA/mrtA) genes of methanogens. Relative amplicon frequencies of microorganisms were quantified by comparing fluorescence intensities of characteristic terminal restriction fragments. SSU ribosomal DNA (rDNA) template ratios in defined template mixtures of the four-membered community were recovered absolutely by PCR-T-RFLP analysis, which demonstrates that the T-RFLP analysis evaluated can give a quantitative view of the template pool. SSU rDNA-targeted T-RFLP analysis of a natural community was found to be highly reproducible, independent of PCR annealing temperature, and unaffected by increasing PCR cycle numbers. Ratios of mcrA-targeted T-RFLP analysis were biased, most likely by PCR selection due to the degeneracy of the primers used. Consequently, for microbial community analyses, each primer system used should be evaluated carefully for possible PCR bias. In fact, such bias can be detected by using T-RFLP analysis as a tool for the precise quantification of the PCR product pool.
Terminal restriction fragment length polymorphism (T-RFLP) fingerprinting (1, 4, 9, 24) is an important tool in microbial ecology. It can be used to monitor the diversity, structure, and dynamics of microbial populations in a statistically adequate manner (22). T-RFLP fingerprinting is based on the PCR amplification of a mixture of genes representing different microorganisms in environmental nucleic acid extracts. Amplicons are digested with restriction enzymes, and only the fluorescently labeled terminal restriction fragments (T-RFs) are visualized by electrophoresis on an automated sequencer.
Frequently, small-subunit (SSU) rRNA genes are used as a phylogenetic marker, and T-RFLP assays to investigate a broad range of different lineages in natural ecosystems have been established, including Bacteria (9, 24, 28, 32), Eukarya (7, 29), and Archaea (8, 33, 48). However, functional guilds of microorganisms (e.g., sulfate reducers or methanotrophs) are often not monophyletic and thus not easily discriminated from closely related but physiologically distinct organisms merely by their SSU rRNA genes (23).
A significant advance has been the fingerprinting of functional marker genes, which allow one to specifically target such functional guilds of microorganisms. Functional markers encode key enzymes of characteristic metabolic pathways and therefore allow the affiliation of microorganisms detected to their likely function in the environment. To date, T-RFLP fingerprinting techniques for marker genes of mercury-resistant bacteria (4), nitrogen fixers (34, 35), denitrifiers (3, 41), ammonium oxidizers (20), methanogens (25), and methane oxidizers (21) have been developed.
However, all of these fingerprinting techniques are PCR based and consequently may be subject to potential PCR bias. PCR-inherent bias, such as preferential amplification of certain templates (PCR selection) (37, 49, 50) and template reannealing with increasing PCR cycle numbers (31, 44, 45), may influence fingerprinting results and has to be considered in interpretation of the data. Consequently, the quantitative (in the sense of relative signal intensities) and qualitative (presence or absence of distinct populations in a given sample) interpretation of fingerprinting data requires careful and adequate controls. Several pioneering studies of the T-RFLP methodology (9, 36) analyzed bias parameters relating to PCR selection and resulted in specific recommendations for the individual PCR assays used. For archaeal templates, the influence of nonspecific priming (40) and the preferential amplification of certain Korarchaeota (5) has been reported.
In this study, we thoroughly evaluated different sources of potential PCR bias and their effects on T-RFLP fingerprinting of methanogenic communities. We used template mixtures of pure methanogen cultures and analyzed these defined “artificial communities” by PCR-T-RFLP, targeting SSU rRNA and the methyl-coenzyme M reductase genes (mcrA/mrtA) to evaluate how given template ratios are recaptured in fingerprinting analysis. Also, we analyzed a natural archaeal community by T-RFLP fingerprinting to verify the reproducibility and reliability of fingerprinting results under various amplification parameters that may relate to potential PCR bias.
MATERIALS AND METHODS
Pure cultures, soil samples, and DNA extraction.
Methanobacterium bryantii DSM 863T, Methanosaeta concilii DSM 3671T, and Methanospirillum hungatei JF1 DSM 864T were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany) and cultivated in standard media as specified by the supplier prior to DNA extraction. Genomic DNA of Methanococcus jannaschii DSM 2661T was provided by Gerrit Buurman (Max-Planck-Institut, Marburg, Germany). Soil was from an Italian rice field near Vercelli that was anoxically incubated in serum bottles for 14 days. DNA was extracted from ≈600 μl of soil slurry aliquots and from pure culture cell material with a bead-beating protocol for cell disruption, followed by consecutive ammonium acetate, isopropanol, and ethanol precipitation steps (19) and polyvinylpyrrolidone purification (2).
DNA quantification.
Precise quantification of diluted DNAs was performed with the PicoGreen double-stranded DNA quantification kit (Molecular Probes, Leiden, The Netherlands) in 96-well microtiter plates on a Fluorolite 1000 fluorescence microtiter plate reader (Dynatech Laboratories, Chantilly, Va.). Measurements were standardized with phage λ DNA (Molecular Probes) in a concentration range between 1 and 50 ng (100 μl well−1).
PCR amplification.
Archaeal SSU rRNA genes were PCR amplified for T-RFLP analysis as described before (26) with the primer combination Ar109f (5′-ACK GCT CAG TAA CAC GT-3′) (16) and a 5′ 6-carboxyfluorescein (FAM)-labeled Ar915r primer (5′-GTG CTC CCC CGC CAA TTC CT-3′) (43) (MWG Biotech, Ebersberg, Germany). mcrA fragments were amplified for T-RFLP analysis with the 5′ FAM-labeled MCRf (5′-TAY GAY CAR ATH TGG YT-3′) and MCRr (5′-ACR TTC ATN GCR TAR TT-3′) primer pair (42) as described recently (25).
Thermal gradient PCR was performed within an annealing temperature range of ±5°C on a Mastercycler gradient thermocycler (Eppendorf, Hamburg, Germany). For amplification of genes from environmental DNA extracts, 30 PCR cycles were used (if not mentioned otherwise), and 25 PCR cycles were used for diluted pure-culture DNA templates. After amplification, amplicons were visualized by standard agarose gel electrophoresis, purified with the MinElute PCR purification kit (Qiagen, Hilden, Germany), reeluted in 25 μl of EB buffer (Qiagen), and quantified by UV photometry.
Real-time PCR.
The SSU rRNA gene content of pure-culture DNA dilutions was quantified with an iCycler iQ real-time PCR detection system (Bio-Rad, Munich, Germany) and amplicon detection via Sybr Green fluorescence. The reactions were set up in MicroAmp eight-tube strips and sealed with optical eight-cap strips (Applied Biosystems, Weiterstadt, Germany). Each reaction mix contained, in a total volume of 50 μl, 1× Sybr Green PCR master mix (Applied Biosystems), 0.3 μM each primer (Ar109f/Ar915r, unlabeled), and 2 μl of standard or unknown DNA template. The thermal protocol for real-time PCR amplification and detection was 10 min of initial denaturation (94°C), followed by 40 amplification cycles of 30 s at 94°C, 30 s at 52°C, and 60 s at 72°C. Real-time PCR runs were set up and data were collected and analyzed with the iCycler optical system interface software (version 2.3.1370; Bio-Rad). As implemented in the iCycler operating system, well factors were collected with a 96-well microtiter plate containing 50-μl aliquots of a 25 nM fluorescein 5-isothiocyanate (FITC) solution before each run. After every run, melting curves were recorded for each PCR to discriminate between specific and unspecific real-time PCR signals.
T-RFLP analysis.
Restriction digests of SSU rDNA amplicons were performed with TaqI (8), and mcrA amplicons were digested with Sau96I as described previously (25). Relative amplicon frequencies were determined as relative signal intensities of T-RFs with peak height analysis or peak area integration. Signals with a peak height below 100 relative fluorescence units (36) or with a peak area contribution below 1% (28) were regarded as background noise and excluded from analysis.
RESULTS AND DISCUSSION
Reproducibility.
For the interpretation of T-RFLP data sets, it is necessary to evaluate the reproducibility of all steps of specific PCR-T-RFLP assays, i.e., the PCR, digestion, and electrophoresis. In several investigations, a high reproducibility of T-RFLP analyses targeting SSU rRNA genes of Bacteria has been shown, e.g., ≈10% absolute peak height variability (36), 1.5% standard deviation of relative peak area (28), or 7% standardized peak height variability (13). Two studies involving PCR-T-RFLP analyses of archaeal SSU rDNA or rRNA found maximum standard deviations of 6% relative peak height (11) and 6% relative peak area (27) upon triplicate DNA and RNA extractions, respectively. In the present study, six replicate DNA extractions from three parallel soil slurry incubations and subsequent SSU rDNA-targeted T-RFLP analysis of the archaeal community yielded an average standard deviation of 1% relative amplicon frequency. The highest absolute standard deviation was 33.4% ± 2.3% relative amplicon frequency, and the highest relative standard deviation was 16% (e.g., 5.1% ± 0.8% relative amplicon frequency).
Only a few studies have looked at PCR-related influences on T-RFLP analysis so far. No significant differences in T-RFLP results were reported for a bacterial T-RFLP system between 25 and 35 PCR cycles (36). To examine the effect of increasing PCR cycle numbers on the archaeal SSU rDNA-targeted T-RFLP system evaluated here, we generated amplicons from identical rice field soil template DNAs with increasing PCR cycle numbers. The relative abundance of T-RFs was stable within standard deviations of maximally 3% between 28 and 45 PCR cycles, and minor peaks did not become more abundant with increasing PCR cycles (data not shown). Therefore, this archaeal T-RFLP system appears to be unaffected by template reannealing. Such a PCR bias would cause less abundant PCR products to become relatively more abundant with increasing PCR cycle numbers (31, 44, 45).
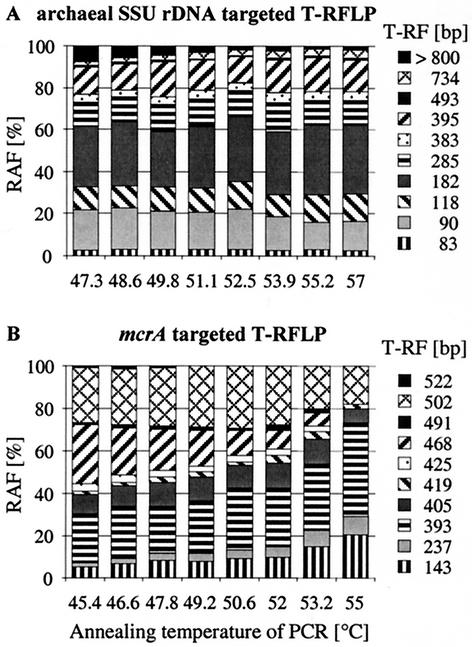
Furthermore, we evaluated the possible effects of selective amplification on fingerprinting results by varying the PCR annealing temperature. Different annealing temperatures may alter primer-binding kinetics in a template mixture, especially if primers with degenerate positions are used. The primer pair used for the amplification of archaeal SSU rRNA genes carries one degenerate position, while the mcrA-targeting primers carry nine degenerate nucleotides altogether. Archaeal SSU rDNA relative amplicon frequencies were virtually not affected by annealing temperature variations (Fig. 1A) within a range from 47.3 to 57°C (within normal standard deviation limits). In contrast, variation of the annealing temperature showed a dramatic effect on mcrA relative amplicon frequencies (Fig. 1B).
FIG. 1.
Effect of various PCR annealing temperatures on the SSU rDNA-targeted (A) and mcrA-targeted (B) T-RFLP analysis of a soil archaeal community. Amplicons were generated from identical soil DNA templates with various PCR annealing temperatures and analyzed by T-RFLP fingerprinting. RAF, relative amplicon frequency.
This PCR was tested in a range between 45.4 and 55°C (few or no products were obtained below or above these temperatures). Most significantly affected was the 468-bp T-RF, which accounted for almost 30% of the relative amplicon frequency at 45.4°C but was below the detection limit at 55°C. The 405- and 502-bp T-RFs first increased slightly and then decreased again in relative amplicon frequency with increasing annealing temperatures but always remained detectable. Others, especially the 143-, 237-, and 393-bp T-RFs, increased significantly in relative amplicon frequency with annealing temperature. We relate these findings to the highly degenerate mrcA primer set used.
It was shown earlier that different binding energies of primer permutations can lead to PCR selection of GC-rich templates (37, 50). Similarly, a reduction in T-RFLP-detected diversity upon an increase in the annealing temperature was found within a model bacterial community, although nondegenerate primers were used (9). In contrast, Osborn and coworkers (36) observed an increasing number of bacterial T-RFs at a higher annealing temperature but related this “apparent anomaly” to electrophoresis detection problems rather than a PCR artifact. Thus, the influence of the annealing temperature on the differential amplification of certain templates cannot be predicted and should be analyzed for each primer system individually.
Sequence conservation of functional marker genes in comparison to SSU rRNA genes is constrained by the genetic code and its variability, especially in the third codon position. Consequently, highly degenerate primers often have to be used for functional marker genes to cover a wide phylogenetic range. This contradicts the potential of PCR-based fingerprinting to obtain an unskewed picture of community structure, and relative community compositions should be inferred carefully.
Defined template mixtures.
The essential question in T-RFLP fingerprinting, how the relative abundance of amplicons (and thus of T-RFs) reflects template ratios, cannot be answered by the analysis of environmental DNA extracts of unknown composition. Therefore, we used artificial DNA template mixtures of defined gene content to evaluate the quantitative potential of archaeal T-RFLP analysis. Genomic DNA from four pure cultures of methanogens (M. bryantii, M. concilii, M. hungatei, and M. jannaschii) was used, because templates in environmental DNA extracts are also genomic and the flanking regions of target genes have been shown to influence amplification efficiency (17). Moreover, genomic templates allow comparison of results obtained via T-RFLP analyses of identical mixtures targeting different marker genes, such as SSU ribosomal DNA (rDNA) and mcrA of methanogens.
Prior to the mixing experiments, the pure-culture DNAs were analyzed by both SSU rDNA- and mcrA-targeted PCR-T-RFLP. This control experiment was conducted to verify the sole appearance of T-RFs in silico predicted from sequence data. Results are summarized in Table 1, and the measured T-RFs of pure cultures were in good agreement with the predicted T-RFs.
TABLE 1.
Measured and predicted in silico T-RFs of SSU rDNA and mcrA/mrtA amplicons of pure cultures of methanogensa
| Organism | Measured T-RF (bp)
|
Predicted T-RF (bp)
|
||||
|---|---|---|---|---|---|---|
| SSU rDNA | mcrA | mrtA | SSU rDNA | mcrA | mrtA | |
| M. bryantii | 90 | 502 | 470 | 93 | 502 | NA |
| M. concilii | 285 | 255 | — | 285 | 256 | — |
| M. hungatei | 395 | 429 | — | 394 | 430 | — |
| M. jannaschii | 789 | 475∗ | 792 | 476 | 473 | |
NA, sequence not available for in silico predicition; —, organisms contain only the mcrA gene, no mrtA gene; ∗, mcrA and mrtA amplicons of M. jannaschii differ by only 3 bp in length and could not be clearly separated in T-RFLP analysis (i.e., form one combined peak with a ≈475-bp length).
For the defined mixtures, the SSU rRNA gene content of freshly diluted working solutions (≈2 ng μl−1, Pico Green measurement) of the genomic DNAs was determined by real-time PCR (18). The measurement was standardized with a 10-fold dilution series of M. jannaschii genomic DNA (1.665 Mbp, two rrn operons [6]), and was found to be reliable between at least 107 and 102 archaeal rrn copies. The average standard deviation of measurements was below 2% of means. For each of the four methanogens, a series with 25%, 40%, 57.1%, and 78.6% SSU rDNA template ratio was prepared, while the remaining three species equally accounted for the residual percentage (25%, 20%, 14.3%, and 7.1% each, respectively). Three entire sets of defined template mixtures were prepared in completely independent mixing procedures (starting from freshly diluted DNA working solutions) to account for pipetting errors and to obtain statistically significant results. Immediately after preparation, the defined genomic DNA mixtures were analyzed by both SSU rDNA- and mcrA-targeted T-RFLP analyses.
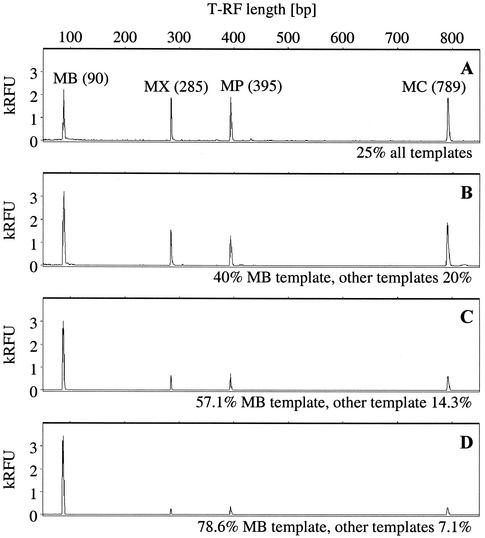
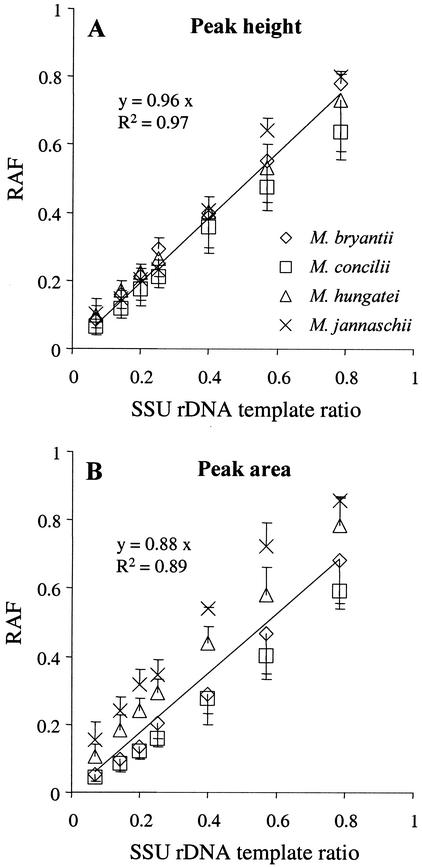
As an example for the T-RFLP analysis of defined template mixtures, electropherograms from a series with increasing M. bryantii SSU rDNA template ratios are shown in Fig. 2. An adequate representation of the defined template ratios was visually apparent in the electropherograms. Precise amplicon ratios were determined by inferring both the peak height and peak area of T-RFs. Summarized PCR-T-RFLP results of the three independent sets of defined template mixtures are shown in Fig. 3. Increasing template ratios were precisely reflected in the relative abundance of T-RFs for the four methanogens tested and in average in absolute correlation when peak height analysis was used (Fig. 3A). Peak area integration, on the other hand, resulted in a significant overrepresentation of M. hungatei and M. jannaschii relative amplicon frequencies (Fig. 3B). Both were characterized by long SSU rDNA T-RFs (395 and 789 bp, respectively), which migrated slowly during electrophoresis, resulting in broad, “fuzzy” T-RF peaks.
FIG. 2.
Electropherograms of T-RFLP analysis of defined template mixtures of M. bryantii (MB), M. concilii (MX), M. hungatei (MP), and M. jannaschii (MC) pure-culture DNA. Shown is a series with increasing M. bryantii template ratios from 25 to 78.6% of SSU rRNA genes (A to D). T-RF lengths of characteristic peaks are indicated in parentheses (in base pairs). RFU, relative fluorescence units.
FIG. 3.
Relationship between SSU rDNA template ratios and relative amplicon frequencies as determined by T-RFLP analysis of defined template mixtures. Relative amplicon frequencies (RAF) were determined by fluorescence peak height analysis (A) or peak area integration (B). Lines indicate best linear fit for combined data sets of each graph, and slopes are given. Average values of separately prepared defined template mixtures are plotted (± standard deviation, n = 3).
We assume that the automated area integration of T-RFs by the Genescan analysis software overestimates the area representation of such fuzzy peaks. This would explain the constant overrepresentation of M. jannaschii and, to a lesser extent, also of M. hungatei template ratios when the relative peak areas of T-RFs were compared. Inconsistencies in the Genescan software peak-calling routine that prevented the use of peak area data have been observed before (9). Consequently, for the evaluation of relative signal intensities in archaeal T-RFLP analysis, peak height comparison should be used.
Defined SSU rDNA template mixtures for PCR and subsequent amplicon ratio analysis have been prepared earlier, but the results were inconsistent. Mostly pairwise mixtures of either genomic or amplicon standard DNAs have been prepared, and post-PCR amplicon ratios were either in good concordance with the prepared template ratios (14, 20, 45-47) or indicated significantly skewed product ratios, depending on the templates or primers used (17, 37, 45, 47). Furthermore, single-species standards have been seeded to natural microbial communities in different ratios, and similar 1:1 relationships between spiked cell numbers (10) or seeded DNA quantities (12) and post-PCR T-RF ratios have been reported. However, in these analyses, it was not tested whether this correlation remained constant, if the seeding had been conducted with different strains, varying in rrn copy number, either separately or simultaneously.
In addition to SSU rDNA as a marker gene, we also analyzed the reliability of mcrA-targeted T-RFLP fingerprinting of methanogenic communities. This was of special interest because earlier investigations showed that the mcrA and mrtA genes of members of the Methanobacteriaceae were of increased relative abundance in T-RFLP fingerprints compared to SSU rDNA-derived relative amplicon frequencies, while members of the Methanomicrobiaceae were underrepresented (25, 38).
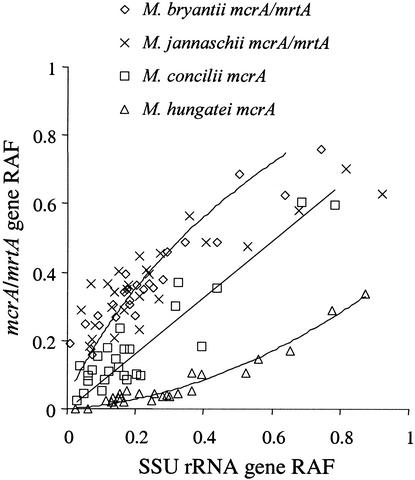
We compared the measured SSU rRNA gene ratios of defined template mixtures to the corresponding mcrA gene ratios. Correlation was low, and pronounced representational differences for the different species analyzed became apparent (Fig. 4). Although increasing ratios were also reflected, the mcrA genes of M. hungatei were strongly underrepresented, while the combined mcrA and mrtA genes of M. bryantii and M. jannaschii were overrepresented. The mcrA genes of M. concilii were of a more consistent representation in both fingerprinting methods. Differences in the respective operon copy numbers may have contributed to the observed distortion.
FIG. 4.
Relationship between SSU rDNA and mcrA/mrtA gene relative amplicon frequencies (RAF) as determined by T-RFLP analysis of defined template mixtures. Lines indicate estimated fits.
Members of the Methanococcales and Methanobacteriales harbor two MCR isoenzymes, encoded by the mcr and mrt operons (39). The α-subunit genes of both operons are detected by T-RFLP analysis (25). On the other hand, M. concilii and M. hungatei are known to contain only one mcr operon; a second isoenzyme has not been detected (42). Given that all four methanogens contain the same rrn operon copy number, this could explain the systematic overrepresentation of M. bryantii and M. jannaschii, but not the dramatic underrepresentation of M. hungatei. Again, a possible explanation for these findings may be the highly degenerate primer set used for the mcrA fingerprinting analyses. The reliability of post-PCR amplicon ratios has earlier been shown to depend on the primer pair used (45), and degenerate primers should be avoided (37, 50). Unfortunately, the complete mcrA and mrtA sequences of M. concilii, M. hungatei, and M. bryantii are not yet available, and therefore the precise primer binding sites cannot be determined. This would be necessary to test the model of PCR selection of mcrA/mrtA templates with GC-rich primer target sites.
Conclusions.
In this study, we showed that the SSU rDNA-targeted PCR system used is a precise and reproducible tool for the determination of archaeal template ratios as analyzed by T-RFLP. A broad range of different SSU rRNA gene ratios for a four-membered community was absolutely reflected in relative amplicon frequencies, as shown by fluorescence peak height comparison, which excludes PCR bias such as PCR selection or template reannealing. For natural communities, variations in neither PCR annealing temperature nor cycle number affected relative amplicon frequencies, which supports the quantitative potential of SSU rDNA-targeted T-RFLP fingerprinting for analysis of archaeal communities. In mcrA-targeted T-RFLP analysis, increasing template ratios were also detected, but representation varied significantly for the four different methanogens tested, probably due to preferential amplification of certain templates caused by the highly degenerate mcrA primer set.
Possible bias involved in PCR should be evaluated carefully for each primer system used if quantitative inferences of relative amplicon frequencies are intended. In fact, our study has shown that T-RFLP analysis can be used as a powerful tool to detect PCR bias of individual PCR assays by the precise quantification of the PCR product pool. But, as noted earlier (30), because of potential PCR bias and largely unknown rrn copy numbers in microbial genomes (15), T-RFLP profiles do not reflect cell counts of natural microbial communities.
Acknowledgments
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) within SFB395 “Interactions, adaptations, and catalytic capabilities of soil microorganisms” and the Max-Planck-Society.
Genomic DNA of Methanococcus jannaschii was a kind gift of Gerrit Buurman. We thank Bianca Wagner for expert technical assistance and Ralf Conrad for continuing support.
REFERENCES
- 1.Avaniss-Aghajani, E., K. Jones, D. Chapman, and C. Brunk. 1994. A molecular technique for identification of bacteria with small subunit ribosomal RNA sequences. BioTechniques 17:144-149. [PubMed] [Google Scholar]
- 2.Berthelet, M., L. G. Whyte, and C. W. Greer. 1996. Rapid, direct extraction of DNA from soils for PCR analysis with polyvinylpolypyrrolidone spin columns. FEMS Microbiol. Lett. 138:17-22. [DOI] [PubMed] [Google Scholar]
- 3.Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in pacific northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruce, K. D. 1997. Analysis of mer gene subclasses within bacterial communities in soils and sediments resolved by fluorescent-PCR-restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 63:4914-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunk, C. F., and N. Eis. 1998. Quantitative measure of small-subunit rRNA gene sequences of the kingdom Korarchaeota. Appl. Environ. Microbiol. 64:5064-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bult, C. J., O. White, G. J. Olsen, L. X. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, et al. 1996. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 7.Casamayor, E. O., R. Massana, S. Benlloch, L. Ovreas, B. Diez, V. J. Goddard, J. M. Gasol, et al. 2002. Changes in archaeal, bacterial, and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 4:338-348. [DOI] [PubMed] [Google Scholar]
- 8.Chin, K. J., T. Lukow, and R. Conrad. 1999. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl. Environ. Microbiol. 65:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clement, B. G., L. E. Kehl, K. L. DeBord, and C. L. Kitts. 1998. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J. Microbiol. Methods 31:135-142. [Google Scholar]
- 10.Clement, B. G., and C. L. Kitts. 2000. Isolating PCR-quality DNA from human feces with a soil DNA kit. BioTechniques 28:640-642, 644-646. [DOI] [PubMed]
- 11.Dollhopf, S. L., S. A. Hashsham, and J. Tiedje. 2001. Interpreting 16S rDNA T-RFLP data: application of self-organizing maps and principal component analysis to describe community dynamics and convergence. Microb. Ecol. 42:495-505. [DOI] [PubMed] [Google Scholar]
- 12.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol. 38:93-113. [DOI] [PubMed] [Google Scholar]
- 16.Grosskopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen, M. C., T. Tolker-Nielsen, M. Givskov, and S. Molin. 1998. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol. Ecol. 26:141-149. [Google Scholar]
- 18.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 19.Henckel, T., M. Friedrich, and R. Conrad. 1999. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horz, H. P., J. H. Rotthauwe, T. Lukow, and W. Liesack. 2000. Identification of major subgroups of ammonia-oxidizing bacteria in environmental samples by T-RFLP analysis of amoA PCR products. J. Microbiol. Methods 39:197-204. [DOI] [PubMed] [Google Scholar]
- 21.Horz, H. P., M. T. Yimga, and W. Liesack. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 67:4177-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitts, C. L. 2001. Terminal restriction fragment patterns: a tool for comparing microbial communities and assessing community dynamics. Curr. Issues Intest. Microbiol 2:17-25. [PubMed] [Google Scholar]
- 23.Liesack, W., P. H. Janssen, F. A. Rainey, N. L. Ward-Rainey, and E. Stackebrandt. 1997. Microbial diversity in soil: the need for a combined approach with molecular and cultivation techniques, p. 375-439. In J. D. van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker Inc., New York, N.Y.
- 24.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lueders, T., K. J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme M reductase α-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 26.Lueders, T., and M. Friedrich. 2000. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lueders, T., and M. W. Friedrich. 2002. Effects of amendment with ferrihydrite and gypsum on the structure and activity of methanogenic populations in rice field soil. Appl. Environ. Microbiol. 68:2484-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukow, T., P. F. Dunfield, and W. Liesack. 2000. Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32:241-247. [DOI] [PubMed] [Google Scholar]
- 29.Marsh, T. L., W. T. Liu, L. J. Forney, and H. Cheng. 1998. Beginning a molecular analysis of the eukaryal community in activated sludge. Water Sci. Technol. 37:455-460. [Google Scholar]
- 30.Marsh, T. L., P. Saxman, J. Cole, and J. Tiedje. 2000. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl. Environ. Microbiol. 66:3616-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathieu-Daudé, F., J. Welsh, T. Vogt, and M. McClelland. 1996. DNA rehybridization during PCR: the ′Cot effect' and its consequences. Nucleic Acids Res. 24:2080-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moeseneder, M. M., J. M. Arrieta, G. Muyzer, C. Winter, and G. J. Herndl. 1999. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:3518-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moeseneder, M. M., C. Winter, J. M. Arrieta, and G. J. Herndl. 2001. Terminal restriction fragment length polymorphism (T-RFLP) screening of a marine archaeal clone library to determine the different phylotypes. J. Microbiol. Methods 44:159-172. [DOI] [PubMed] [Google Scholar]
- 34.Noda, S., M. Ohkuma, R. Usami, K. Horikoshi, and T. Kudo. 1999. Culture-independent characterization of a gene responsible for nitrogen fixation in the symbiotic microbial community in the gut of the termite Neotermes koshunensis. Appl. Environ. Microbiol. 65:4935-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohkuma, M., S. Noda, and T. Kudo. 1999. Phylogenetic diversity of nitrogen fixation genes in the symbiotic microbial community in the gut of diverse termites. Appl. Environ. Microbiol. 65:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 37.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramakrishnan, B., T. Lueders, P. F. Dunfield, R. Conrad, and M. W. Friedrich. 2001. Archaeal community structures in rice soils from different geographical regions before and after initiation of methane production. FEMS Microbiol. Ecol. 37:175-186. [Google Scholar]
- 39.Reeve, J. N., J. Nölling, R. M. Morgan, and D. R. Smith. 1997. Methanogenesis: genes, genomes, and who's on first? J. Bacteriol. 179:5975-5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reysenbach, A. L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scala, D. J., and L. J. Kerkhof. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Springer, E., M. S. Sachs, C. R. Woese, and D. R. Boone. 1995. Partial gene-sequences for the α-subunit of methyl-coenzyme M reductase (MCRI) as a phylogenetic tool for the family Methanosarcinaceae. Int. J. Syst. Bacteriol. 45:554-559. [DOI] [PubMed] [Google Scholar]
- 43.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, England.
- 44.Suzuki, M., M. S. Rappe, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trotha, R., U. Reichl, F. L. Thies, D. Sperling, W. Konig, and B. Konig. 2002. Adaption of a fragment analysis technique to an automated high-throughput multicapillary electrophoresis device for the precise qualitative and quantitative characterization of microbial communities. Electrophoresis 23:1070-1079. [DOI] [PubMed] [Google Scholar]
- 48.van der Maarel, M. J. E. C., R. R. E. Artz, R. Haanstra, and L. J. Forney. 1998. Association of marine archaea with the digestive tracts of two marine fish species. Appl. Environ. Microbiol. 64:2894-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 50.Wagner, A., N. Blackstone, P. Cartwright, M. Dick, B. Misof, P. Snow, G. P. Wagner, J. Bartels, M. Murtha, and J. Pendleton. 1994. Surveys of gene families with polymerase chain reaction-PCR selection and PCR drift. Syst. Biol. 43:250-261. [Google Scholar]