Abstract
The purpose of this study was to investigate the mechanism by which phytoremediation systems promote hydrocarbon degradation in soil. The composition and degradation capacity of the bulk soil microbial community during the phytoremediation of soil contaminated with aged hydrocarbons was assessed. In the bulk soil, the level of catabolic genes involved in hydrocarbon degradation (ndoB, alkB, and xylE) as well as the mineralization of hexadecane and phenanthrene was higher in planted treatment cells than in treatment cells with no plants. There was no detectable shift in the 16S ribosomal DNA (rDNA) composition of the bulk soil community between treatments, but there were plant-specific and -selective effects on specific catabolic gene prevalence. Tall Fescue (Festuca arundinacea) increased the prevalence of ndoB, alkB, and xylE as well as naphthalene mineralization in rhizosphere soil compared to that in bulk soil. In contrast, Rose Clover (Trifolium hirtum) decreased catabolic gene prevalence and naphthalene mineralization in rhizosphere soil. The results demonstrated that phytoremediation systems increase the catabolic potential of rhizosphere soil by altering the functional composition of the microbial community. This change in composition was not detectable by 16S rDNA but was linked to specific functional genotypes with relevance to petroleum hydrocarbon degradation.
Reduction of hydrocarbons in soil is enhanced in the presence of plants (3), but the mechanisms by which plants enhance hydrocarbon removal are not completely understood. The uptake of hydrocarbons from contaminated soils by plants (phytoaccumulation) is limited by the high lipophilicity of hydrocarbons (24), and there is little data available on plant-induced sequestration (phytostabilization) of hydrocarbons in soil (20). The mechanism responsible for the phytoremediation of contaminated soil is thought to be an increase in microbial activity. Supporting this hypothesis, the population levels of contaminant-degrading bacteria and the potential of soil to degrade contaminants typically increases during phytoremediation (18, 22).
We hypothesize that there may be two mechanisms by which plants increase catabolic activity in the rhizosphere or the bulk soil. Increases in catabolic activity may result from an enhancement of general microbial activity. For example, plants sometimes increase catabolic activity in rhizosphere soil (10, 18) independent of contaminants. This suggests that the enhancement of catabolic activity may simply be the result of microbial activity increasing due to the release of plant lysates and root exudates by what is commonly termed the rhizosphere effect. Additionally, increases in catabolic activity may result from the proliferation of specific microbial groups as the microbial community size increases due to the rhizosphere effect. This is the reasoning behind the use of microbial inoculants that stimulate phytoremediation (7) and also alter the diversity of the root-associated community (27). In this scenario, the microbial community has little inherent ability to degrade hydrocarbons, and it requires either the selective enrichment or addition of specific microbial species before significant remediation activity is observed.
In this study we assessed the impact of phytoremediation on the composition of the bulk soil microbial community and the ability of that community to degrade hydrocarbons. We postulated that phytoremediation treatments would have an effect beyond that of the rhizosphere and would alter bulk soil microbial processes in such a way as to promote hydrocarbon degradation. The catabolic genes encoding alkane monooxygenase (alkB) (36), naphthalene dioxygenase (ndoB) (31), and catechol-2,3-dioxygenase (xylE) (16) were used to assess the prevalence of bacteria involved in petroleum hydrocarbon degradation. The potential of the indigenous soil microbial population to mineralize representative polycyclic aromatic hydrocarbons (PAHs) was also assessed. The purpose of this study was to determine if phytoremediation treatments increased the catabolic potential of bulk soil by altering the taxonomic structure of the soil microbial community or by stimulating a specific functional genotype in the bulk soil microbial community. It is unlikely that these processes are mutually exclusive, but we wished to determine which process dominates during the phytoremediation of aged hydrocarbons in soil.
MATERIALS AND METHODS
Site description.
The phytoremediation study site was located at the Department of Defense National Test Site, Port Hueneme, Calif. The ability of this phytoremediation project to reduce contaminant concentrations and toxicity was previously documented (4). The study site, which was approximately 20 by 35 m, was divided into 12 plots in a randomized block design. The soil was contaminated with 1.5 g of total petroleum hydrocarbons kg−1 that originated from diesel fuel and heavy oil use over a 20-year period. Treatment plots with four replicates consisted of plant mixture 1 (grasses and legumes: Bromus hordeaceous, Festuca arundinacea, Trifolium fragiferum, Trifolium hirtum, and Vulpia microstachys), plant mixture 2 (native grasses: Bromus carinatus, Elymus glaucus, Festuca ruba, Hordeum californicum, Leymus triticoides, and Nassella pulchra), and an unvegetated control plot. The plant mixtures were selected for their suitability to the northern California climate and for their germination ability in the presence of hydrocarbons. Plants were seeded in August 1997. Plots were sampled in July 1998, in January, April, and August 1999, and in January 2000. Soil samples were collected by using soil augers from the 0- to 6-cm region of the soil for nonvegetated samples. Plant samples were collected by carefully digging around the roots of plants and then removing the plants, leaving the major roots intact. To investigate plant species dependence, two plants, Tall Fescue and Rose Clover, were separately harvested and their major roots and adhering soils were placed in sterile PVC bags for analysis in the laboratory.
The soil at the field site had a pH of 7.2 and an organic matter content of 2.1%, and it was composed of 59% sand, 26% silt, and 15% clay. The site was irrigated as needed, and all treatments were fertilized with 16 g of urea m−2 and 16 g of (NH3)2PO4 m−2 every 3 months. MDS Harris (Lincoln, Neb.) analyzed available phosphorus and inorganic N (NH4+ and NO3−) determined in 2 M KCl extracts in the soil.
Contaminant analysis.
The following procedure was adapted from Schwab et al. (25). Target contaminant extraction from soil was performed by sieving approximately 3 g of air-dried soil with a No. 10 (2-mm pore size) sieve and placing the soil into a tared scintillation vial. A representative moisture content for this sample was recorded. Then 10 ml of solvent (50:50 mixture of methylene chloride:acetone by volume) was combined with 100 μl of tetracosane in methylene chloride as a matrix spike. The vial was then capped and shaken for 30 min. The samples were then centrifuged in a Sorvall J18 centrifuge (Dupont Instruments) for 10 min at 2,000 rpm, and the supernatant was poured off into a separate container. This process was repeated two more times, excluding the addition of the tetracosane matrix spike. For every set of samples, at least 10% of the samples were blanks and an additional 15% were replicates. Target contaminant concentrations were determined by placing 1.5 ml of the supernatant into a gas chromatograph (GC) vial, adding 5 μl of androstane (1,000 mg/liter) in methylene chloride as an internal standard, securely capping, and analyzing this supernatant-androstane mix by gas chromatography (Hewlett-Packard 5890A GC). The GC was interfaced with a Hewlett-Packard Vectra XM2 computer and Chem Station for integration and sampling programming. The GC was equipped with a splitless capillary liner and a flame ionization detector. The column was a J&W DP-TPH, 30 m in length with an inside diameter of 0.32 mm and a stationary-phase film thickness of 0.25 μm. Attached to the column was a guard column constructed of a 2-m-long fused silica tube. A Merlin microseal septum was used to inhibit contamination between samples. Hydrogen was the carrier gas and the source of fuel for the detector, and N2 was the auxiliary gas with air as the makeup gas. The injection volume for all samples was 2 μl, and the injection temperature was 250°C. The total run time was approximately 30 min. The initial temperature was 40°C, maintained for 6 min after injection. The temperature was taken to 300°C at the rate of 10°C/min to 300°C and was held for 5 min.
Plant biomass analyses.
Above-ground biomass was measured in two random locations in each vegetated subplot by cutting the biomass in a 0.5- by 0.5-m frame to within 2.5 cm above the ground. The biomass was then dried and weighed. For quantification of root development, soil cores were taken at four random locations in each vegetated subplot. Root samples were taken from site soil by using a 15-cm-diameter core (300 cm3 total soil volume). Samples were stored in sealed bags and were immediately placed on ice for shipment back to the laboratory. Samples were processed by placing each sample into a 1-liter container. Sodium hexametaphosphate was added to approximately half the volume of the container, and then the container was filled to volume with hot tap water. Samples were hand shaken to agitate soil and were allowed to soak overnight. The following day, each sample was rinsed well to remove soil particles and was suspended in water. Roots and debris were carefully separated by hand. Cleaned root samples were stored in a mixture of 25%/75% (vol/vol) ethanol/water. Root samples were scanned by using the WinRHIZO system (Regent Instruments, Inc., Quebec, Canada) on an Epson 400 scanner. Roots were spread into a single layer on a transparent tray and then were scanned and analyzed automatically by the software. Total surface area was determined by the software and was standardized to surface area/volume of soil. Values for 0- to 30-cm-diameter cores were combined to obtain total surface area/volume for each sample.
Potential of soil to mineralize hexadecane, naphthalene, or phenanthrene.
The potential of the soil to mineralize 14C-labeled hexadecane (C-16 n-alkane), naphthalene, or phenanthrene (both PAHs) was assessed as an indicator of the potential of soil microorganisms to degrade hydrocarbons. Soil (20 g) was placed in 100-ml serum bottles and was amended with ca. 100,000 dpm of either 2 mg of hexadecane kg−1 labeled with 14C at the C-1 position (Sigma, Oakville, Ontario, Canada) with a specific activity of 4.1 mCi/mmole, 2 mg of naphthalene kg−1 uniformly labeled (99% purity; Sigma Chemicals) or 2 mg of phenanthrene kg−1 labeled with 14C at the C-9 position (99% purity; Sigma Chemicals) with a specific activity of 46.9 mCi/mmole. One milliliter of 0.5 N KOH was added to a small test tube placed inside the microcosm to trap 14CO2, and the microcosm crimp was sealed. After 2 weeks of incubation at room temperature, the KOH was removed, 1 ml of 0.5 N KOH was used to wash the inside of the small test tube, and the resulting 2 ml of KOH was transferred to a scintillation vial. Radioactivity was determined in a Tri-Carb liquid scintillation counter model 2100TR (Packard Instruments Co., Meriden, Conn.). Selected microcosms were fertilized with a commercial fertilizer containing 20% (wt/wt) inorganic nitrogen (ammonium nitrate), 20% (wt/wt) organic nitrogen (urea), and 20% (wt/wt) phosphorous (potassium phosphate) to a final concentration of 1,250 mg kg−1 of soil (wet weight) (37).
Catabolic gene probe analysis of the cultured and noncultured community.
Bacteria were extracted from the root interior, rhizosphere, and bulk soil and were resuspended in 0.1% (wt/vol) tetrasodium pyrophosphate (pH 7.0) (28). A 0.1-ml aliquot of serial dilutions in tetrasodium pyrophosphate (10−2, 10−3, and 10−4) was spread plated onto triplicate plates containing (per liter of tap water) 250 mg of yeast extract (Becton Dickinson, Cockeysville, Md.), 250 mg of tryptone (Difco Laboratories, Detroit, Mich.), 250 mg of soluble starch (Anachemia, Montreal, Quebec, Canada), and 15 g of granulated agar (YTS 250 medium; Becton Dickinson). Bacterial colonies were counted after incubation at room temperature (21 to 24°C) for 2 weeks and then were lifted onto nylon membranes. Cells adhering to membranes were lysed and the DNA was denatured, fixed, and cross-linked to the membrane and hybridized to the alkB, ndoB, or xylE gene probes (9).
Total microbial community DNA was extracted from bulk soil by chemical lysis (9) and was purified on polyvinylpolypyrrolidone spin columns (5), and 100 ng of the purified extract was dot blotted in triplicate on Zeta-Probe membranes (Bio-Rad Laboratories, Hercules, Calif.) (30). The concentration of total microbial community DNA applied to the membranes was determined by agarose gel electrophoresis. Scintillation counting of cut dot blot membranes after overnight hybridization (65°C) with 32P-labeled probes was performed with a Tri-Carb scintillation counter. Standard curves, constructed with total genomic DNA of Pseudomonas oleovorans (ATCC 29347) for alkB, Pseudomonas putida strain G7 for ndoB (ATCC 17484), and P. putida mt-2 (ATCC 33015) for xylE were used to estimate the amount of bound probe. The response values, termed genome equivalents, were expressed as ng of genomic DNA/100 ng of total community DNA.
Microbial community composition by DGGE.
Microbial community DNA was extracted from the bulk soil of each treatment as described above and was amplified with eubacterial primers (8F-GC and 519R [6]) in triplicate. A hot-start, touchdown program (annealing temperature of 65°C, touching down to 55°C in 10 cycles followed by 15 cycles at 55°C) was used to amplify community DNA as previously described (29). Triplicate reactions of DNA from the same soil extraction were used to minimize the stochastic biases present in PCRs. Reaction mixtures were concentrated 7.5 times by ethanol precipitation and were loaded onto a 6% polyacrylamide gel containing a 35 to 65% urea-formamide denaturing gradient. The denaturing gradient was prepared and allowed to polymerize for 1.5 h. A spacer gel (6% acrylamide, 0% denaturant, approximately 7.5 mm in height) into which the 20-well comb was inserted was applied on top of the denaturant gel to minimize denaturant gradient disturbance during comb insertion and was allowed to polymerize for 0.5 h. Wells were washed with buffer (40 mM Tris-acetate, 1 mM EDTA, pH 8.0), and approximately 350 ng of amplified PCR product was loaded per lane and run at 80 V for 16 h at 60°C. The resulting gel was stained in 1 mg of ethidium bromide liter−1 in Tris-acetate-EDTA buffer and was photographed. Banding patterns were analyzed by using a ChemiImager (Alpha Innotech, Mississauga, Ontario, Canada), and specific bands were removed by using a scalpel. DNA fragments from the denaturing gradient gel electrophoresis (DGGE) were reamplified with primers 8F and 519R and were sequenced as previously described (12). Briefly, DNA from DGGE bands was amplified with a hot-start program that included heating the PCR cocktail at 96°C for 5 min, after which Taq DNA polymerase was added at 80°C followed by 30 cycles of 1 min at 94°C, 30 s at 60°C, and 30 s at 72°C. The reamplified bands were purified by using the QIAQuick PCR purification kit (Qiagen, Mississauga, Ontario, Canada) and were sequenced by using the ABI Prism dye terminator cycle sequencing ready reaction kit (Perkin Elmer, Montreal, Quebec, Canada) and the ABI Prism 377 automated fluorescence sequencer (Applied Biosystems, Foster City, Calif.). Sequences were compared to those in the GenBank databases by using the BLAST algorithm (1).
Nucleotide sequence accession numbers.
Sequences were submitted to GenBank and were assigned accession numbers AF417499 to AF417506.
RESULTS
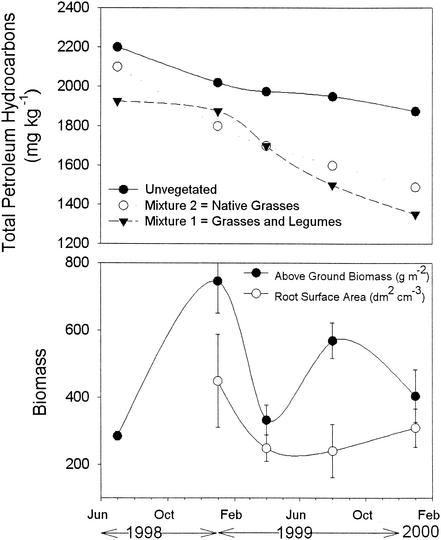
The phytoremediation treatments successfully decreased total petroleum hydrocarbon (TPH) concentrations in bulk soil by 30% after 2 years (Fig. 1). There was little difference in TPH degradation between the two types of phytoremediation systems used, i.e., native grasses or a grass-legume mixture. Both systems degraded approximately 38 mg of TPH kg−1 month−1 (σ = 3.7 mg of TPH kg−1 month−1), which is double that observed for the nonvegetated cells: 19 mg of TPH kg−1 month−1 (σ = 3.4 mg of TPH kg−1 month−1). If this degradation rate continued, one would expect complete removal of TPH from the phytoremediation cells within 5 years compared to over 10 years for the nontreated cells. There was little difference between the above-ground biomass or root surface area between the two phytoremediation treatments, so the results are combined for these treatments. These plant parameters did not display a temporal trend with the above-ground biomass displaying the typical trends expected of the California ecosystem, fluctuating around 460 g m−2. Below-ground root surface area remained relatively constant at around 312 dm2 cm−3.
FIG. 1.
Decrease in TPHs in soil during the phytoremediation trial and above-ground biomass and root surface area of planted treatments over the 2-year phytoremediation period. The least significant difference for total petroleum hydrocarbons was 95 mg kg−1. For plant biomass parameters, results are combined for both plant mixtures and error bars represent the standard errors of the means.
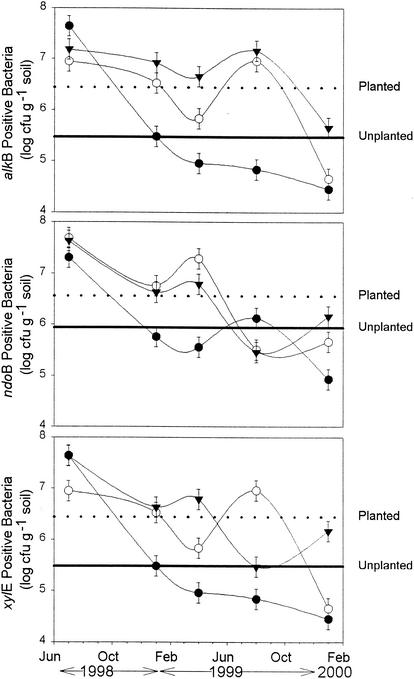
Supporting the chemical analysis, more alkB-, ndoB-, or xylE-positive bacteria were isolated from the planted treatments compared to controls for the majority of the sampling times (Fig. 2). The trend was at its maximum during the summer of 1999, when alkB and xylE gene-positive bacteria were as much as two orders of magnitude greater in the planted treatments than in the nonplanted controls. However, by January 2000 this difference had decreased to less than one order of magnitude. In all treatments there was a significant (P < 0.01) decrease over time in the number of bacteria containing these catabolic genes. For all sampling dates except June 1998 and January 2000, alkB and ndoB prevalence in the cultured community was significantly (P < 0.05) greater in the planted versus nonplanted cells. The prevalence of xylE was only significantly (P < 0.05) greater for January and August 1999. There was no corresponding decrease in the total heterotrophic population, as numbers remained between 4 × 108 to 8 × 108 CFU g−1 of soil and 2 × 107 to 10 × 107 CFU g−1 of soil throughout the study for the planted and nonplanted treatments, respectively. Despite this temporal decrease in the number of catabolic genotypes isolated, the planted treatment cells had, on average, 1 log unit more bacteria containing the alkB or xylE genes and 0.6 log units more bacteria containing ndoB genes. This corresponds to an increase in the total heterotrophic population between the nonplanted cells (7 × 107 CFU g−1) and the planted cells (4 × 108 CFU g−1).
FIG. 2.
Number of soil bacteria isolated on YTS 250 medium from an unvegetated mixture (•), grasses-legume mixture 1 (▾), and native grasses mixture 2 (○) that contained a gene involved in hydrocarbon degradation, alkB, ndoB, or xylE. Error bars (n = 4) represent one standard error and may be obscured by the data points. The dashed and solid lines indicate the gene prevalence in the planted and unplanted treatments averaged over all the sampling times.
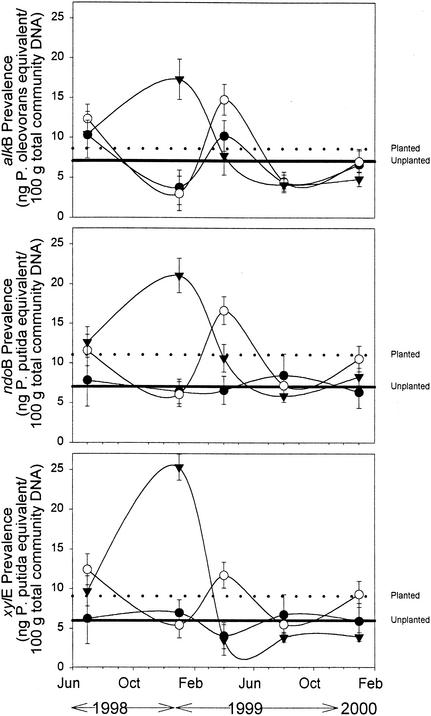
In the uncultured community, catabolic gene prevalence in the phytoremediation treatments peaked in January and April 1999 (Fig. 3). There were significant (P < 0.001) treatment-time interactions for the dot blot results, with native grass mixture 2 peaking in January 1999 and grass-legume mixture 1 peaking in April 1999. For the January and April 1999 samples, ndoB and xylE prevalences were significantly (P < 0.05) greater in the planted versus nonplanted cells. The prevalence of alkB was only significantly greater in planted cells for the January 1999 sampling date. In general, the prevalence of catabolic genotypes in the total extracted community DNA declined with time. When both planted treatments were combined and the last two sampling points were averaged, the prevalence of catabolic genotypes was lower (5, 8, and 6% for alkB, ndoB, and xylE, respectively) compared to the average of the first three sampling points (11, 13, and 11 for alkB, ndoB, and xylE, respectively). In contrast, the nonplanted treatments did not show a similar trend, remaining at approximately 7% for the duration of the experiment. The results with total community DNA had a much lower degree of precision, with a coefficient of variation, which is the standard deviation expressed as a percentage of the mean, of 24% compared to a coefficient of variation of 5% for the colony lifts with 37,000 colonies probed. This difference in precision may be related to the selective properties of culturing in which only a small portion of the community is being monitored.
FIG. 3.
Prevalence of alkB, ndoB, and xylE genes involved in hydrocarbon degradation in total microbial community DNA extracted from soil of different phytoremediation treatments: unvegetated soil (•), grass-legume mixture 1 (▾), and native grasses mixture 2 (○). Error bars represent one standard error. The dashed and solid lines indicate the gene prevalence in the planted and unplanted treatments averaged over all the sampling times.
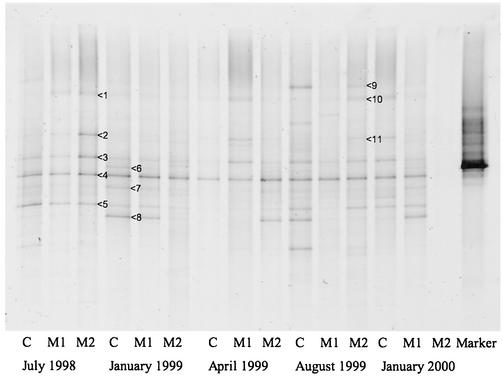
The diversity of the total microbial community in the bulk soil, as assessed by DGGE of 16S ribosomal DNA, was not altered by the phytoremediation treatments. For the first two sample periods there was little difference between replicates of the same treatment taken at the same sampling time. Thus, results are presented from one replicate of each treatment taken over the 2-year period (Fig. 4). No particular band present in the DGGE was only present in a particular treatment, with the majority of bands being present in almost all lanes, except band 9 (93% identity with AF058380 [35]), which was found only three times out of a maximum of 15. The remainder of the sequences obtained from the DGGE gel were closely related to sequences obtained during other experiments investigating contaminant degradation (band 2 had 92% identity with AB013416, from Aquaspirillum spp. [33]; band 4 had 98% identity with AB013429, from an unidentified β-proteobacterium [33]; and band 5 had 95% identity with AJ233535, from an uncultured eubacterium [19]) or bacteria closely linked to plant-root systems (band 1 had 98% identity with AJ232881, from Flavobacterium indologenes [14]; band 6 had 99% identity with X67035, which is from Pseudomonas, Burkholderia, or Ralstonia solanacearum [26], and band 7 had 85% identity with U28230 from B. solanacearum strain R633 [34]). Band 10 had the same sequence as band 1, and band 11 had the same sequence as band 2. These results suggest that the bulk soil community of planted versus nonplanted treatment was highly similar and remained relatively stable over the study period.
FIG. 4.
16S-DGGE analysis of total community DNA present in bulk soil from the unvegetated soil (C, control), soil planted with grasses or legumes (M1, mixture 1), and soil planted with native grasses (M2, mixture 2) over a 2-year period. Arrows indicate the band sequenced, and their closest identities are provided in the text.
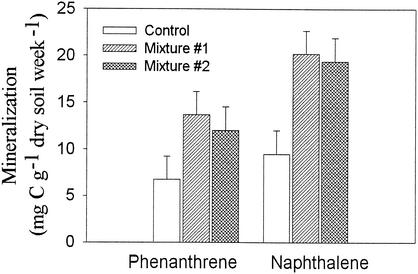
In the bulk soil of the control and mixture 1, the potential to mineralize hexadecane was similar if the microcosms were not fertilized. Similarly, the mineralization potential of mixture 2 was only 84% that of the control soil (data not shown). When microcosms were fertilized, the planted treatments mineralized significantly more hexadecane and phenanthrene than the unplanted control (Fig. 5). This suggests that the hydrocarbon-degrading populations were higher in the planted soils but that they were nutrient limited. Soil chemical analysis indicated that, despite repeated fertilization of all treatments, the available N and P were significantly lower in the planted treatments than in the nonplanted treatments. Grass-legume mixture 1 contained 7.5 μg of N kg−1 and 13 μg of P kg−1, the native grasses mixture 2 contained 3.8 μg of N kg−1 and 13 μg of P kg−1, and the nonplanted control contained 18 μg of N kg−1 and 23 μg of P kg−1 24 months after planting.
FIG. 5.
Potential of fertilized soil to mineralize phenanthrene or hexadecane in soil after 7 days of incubation at room temperature. Soil was fertilized with commercial fertilizer (20:20:20) to a final concentration of 250 mg of N/kg of soil (wet weight) and was spiked with C14-labeled phenanthrene or hexadecane. Error bars represent one standard error.
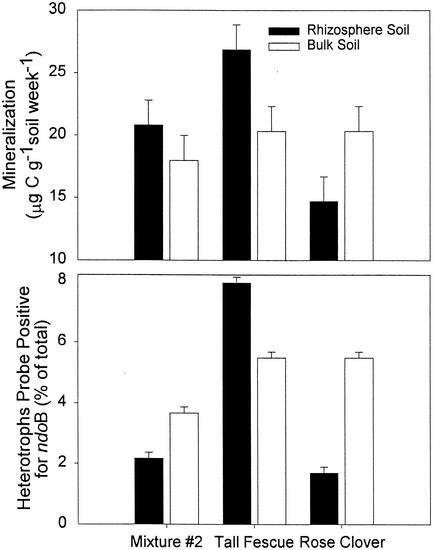
We also assessed plant-specific effects by harvesting selected species from the planted treatment plots and assessing their mineralization potential as well as the prevalence of catabolic genes. Mineralization of naphthalene in Tall Fescue (F. arundinacea) rhizosphere soil was significantly (P < 0.05) greater than naphthalene mineralization in the bulk soil (Fig. 6). In contrast, Rose Clover (T. hirtum) significantly (P < 0.05) decreased naphthalene mineralization in rhizosphere soil compared to that in bulk soil. For the three dates examined, July 1998, January 1999, and April 1999, there were no treatment-time interactions with each plant consistently increasing or decreasing rhizosphere naphthalene mineralization compared to that for bulk soil. The increase in Tall Fescue rhizosphere soil naphthalene mineralization was reflected by a 45% increase in the prevalence of ndoB-positive bacteria in the rhizosphere. Similarly, the decrease in naphthalene mineralization in Rose Clover rhizosphere soil was reflected by a 67% decrease in the prevalence of ndoB-positive bacteria in the rhizosphere. The number of culturable rhizosphere bacteria was similar in both plant species, averaging 1.3 × 109 CFU g−1 of rhizosphere soil for the duration of the experiment. This pattern was not observed with mixture 2, in which mineralization activity increased despite decreases in catabolic gene prevalence.
FIG. 6.
Mineralization of 14C-naphthalene in the bulk (open bar) or rhizosphere (solid bar) soil and prevalence of ndoB in the cultured community of the rhizosphere and bulk soil of specific plant species. Plant species-specific rhizosphere samples were collected on July 1998, January 1999, and April 1999 with four replicates per treatment for each sampling period (n = 12). Error bars indicate the standard error of the estimate for mineralization, which is based on the normal distribution. The standard deviation for catabolic gene prevalence is based on the binomial distribution with 7,476 and 22,338 colonies probed for the rhizosphere and bulk soils, respectively.
DISCUSSION
This study demonstrated that phytoremediation systems that substantially decrease aged TPHs in soil do so by increasing bacteria containing hydrocarbon catabolic genes in the bulk and rhizosphere soil. Both planted treatments increased numbers of bacteria harboring specific catabolic genes in the bulk soil. In the case of Tall Fescue, the catabolic activity of rhizosphere soil was stimulated, and this increase in catabolic activity corresponded to a specific alteration in microbial community composition, i.e., specific catabolic genes increased in prevalence. This stimulation of catabolic activity was not directly related to increases in total heterotrophic numbers but instead was closely linked to an increase in ndoB-positive organisms. In contrast, Rose Clover reduced phenanthrene mineralization activity and ndoB-positive organisms compared to those in the bulk soil. Most studies investigating the promotion of catabolic activity focus on herbicide degradation, but in one study rhizosphere soil from alfalfa grown in a greenhouse had higher pyrene mineralization activity than bulk soil (23). Similarly, in this study increased mineralization of naphthalene was observed in the rhizosphere soil compared to that in the bulk soil. However, it should be noted that the reverse was observed with Rose Clover, confirming that plant-specific interactions are important during phytoremediation. Not only was the effect of phytoremediation systems evident in rhizosphere soil, but phytoremediation treatments also doubled phenanthrene and hexadecane mineralization in the bulk soil of planted treatment cells compared to that of nonplanted controls under field conditions. This confirms the results of earlier greenhouse studies examining mineralization in the rhizosphere (23) and also illustrates that the beneficial effect of plants on hydrocarbon degradation extends beyond the confines of the rhizosphere to the bulk soil.
Previous studies showed that the number of cultured organisms capable of using aged crude oil as a sole carbon source was higher in the rhizosphere of plants than in bulk soil (18). This rhizosphere effect was more pronounced in plants growing in contaminated compared to noncontaminated soil (22). Further, community-level physiological profiles of rhizosphere communities involved in the phytoremediation of PAH have an increased preference for carboxylic or organic acid substrates compared to those in the bulk soil community (11). We also observed an increase in the prevalence of organisms capable of contaminant degradation by using catabolic gene probes, not only in rhizosphere soil but also in the bulk soil associated with phytoremediation treatments. Since many of these catabolic genes can be carried by plasmids, it is not clear from our study if the number of organisms containing the genetic elements increased or if, instead, the number of genetic elements present in the community increased through transconjugation. Changes in the taxonomic diversity of the bulk soil community were not detectable by 16S ribosomal DNA DGGE, which may be related to the low sensitivity of DGGE analysis when eubacterial primers are used (15).
The promotion of catabolic activity by plants was strongly modulated by site factors, such as available nutrients. For example, in unfertilized microcosms, mixture 2 mineralized 17% less hexadecane than the control or grass-legume mixture 1 soil, and it had lower nitrogen levels of 21 and 51%, respectively, compared to the control or grass-legume mixture 1 soil. However, when the microcosms were fertilized, this trend was reversed and both phytoremediation treatments significantly outperformed the control soil. This shows that decreases in available nutrients nullified the beneficial impact of phytoremediation on hydrocarbon degradation. Microbial communities in petroleum-contaminated soils commonly have the potential to degrade hydrocarbons but require nutrients for this potential to be realized (37). Furthermore, the influence of plants on microbial diversity is dependent on the nutrient status of the soil and the indigenous microbial community. Our results suggest that phytoremediation systems can be considered as a method of increasing the potential of soil to degrade hydrocarbons but that other site management techniques, such as increased fertilization, will be required to realize this potential.
The effectiveness of the phytoremediation system was plant species dependent, with Tall Fescue stimulating catabolic activity and Rose Clover depressing it. Other investigators have also found a strong species dependence on the ability of phytoremediation systems to promote hydrocarbon degradation (13, 38). This may be due to alterations in root exudate patterns (8) but may also be due to differences in root architecture (2). Tall Fescue has a fibrous root system, whereas Rose Clover has a coarse, woody root system. The alteration of microbial diversity by plants is not only dependent on nutrient status but is also known to be dependent on the genetic composition of the plant (32) as well as the diversity of organisms already present in the soil (17). Typically, plant-dependent changes in microbial functionality are the result of some form of communication between the associated microorganisms and the plant. For example, bacterial products, such as lumichrome, stimulate root respiration and thereby increase the availability of root exudates for bacteria (21). This effect of lumichrome has been demonstrated with alfalfa, which is a plant shown to promote pyrene degradation in the rhizosphere (21). A present challenge in phytoremediation research is to identify the appropriate plant species that can beneficially alter microbial diversity for a specific soil contamination scenario or, alternatively, that is susceptible to manipulation by the appropriate bacterial inoculant.
Acknowledgments
We thank A. Mihoc and S. Labele for their technical assistance with the microcosms and R. Kim and S. Smith for providing us with samples from their field trial. Critical reading of the manuscript by B. Smets, N. Boon, D. Seghers, T. van de Wiele, and W. Dejonghe is greatly appreciated.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aprill, W., and R. C. Sims. 1990. Evaluation of the use of prairie grasses for stimulating polycyclic aromatic hydrocarbon treatment in soil. Chemosphere 20:253-265. [Google Scholar]
- 3.Banks, M. K., E. Lee, and A. P. Schwab. 1999. Evaluation of dissipation mechanisms for benzo[a]pyrene in the rhizosphere of tall fescue. J. Environ. Qual. 28:294-298. [Google Scholar]
- 4.Banks, M. K., A. P. Schwab, B. Liu, P. Kulakow, J. S. Smith, and R. Kim. The effect of plants on the degradation and toxicity of petroleum contaminants in soil: a field assessment. In D. Tsao (ed.), Advances in biochemical engineering/bio/technology, special volume: phytoremediation, in press. Springer-Verlag, New York, N.Y. [DOI] [PubMed]
- 5.Berthelet, M., L. G. Whyte, and C. W. Greer. 1996. Rapid, direct extraction of DNA from soils for PCR analysis using polyvinylpolypyrrolidone spin columns. FEMS Microbiol. Lett. 138:17-22. [DOI] [PubMed] [Google Scholar]
- 6.Coates, J. D., D. J. Ellis, E. L. Blunt-Harris, C. V. Gaw, E. E. Roden, and D. R. Lovley. 1998. Recovery of humic-reducing bacteria from a diversity of environments. Appl. Environ. Microbiol. 64:1504-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowley, D. E., M. V. Brennerova, C. Irwin, V. Brenner, and D. D. Focht. 1996. Rhizosphere effects on biodegradation of 2,5-dichlorobenzoate by a bioluminescent strain of root-colonizing Pseudomonas fluorescens. FEMS Microbiol. Ecol. 20:79-89. [Google Scholar]
- 8.Fletcher, J. S., and R. S. Hegde. 1995. Release of phenols by perennial plant roots and their potential importance in bioremediation. Chemosphere 31:3009-3016. [Google Scholar]
- 9.Fortin, N., R. R. Fulthorpe, D. G. Allen, and C. W. Greer. 1998. Molecular analysis of bacterial isolates and total community DNA from kraft pulp mill effluent treatment systems. Can. J. Microbiol. 44:537-546. [PubMed] [Google Scholar]
- 10.Haby, P. A., and D. E. Crowley. 1996. Biodegradation of 3-chlorobenzoate as affected by rhizodeposition and selected carbon substrates. J. Environ. Qual. 25:304-310. [Google Scholar]
- 11.Heinonsalo, J., K. S. Jorgenson, K. Haahtela, and R. Sen. 2000. Effects of Pinus sylvestris root growth and mycorrhizosphere development on bacterial carbon source utilization and hydrocarbon oxidation in forest and petroleum-contaminated soils. Can. J. Microbiol. 46:451-464. [DOI] [PubMed] [Google Scholar]
- 12.Juck, D., T. Charles, L. G. Whyte, and C. W. Greer. 2000. Polyphasic microbial community analysis of petroleum hydrocarbon-contaminated soils from two northern Canadian communities. FEMS Microbiol. Ecol. 33:241-249. [DOI] [PubMed] [Google Scholar]
- 13.Liste, H.-H., and M. Alexander. 2000. Plant-promoted pyrene degradation in soil. Chemosphere 40:7-10. [DOI] [PubMed] [Google Scholar]
- 14.Marilley, L., G. Vogt, M. Blanc, and M. Aragno. 1998. Bacterial diversity in the bulk soil and rhizosphere fractions of Lolium perenne and Trifolium repens as revealed by PCR restriction analysis of 16S rDNA. Plant Soil 198:219-224. [Google Scholar]
- 15.Muyzer, G., E. C. de-Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakai, C., H. Kagamiyama, M. Nozaki, T. Nakazawa, S. Inouye, Y. Ebina, and A. Nakazawa. 1983. Complete nucleotide sequence of the metapyrocatechase gene on the TOI plasmid of Pseudomonas putida mt-2. J. Biol. Chem. 258:2923-2928. [PubMed] [Google Scholar]
- 17.Nehl, D. B., S. J. Allen, and J. F. Brown. 1997. Deleterious rhizosphere bacteria: an integrating perspective. Appl. Soil Ecol. 5:1-20. [Google Scholar]
- 18.Nichols, T. D., D. C. Wolf, H. B. Rogers, C. A. Beyrouty, and C. M. Reynolds. 1997. Rhizosphere microbial populations in contaminated soils. Water Air Soil Pollut. 95:165-178. [Google Scholar]
- 19.Nogales, B., E. R. B. Moore, W. R. Abraham, and K. N. Timmis. 1999. Identification of the metabolically active members of a bacterial community in a polychlorinated biphenyl-polluted moorland soil. Environ. Microbiol. 1:199-212. [DOI] [PubMed] [Google Scholar]
- 20.Olson, P. E., and J. Fletcher. 1999. Field evaluation of mulberry root structure with regard to phytoremediation. Bioremediation 3:27-33. [Google Scholar]
- 21.Phillips, D. A., C. M. Joseph, G. P. Yang, R. E. Martinez, J. R. Sanborn, and H. Volpin. 1999. Identification of lumichrome as a Sinorhizobium enhancer of alfalfa root respiration and shoot growth. Proc. Natl. Acad. Sci. USA 96:12275-12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radwan, S. S., H. Al-Awadhi, N. A. Sorkhoh, and I. M. El-Nemr. 1998. Rhizosphere hydrocarbon-utilizing microorganisms as potential contributors to phytoremediation for the oily Kuwaiti desert. Microbiol. Res. 153:247-251. [Google Scholar]
- 23.Reilley, K. A., M. K. Banks, and A. P. Schwab. 1996. Dissipation of polycyclic aromatic hydrocarbons in the rhizosphere. J. Environ. Qual. 25:212-219. [DOI] [PubMed] [Google Scholar]
- 24.Schwab, A. P., A. A. Al-Assi, and M. K. Banks. 1998. Adsorption of naphthalene onto plant roots. J. Environ. Qual. 27:220-224. [Google Scholar]
- 25.Schwab, A. P., J. Su, S. Wetzel, S. Pekarek, and M. K. Banks. 1999. Extraction of petroleum hydrocarbons from soil by mechanical shaking. Environ. Sci. Technol. 33:1940-1945. [Google Scholar]
- 26.Shiomi, Y., M. Nishiyama, T. Onizuka, and T. Marumoto. 1999. Comparison of bacterial community structures in the rhizoplane of tomato plants grown in soils suppressive and conducive towards bacterial wilt. Appl. Environ. Microbiol. 65:3996-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siciliano, S. D., and J. J. Germida. 1998. Biolog analysis and fatty acid methyl ester profiles indicate that pseudomonad inoculants that promote phytoremediation alter the root-associated microbial community of Bromus biebersteiniii. Soil Biol. Biochem. 30:1717-1723. [Google Scholar]
- 28.Siciliano, S. D., and J. J. Germida. 1999. Taxonomic diversity of bacteria associated with the roots of field grown transgenic Brassica napus cv. Quest, compared to the non-transgenic B. napus cv. Excel and B. rapa cv. Parkland. FEMS Microbiol. Ecol. 29:263-272. [Google Scholar]
- 29.Siciliano, S. D., P. Gong, G. I. Sunahara, and C. W. Greer. 1999. Assessment of 2,4,6-trinitrotoluene toxicity in field soils by pollution induced community tolerance (PICT), denaturing gradient gel electrophoresis (DGGE) and seed germination assay. Environ. Toxicol. Chem. 19:2154-2160. [Google Scholar]
- 30.Siciliano, S. D., R. Roy, and C. W. Greer. 2000. Reduction in denitrification activity in field soils exposed to long term contamination by 2,4,6-trinitrotoluene (TNT). FEMS Microbiol. Ecol. 32:61-68. [DOI] [PubMed] [Google Scholar]
- 31.Simon, M. J., T. D. Osslund, R. Saunders, B. D. Ensley, S. Suggs, A. Harcourt, W. Suen, D. L. Cruden, D. T. Givson, and G. L. Zylstra. 1993. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene 127:31-37. [DOI] [PubMed] [Google Scholar]
- 32.Smith, K. P., J. Handelsman, and R. M. Goodman. 1999. Genetic basis in plants for interactions with disease-suppressive bacteria. Proc. Natl. Acad. Sci. USA 96:4786-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suyama, T., Y. Tokiwa, P. Ouichanpagdee, T. Kanagawa, and Y. Kamagata. 1998. Phylogenetic affiliation of soil bacteria that degrade aliphatic polyesters available commercially as biodegradable plastics. Appl. Environ. Microbiol. 64:5008-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taghavi, M., C. Hayward, L. I. Sly, and M. Fegan. 1996. Analysis of the phylogenetic relationships of strains of Burkholderia solanacearum, Pseudomonas syzygii, and the blood disease bacterium of banana based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 46:10-15. [DOI] [PubMed] [Google Scholar]
- 35.Tanner, M. A., B. M. Goebel, M. A. Dojka, and N. R. Pace. 1998. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl. Environ. Microbiol. 64:3110-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Beilen, J. B., M. G. Wubbolts, and B. Witholt. 1994. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 5:161-174. [DOI] [PubMed] [Google Scholar]
- 37.Whyte, L. G., L. Bourbonnière, C. Bellerose, and C. W. Greer. 1999. Bioremediation assessment of hydrocarbon contaminated soils from the high Arctic. Bioremediation J. 3:69-79. [Google Scholar]
- 38.Wiltse, C. C., W. L. Rooney, Z. Chen, A. P. Schwab, and M. K. Banks. 1998. Greenhouse evaluation of agronomic and crude oil-phytoremediation potential among alfalfa genotypes. J. Environ. Qual. 27:169-173. [Google Scholar]